Abstract
We hypothesized that odontoblasts release exosomes as well as dental pulp cells and focused on the exosome membrane marker CD63. Odontoblasts are well-differentiated mesenchymal cells that produce dentin. Dental pulp, a tissue complex formed with odontoblasts, releases exosomes to epithelial cells and stimulates their differentiation to ameloblasts. However, the localization of CD63 in differentiated odontoblasts is poorly understood. Therefore, herein, we aimed to reveal the expression of CD63 in odontoblasts during tooth development. We first investigated the localization of CD63 in mouse incisors and molars using immunofluorescence. In adult mouse incisors, the anti-CD63 antibody was positive in mature odontoblasts and dental pulp cells but not in pre-odontoblasts along the ameloblasts in the apical bud. Additionally, the anti-CD63 antibody was observed as a vesicular shape in the apical area of odontoblast cytosol and inside Tomes’ fibers. The anti-CD63 antibody-positive vesicles were also observed using immunoelectron microscopy. Moreover, during mouse mandibular molar tooth morphogenesis (E16 to postnatal 6 weeks), labeling of anti-CD63 antibody was positive in the odontoblasts at E18. In contrast, the anti-CD63 antibody was positive in the dental pulp after postnatal day 10. Furthermore, anti-CD63 antibody was merged with the multivesicular body marker Rab7 in dental pulp tissues but not with the lysosome marker Lamp1. Finally, we determined the effect of a ceramide-generation inhibitor GW4869 on the mouse organ culture of tooth germ in vitro. After 28 days of GW4869 treatment, both CD63 and Rab7 were negative in Tomes’ fibers, but were positive in control odontoblasts. These results suggest that CD63-positive vesicular organelles are important for mouse tooth morphogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The tooth germ is a primordium in the maxilla and mandible that develops into teeth. From the cap to the bell stage, odontoblasts are differentiated from the dental papilla lined with ameloblasts, and they participate in dentin formation. Odontoblasts are present in the dental pulp as dentin-producing columnar cells and are involved in the lifelong accumulation of dentin matrix. Dentin matrix is released from the odontoblasts via matrix vesicles, which possess a lipid bilayer and are released from the plasma membrane via budding, like osteoblasts in bone tissues. Released matrix vesicles attach to collagen fibers and provoke the deposition of calcium phosphate around themselves via calcium pumps and phosphatases in the vesicle membrane in vivo (Stratmann et al. 1997) and in vitro (Chaudhary et al. 2016).
Extracellular vesicles (EVs) are lipid-bound vesicles that are directly released from the plasma membrane via budding (microvesicles) or the secretion of stored small vesicles from multivesicular bodies (exosomes) (Veziroglu and Mias 2020). The well-known associated proteins of EVs are tetraspanins, including CD63, CD9, and CD81 (Veziroglu and Mias 2020). The lipid membranes of EVs also contain ceramide, a bioactive sphingolipid. Inhibition of ceramide production using the neutral sphingomyelinase inhibitor, GW4869, is a useful method for confirming the release ability of EVs in cells (Catalano 2020). EVs, especially exosomes, often load nucleic acids (microRNAs and mRNAs) or content proteins (Tsg101, annexins, and heat shock proteins). Released EVs fuse with target cell membranes and import nucleic acids or content proteins. Therefore, exosomes are important for the regulation of gene expression and the physiological functions of target cells.
Vesicular organelles are well developed in the odontoblasts for matrix secretion or endocytosis (Sasaki and Garant 1996; Garant et al. 1968). Previous morphological studies have recognized specific vesicular organelles and secretory granules in the cell body and Tomes’ fiber of odontoblasts. Pro-collagen-containing granules, endosomal vesicles, lysosome-like dense vesicles, and multivesicular bodies (MVBs) have been reported in the cell bodies of odontoblasts using transmission electron microscopy (TEM). Additionally, collagen-containing cylindrical granules and coated vesicles have been identified in Tomes’ fibers using TEM and immunofluorescence. MVBs in odontoblasts are involved in the endocytosis of extracellular proteins (Nanci et al. 1996). Thus, odontoblasts can be considered as manipulators of vesicles for various functions inside the cell body and Tomes’ fiber. CD63 is one of the most well-known tetraspanin proteins that localizes in the exosome membrane, MVB, or lysosomes (Nieuwenhuis et al. 1987; Perez-Hernandez et al. 2013). Using immunofluorescence, co-localization between CD63 and other proteins on organelle membrane proteins has been observed in various tissues (Veziroglu and Mias 2020), especially in ameloblasts at the developmental stage of incisors (Shapiro et al. 2007). Stem cells in the dental pulp are also reported to express CD63-positive exosomes (Swanson et al. 2020). However, the localization of CD63 in differentiated odontoblasts is poorly understood.
Therefore, in the present study, we aimed to elucidate the presence of CD63 during odontoblast differentiation and dentinogenesis. Our findings suggest that a substance that inhibits exosome release induces the accumulation of intracellular vesicles in primary dental pulp cells.
Materials and methods
Animals
All human and animal studies were approved by the appropriate ethics committee and were performed in accordance with ethical standards stated in the 1964 Declaration of Helsinki and its later amendments. Female Institute of Cancer Research (ICR) mice at 6 weeks or 14 days of gestation (E14) (Figs. 1, 3, 4, 5, 6) and female C57BL/6n mice (Fig. 2) were purchased from Sankyo Laboratory Service Corporation (Tokyo, Japan). All mice were housed under normal conditions. Mandible tissues from embryonic day 16 (E16), E18, and postnatal day 0, 2, 5, 8, 10, 15, and 18, and 6 weeks were examined in the experiments mentioned below. Kidney tissues were extracted at 6 weeks and examined in the experiments described below.
Immunolocalization of CD63 in mouse incisors. Immunofluorescence staining revealed a punctate appearance of CD63 in mature and labial odontoblasts. a Macroscopic photograph of the decalcified mandible bone. The incisor occupies the center of the mandible (dotted line). b Hematoxylin–eosin staining against mandible cryosections. The asterisk indicates the apical bud of the incisor (pre-odontoblast region), and the arrow indicates the mature odontoblast region. c The same series of cryosections were prepared with the anti-CD63 antibody (red), Alexa 488-conjugated phalloidin (green), and DAPI (nuclear). Enlarged images are shown in (d) and (e). d Puncture anti-CD63 antibody reaction was observed in the odontoblast layer and dental pulp cells. A puncture reaction was also observed in Tomes’ fiber (odontoblast process, arrowhead, and inset). e In the apical bud region, pre-odontoblasts were not positive for the anti-CD63 antibody. f and g The same antibody reaction was confirmed in the kidney. h Negative control for kidney tissues. i High magnification photograph of Tomes’ fiber and odontoblast cell body. Bar: c, 100 µm, d, e and i, 20 µm
Ultrastructure of anti-CD63 positive vesicles in odontoblasts of mouse incisors. a Representative image of odontoblasts with anti-CD63 antibody stained with diaminobenzidine observed under a transmission electron microscope. Anti-CD63 positive vesicles were observed (arrowhead). The cells possess adherence junction (arrow). b Anti-CD63 positive vesicles were observed even without acetate uranyl-lead citrate staining (arrowhead). Scale bar: 500 nm
Antibodies
Primary antibodies were purchased and diluted; the antibody dilution ratio is presented against the protein concentration. The following primary antibodies were used: anti-CD63 antibody (1:200 for immunofluorescence, 1:2000 for western blotting; BD Pharmingen, CA, USA), anti-Rab7 antibody (1:4500; Abcam, Cambridge, UK), anti-Tsg101 antibody (1:100; Abcam), anti-DMP-1 antibody (1:500, R&D Systems, MN, USA), anti-biglycan antibody (1:450; Proteintech, IL, USA), anti-collagen alpha 1 antibody (1:100; Novus Biologicals, CO, USA), anti-LAMP-1 antibody (1:100; Sigma-Aldrich, MO, USA), and anti-EEA1 antibody (1:100; MBL, Aichi, Japan). The following secondary antibodies were used: anti-rat IgG conjugated Alexa 488 or 594, anti-mouse conjugated Alexa 594, anti-rabbit IgG conjugated Alexa 488 and 594, anti-sheep IgG conjugated Alexa 568 (1:200; Thermo Fisher Scientific, MA, USA), and horseradish peroxidase (HRP)-conjugated anti-rat IgG antibody (for immunoelectron microscopy; 1:100, for Western blotting; 1:10,000; Cytiva, Penzberg, Germany).
Microscopic observation
We applied all immunofluorescence samples to laser scanning microscopy (LSM; Nikon A1, Tokyo, Japan). The LSM was equipped with an objective lens: 4 (Plan Apo, NA 0.13), 10 (Plan Apo, NA 0.45), 40 (Plan Apo, NA 0.95) and 60 (Plan Apo VC, NA 1.4, Oil immersing lens) magnifications. All image data pixels were 1024 × 1024. Laser scanning was operated using NIS Elements software. For image acquisition, the pinhole diameter was set to 1.2 mm, according to laser wavelength which was calculated automatically by NIS Elements software. In addition, laser power and detector gain were limited to 4–15% and 90–120, respectively. Z-axis scanning was not applied.
Immunofluorescence observation
For the immunofluorescence studies, longitudinally divided kidney tissue and mandible tissues were fixed with 4% paraformaldehyde (Merck) in phosphate-buffered saline (PBS; 138 mM NaCl, 8.1 mM Na2HPO4, 1.9 mM NaH2PO4) at 4 °C for 16 h and decalcified with 10 mM Tris-buffered 10% ethylenediaminetetraacetic acid (EDTA) at 4 °C for 14–21 days. After embedding in optimal cutting temperature (OCT) compound (Sakura Finetek Japan, Tokyo, Japan), the specimens were rapidly frozen in isopentane (Fujifilm Wako Chemicals, Osaka, Japan), and they were cooled in ethanol with a dry ice pellet. Frozen sagittal mandible sections were sliced into 4 μm-thick sections, and subsequently pasted onto a Silane III-coated glass slide (Muto Pure Chemicals, Tokyo, Japan). After air drying for 30 min, the specimens were washed twice with PBS and blocked with PBS containing 0.1% bovine serum albumin and 0.05% goat IgG for 30 min. Specimens were treated with or without (for negative control) primary antibodies (concentration is shown in Table 1) overnight, washed three times with PBS, and then treated with secondary antibodies (as described above), Alexa 488-conjugated phalloidin (for cortical actin staining, Thermo Fisher Scientific), and 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, for nuclear staining, Dojindo Laboratories) for 60 min. PBS-washed specimens were mounted with coverslips and Aqua-Poly/Mount (Polysciences, PA, USA). The sections were observed under a laser scanning microscope (Nikon Corporation, Tokyo, Japan).
Immunoelectron microscopic observation
For the immunoelectron microscopic studies, mandible tissues were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4 °C for 16 h. Incisor tissues were extracted from the fixed mandible and dissected to 3–4 mm-thick slices with fine blade. Tissues were rinsed with PBS twice and dipped in 1% H2O2-methanol solution for 15 min, then blocked with PBS containing 0.1% bovine serum albumin and 0.05% goat IgG for 30 min. Specimens were subsequently incubated with primary antibodies (concentrations are described under the subheading “Antibodies” in this section) for 2 h, washed three times with PBS, and then treated with secondary antibodies (HRP-conjugated anti-rat antibody) for 2 h. Next, the PBS-washed specimens were stained with diaminobenzidine (KPL, MD, USA) for 5 min, washed twice with PBS, and subjected to osmication for 1 h. Post-osmication, the specimens were dehydrated with ethanol, treated with propylene oxide, and embedded in EPON 812 (TAAB Laboratories Equipment Ltd, UK). After ultrathin sectioning, slices were attached to a polyvinyl formal-covered nickel grid and subjected to uranyl acetate-lead citrate staining. Finally, the sections were observed under a transmission electron microscopy (H-7600, Hitachi, Tokyo, Japan).
Western blotting
Proteins were collected from the kidney, dental papilla (P0) and dental pulp (P15) of molar teeth or incisor dental pulp (6 weeks), immersed in 15% weight/volume of lysis buffer according to the manufacturer's instructions [25 mM Tris (pH: 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, 0.1% Triton X-100, 1 tablet of complete Mini in 10 mL], and mechanically homogenized 20 times with a plastic pestle in a 1.5-mL plastic tube on ice. Protein concentration was measured using a BCA assay kit (Thermo Fisher Scientific), and protein lysates were mixed with 4× sample buffer at room temperature for 30 min. Protein (20 μg) was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel (FasctCast 12% gel, Bio-Rad, CA, USA). Proteins were transferred onto polyvinylidene difluoride (PVDF, Thermo Fisher Scientific) membranes with a Trans-Blot Turbo System (Bio-Rad) in a standard transfer setting. The membranes were blocked with 2% ECL blocking agent (Cytiva) in Tris-buffered salt solution with Tween 20 (TBST; 0.05 M Tris–HCl in pH 7.4, 0.15 M CaCl, 0.1% Tween 20 purchased in Sigma-Aldrich) for 60 min and incubated with anti-CD63 antibody overnight. After washing twice with TBST for 15 min each, the membranes were reacted with an HRP-conjugated anti-rat IgG antibody for 1.5 h. Following this, the membranes were washed with TBST, and blots were detected with an ECL prime western blotting detection reagent (Cytiva).
Reverse transcription and gene amplification using polymerase chain reaction (PCR)
Total RNA was isolated from 2–3 mouse mandibular incisors and kidney tissues using TRIzol (Thermo Fisher Scientific), isopropyl alcohol, and 75% ethanol using standard techniques. Primers were synthesized using Primer3. Cross-reactions to similar sequences were neglected using the Basic Local Alignment Search Tool search. Oligopolymerization of primers was produced by Eurofins Genomics (Tokyo, Japan). Total RNA (1 μg) was subjected to reverse transcription with VILO Master Mix (Thermo Fisher Scientific) and amplified using Takara Ex Taq DNA polymerase (Takara Bio, Shiga, Japan). Amplified genes of interest were assessed using fluorescence imaging and Midori Green Direct staining (Nippon Genetics, Tokyo, Japan) with 2% agarose (Takara Bio) gel electrophoresis. The primers used are listed in Table 1.
Organ culture of tooth germ
At E16, the first molar tooth germs were dissected from the mandible and cultured on 0.45 μm pore-size filter (Millipore, Bedford, MA, USA) supported by a stainless-steel mesh with Fitton-Jackson modified BGJb medium supplemented with 10% fetal bovine serum, 100 mM ascorbic acid, antibiotic–antimycotic reagent (Thermo Fisher Scientific), and 50 mM β-glycerophosphate (Sigma-Aldrich) under 5% CO2. Following this, 50 μM GW4869 (Tocris, MN, USA) or 0.1% dimethyl sulfoxide (Fujifilm Wako Chemicals, for control group) was added to the culture medium for 28 days (Ohki et al. 2021). For immunofluorescence imaging, specimens were fixed with 4% paraformaldehyde overnight and then decalcified with 10% EDTA for 21 days. Specimens were applied to cryosections, stained with antibodies, and observed using laser scanning microscopy as previously described in “Immunofluorescence observation.” The specimens were also subjected to conventional hematoxylin–eosin staining (Muto Pure Chemicals).
Measurement of dentin thickness
For the measurement of whole dentin and pre-dentin thickness, photographic data of tooth germ organ culture stained with hematoxylin–eosin (three individual samples each, control and GW4869) were taken in ×100 magnification and printed on paper using full-color ink. Next, whole dentin and pre-dentin thickness were measured with a micrometer caliper. For elimination of diagonally cut areas, the region of enamel-coated dentin was collected. In each enamel-coated region, 5–6 representative points were collected and measured. The actual thickness of dentin was calculated from the scale bar.
Results
CD63-positive vesicles are localized in mouse incisor odontoblasts
First, we observed the localization of CD63 in mouse incisor odontoblasts using an anti-CD63 antibody. The mouse incisor in the mandible (Fig. 1a, dotted line) was decalcified and cryosectioned. Hematoxylin–eosin staining was confirmed for the cryosections of incisors, pre-odontoblasts in tooth-producing terminal (apical bud, *), and mature odontoblasts (arrow) (Fig. 1b). We then analyzed the reaction of the anti-CD63 antibody in mouse incisors. Mouse incisors produce enamel and dentin on the labial side whereas the lingual side can only produce dentin. Moreover, mouse incisors produce hard tissues from the ends of incisors, the so-called apical bud (also the cervical loop), during the entire lifespan of mice. In our study, the anti-CD63 antibody was negative in the pre-odontoblast cell area (Fig. 1c and d) near the ameloblasts of the apical bud. However, mature odontoblasts were positive for the anti-CD63 antibody, which was observed as a diffuse and small vesicle-like pattern (Fig. 1e). To determine the cell shape of odontoblasts, filamentous cortical actin was stained with Alexa-488 (green)-conjugated phalloidin. Phalloidin staining can depict the branching of odontoblasts in dentinal tubules (Mahdee et al. 2018). Anti-CD63 antibody-positive vesicles stained with Alexa 594 (red) in odontoblasts were also observed in the odontoblast branching processes surrounded by phalloidin (Fig. 1e, arrowhead). These findings clearly indicate that odontoblasts possess anti-CD63 antibody-positive vesicles in the cytoplasmic area, especially in the apical portion and Tomes’ fibers surrounded by cortical F-actin (Fig. 1i, arrowhead). The reactivity of anti-CD63 antibody was confirmed in the microvilli region of kidney renal tubule cells (Fig. 1f and g), similar to previous findings (Schulze et al. 2017).
Next, we observed the ultrastructure of vesicles stained with anti-CD63 antibody in odontoblast using immunoelectron microscopy. Anti-CD63 antibody-positive small and large vesicles in the cell body of odontoblast were stained black with diaminobenzidine (Fig. 2a, arrowhead). Large vesicles were found to have multilamellar or multivesicular structure. Based on the presence of adherence junctions (arrow), we could distinguish the odontoblast from dental pulp cells. The anti-CD63 antibody-positive vesicles were distinctly stained with diaminobenzidine even without uranyl acetate-lead citrate staining (Fig. 2b, arrowhead). These data indicate that CD63 localizes in large and small vesicles.
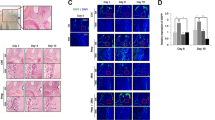
Moreover, we determined the expression and localization of CD63 during molar tooth morphogenesis using the anti-CD63 antibody according to the developmental stage of tooth germ (Gaete et al. 2004) (Fig. 3a–e). At E16, the tooth germ was at the cap stage. Dental papilla does not produce dentin; however, the anti-CD63 antibody was negative for odontoblasts (Fig. 3a) whereas the anti-CD63 antibody was positive for periosteal (Fig. 3b), similar to previous results (Joyner et al. 1997). At E18, pre-dentin production had already started and CD63 antibody labeling was positive for the cusp region of dental papilla (Fig. 3b). From P5 to P15, anti-CD63-positive vesicles were observed in odontoblasts at P5, and the vesicles were localized at the tip of Tomes’ fiber (Fig. 3c–e). Interestingly, the anti-CD63 antibody was also present in the dental pulp tissue after P10 (Fig. 3d). These data suggest that CD63 expression in odontoblasts starts at the same time as dentin formation in the molar tooth.
Protein expression pattern in the molar tooth of mandible. The first molar tooth expressed CD63 from E18 until the eruption stage. [“] denotes an enlarged photograph of the cusp region and [‘“] denotes an enlarged photograph of the root region. a CD63 was not observed in the E16 tooth germ. b CD63 was only observed in odontoblasts at the cusp region of E18 tooth germ. The pre-odontoblasts in the terminal region were negative. c CD63 was observed in both the cusp and root odontoblasts. d and e CD63 was observed in the cusp, root, and dental pulp regions. Bar: a–e, 50 µm, other data, 20 µm
Co-localization of CD63 and related organelle proteins in odontoblasts in developing tooth germ
We analyzed the co-localization of CD63 with other organelle marker proteins in odontoblasts. Tsg101, which sorts ubiquitinated proteins in MVBs to exosomes, was co-localized with CD63 in odontoblasts (Fig. 4a–c). Additionally, CD63 was merged with Rab7 small GTPase, an MVB marker at P5 (Fig. 4d–f). Rab7 was positive in the cytoplasmic area of crown odontoblasts above the nucleus (Fig. 4d), and the position has been found to be enriched with MVBs using TEM (Sasaki and Garant 1996).
CD63 was merged with Tst101 and Rab7 but not with other organelle proteins. Molar tooth cryosections (P10 except d and f) were prepared with the anti-CD63 antibody (green), Alexa 488-conjugated phalloidin (red), and DAPI (nuclear). a–c Tsg101 and CD63, d–f Rab7, and CD63 were merged. d P5, e P10, and f P15. On the other hand, EEA1 (g, inset: endothelial cells in dental pulp), LAMP1 (h, inset: ameloblast), Cola1 (i, inset: bone tissue around tooth germ), DMP-1 (j), and biglycan (k) did not merge with CD63. Bar: 20 µm
EEA1 (early endosome marker, Fig. 4g, inset: endothelial cells in dental pulp) or LAMP1 (lysosome marker, Fig. 4h, inset: ameloblast) did not merge with CD63-positive intracellular vesicles. Moreover, CD63 did not co-localize with collagen alpha 1 (major dentin collagen fiber, Fig. 4i, inset: bone tissue around tooth germ), DMP-1 (major peritubular dentin protein, Fig. 4j), or biglycan (pre-dentin proteoglycan, Fig. 4k). These data suggest that odontoblasts’ CD63 is present in the exosomes or MVB-related vesicular organelles and is not involved in other types of membrane trafficking or dentin matrix secretion.
Exosome-related genes and proteins are expressed during the tooth morphogenesis of dental papilla to dental pulp, including odontoblasts
Morphological observations demonstrated that CD63 was localized in odontoblasts and dental pulp cells, and RT-PCR and western blotting revealed the gene and protein expression of exosome-related genes in odontoblasts and pulp cells.
Dental pulp tissue, including odontoblasts, was extracted from mouse incisors and subjected to conventional RT-PCR. The amplified cDNA is shown in Fig. 5a. The kidney was used as the positive control, and the spleen was collected as the negative control of exosome-related genes. Cd9, Cd63, Cd81, Alix, and Tsg101 are exosome-related genes. Dentin sialophosphoprotein (Dspp) is an odontoblast or dental pulp marker used as a positive control of tissues. In kidney (K) and dental pulp (D), the band of exosome-related genes was positive, whereas the spleen (S) showed weak (CD9, CD63, CD81) or negative bands (Alix, Tsg101) (Fig. 5a). In DSPP, only the dental pulp tissue was positive. These data suggest that dental pulp tissues express exosome-related genes.
Gene expression and protein expression of CD63 and CD63-related genes. a Expression of CD63 mRNA and related gene mRNA was determined using RT-PCR. b Protein expression was recognized as a band at 40–50 kDa using western blotting. K: kidney; D: incisor dental pulp. c The amount of protein expression increased from P0 to P15 in the dental pulp of molar teeth
Previous reports have shown that the molecular weight of non-glycosylated CD63 is approximately 25 kDa (Tominaga et al. 2014). Glycosylated CD63 is detected in the 40–50 kDa band or a higher band up to 70–100 kDa. In dental pulp tissue (D), the anti-CD63 antibody band appeared at 40–50 kDa, almost the same as kidney (K) tissues (Fig. 5b). Additionally, as reported previously, CD63 was observed in whole dental pulp tissues at P10 and was not expressed around P0 (Fig. 3c). To support the immunofluorescence data, we examined protein lysates of dental pulp tissues extracted from P0 and P15 molar teeth. At P0, CD63 band did not appear at any molecular weight (Fig. 5c). At P15, a positive band of CD63 was observed in the 40–50 kDa area, similar to the adult incisor dental pulp tissues. These findings proved that the CD63 protein was expressed in the incisor and molar dental pulp tissues, including odontoblasts, and was glycosylated as a positive control tissue.
Neutral sphingomyelinase 2 (nSMase) inhibitor abolishes CD63-positive vesicles in the Tomes’ fibers of tooth germ organ culture
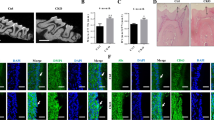
To determine whether CD63-positive intracellular vesicles are involved in dentinogenesis, we investigated the morphological effect of ceramide production blocking reagent on the organ culture of dental pulp cells using GW4869, an inhibitor of nSMase (Menck et al. 2017). nSMase is an enzyme that can produce ceramide, and is involved in vesicle formation. To determine the effect of the drug on dentinogenesis, we applied 50 μM of GW4869 and observed the co-localization of CD63 and Rab7 (Fig. 6a–f). After 28 days of culture without GW4869, dentin (arrow) and pre-dentin (arrowhead) were formed around the odontoblasts with dental pulp (Fig. 6a and inset). The formed dentin (arrowhead) appeared thinner in the tooth germ cultured with GW4869 for 28 days than in the control (Fig. 6b and inset). In the tooth germ cultured with GW4869, both the whole dentin thickness and pre-dentin thickness were thinner than the control. In contrast, there was no significant difference observed between the ratio of pre-dentin to whole dentin thickness. Moreover, in the cultured odontoblasts, CD63 and Rab7 were positive in the cytoplasm and Tomes' fibers as native tissues (Fig. 6d). Both CD63 and Rab7 were not detected in Tomes’ fibers (Fig. 6g). Taken together, GW4869 blocked the recruitment of CD63 and Rab7-loaded vesicles to Tomes’ fibers of odontoblasts and partially hindered dentinogenesis, especially in the pre-dentin.
Ceramide-producing inhibitor decreases pre-dentin formation and alters protein localization in the tooth germ of organ culture. a and b Hematoxylin–eosin staining of tooth germ cultured without (a) or with (b) GW4869 for 28 days. The inset shows the pre-dentin region. c and d Thickness of whole dentin and pre-dentin. e Ratio of pre-dentin thickness to whole dentin thickness. g and f Both CD63- and Rab7-stained intracellular vesicles were negative in GW4869-treated tooth germ (g). h and i: Negative controls. Bar: 20 µm
Discussion
In our study, probing with the anti-CD63 antibody revealed that CD63 was localized in vesicle-like structures in mouse incisors and molar teeth. Kidneys and membranous bone in the developmental stage also showed positive results, as reported previously (Schulze et al. 2017; Joyner et al. 1997). These results suggest that the anti-CD63 antibody reaction in the present study indicates CD63 tetraspanin protein localization in odontoblasts and pulp cells. Moreover, the presence of CD63 was also confirmed with immunoelectron microscopy. The antibody-positive vesicle was multilamellar or multivesicular in odontoblasts (Fig. 2a). These data suggest that CD63 localizes in intracellular vesicles in odontoblasts.
In mouse incisors, CD63 was localized in the secretory and maturation stages of odontoblasts; however, it was not present in the pre-odontoblasts at the apical bud area (Fig. 3). These data suggest that CD63-positive vesicles are expressed in mature odontoblasts and daughter cells, in addition to mature odontoblast cells. Thus, the presence of CD63-positive vesicles depends on the differentiation status. Furthermore, CD63 was observed after postnatal day 10 in dental pulp cells. In P10, root formation begins in teeth and dentinogenesis in the tooth crown area slows. These data suggest that a decline in dentinogenesis in odontoblasts stimulates the expression of CD63 in the dental pulp.
It was previously reported that human odontoblasts are negative for immunohistochemistry with the anti-CD63 antibody and positive for caries-infected dentin-pulp complex tissues (Wang et al. 2019). In contrast, our data showed that immunofluorescence with anti-CD63 antibody was positive in healthy odontoblasts and dentin in mice (Figs. 1, 2, 3, 4). We speculated that the difference in CD63 expression between mice and humans was due to species-specific patterns. Such difference is useful for investigating the functional behavior of CD63 in mesenchymal cells in the tooth germ.
Similar to our morphological findings of CD63 in odontoblasts, CD81 has been observed on matrix vesicle-associated proteins (Chaudhary et al. 2016). CD81 is present in a vesicular pattern in the apical region of odontoblasts (Sasaki et al. 2008), and it releases matrix vesicles from odontoblast-derived immortal cell lines (Chaudhary et al. 2016). We have not yet tested the anti-CD81 antibody in mouse teeth. On obtaining an appropriate antibody for CD81, double immunofluorescence staining of tooth tissue will be performed. Co-localization of tetraspanin proteins (CD9, CD63, and CD81) has been reported in the plasma membrane or extracellular vesicle membrane (Nydegger et al. 2006; Gori et al. 2020). It is conceivable that CD63 and CD81 might be localized in the same vesicle membrane and facilitate matrix vesicle release. Further experiments are needed to clarify the cooperation between CD63 and CD81 in odontoblasts.
Tsg101 is a transmembrane protein that loads ubiquitinated proteins into viral particles or exosome cargo. Tsg101 has often been reported to co-localize with CD63 in multivesicular bodies or exosomes (Veziroglu and Mias 2020). In our study, Tsg101 was partially co-localized with CD63 in the apical area of odontoblasts in a vesicular shape. Therefore, CD63 is speculated to function with Tsg101 for loading cargo onto the vesicles in odontoblasts. Moreover, Rab7 was partially co-localized with CD63 in the cell body and Tomes’ fibers of odontoblasts. Rab7 plays a central role in endosomal maturation in MVBs (Huotari and Helenius 2011). These results suggest that CD63 is localized in MVBs and plays an important role in recycling proteins or releasing exosomes via MVBs.
A previous study reported that CD63 is localized in the lysosomes (Schröder et al. 2009). However, our results showed that the anti-CD63 antibody was not co-localized with the lysosome marker, anti-LAMP1 antibody (Fig. 4). Additionally, the endosomal marker, anti-EEA1 antibody, did not merge with CD63 in odontoblasts. Early endosomes and lysosomes should be excluded from the candidate organelles of CD63 localization.
Odontoblasts release dentin matrix proteins via membrane-surrounded vesicles by budding from the plasma membrane. Type I collagen is released into the scaffold of intertubular dentin matrix whereas DMP-1 is a peritubular protein that is released into the inner walls of dentin tubule (Goldberg et al. 2011; Massa et al. 2005). In our immunofluorescence results, antibodies against the dentin matrix-related proteins did not merge with the anti-CD63 antibody (Fig. 4). Additionally, biglycan, the proteoglycan accumulated in pre-dentin (Orsini et al. 2007; Yamakoshi et al. 2005), was not co-localized with CD63 (Fig. 4). These data suggest that CD63 does not work with matrix vesicle formation or the release of dentin matrix.
A positive band was detected against the anti-CD63 antibody reaction using western blotting. In our study, CD63 was glycosylated as a positive control in kidney tissues. CD63 is known to be glycosylated; however, the specific function of glycosylation is still unknown. In cancerous cell lines, glycosylation of CD63 provokes its transfer from the intracellular fraction to the plasma membrane in HIV-infected cells (Chen et al. 2018) or co-localization with MDR1 and acceleration of malignancy in breast cancer cells (Tominaga et al. 2014). Taken together, glycosylation of CD63 in odontoblasts might be involved in the activation of an unknown physiological mechanism. In vitro studies have revealed that glycosylation of CD63 is necessary for its translocation to the plasma membrane (Tominaga et al. 2014). Further studies are needed to clarify whether CD63 requires glycosylation for translocation to odontoblasts.
Using the sphingomyelinase inhibitor, GW4869, anti-CD63 antibody-positive vesicles were not observed in the Tomes’ fibers of tooth germ organ culture. The positive reaction of the anti-Rab7 antibody was also absent in GW4869-treated Tomes’ fibers of tooth germ. These results suggest that ceramide formation is related to CD63-loaded intracellular vesicles in odontoblasts. GW4869 blocks ceramide synthesis from sphingomyelin by inhibiting neutral sphingomyelinase activity in the sphingomyelinase pathway. Ceramide is required for MVB formation, exosome formation, and lipid raft maintenance (Catalano and O’Driscoll 2020). It is considered that GW4869 blocked CD63- and Rab7-loaded vesicle formation via a reduction of ceramide formation. Additionally, GW4869 is a well-known inhibitor of exosome release; thus, CD63-loaded vesicles are presumed exosomes. Further studies are needed to consider CD63-loaded intracellular vesicles as exosomes.
Conclusion
Taken together, our results demonstrated that odontoblasts contain CD63-loaded vesicles. The results of our study indicate their physiological and pathological roles in dentinogenesis and homeostasis of the dentin-pulp complex.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Catalano M, O’Driscoll L (2020) Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles 9:1703244. https://doi.org/10.1080/20013078.2019.1703244
Chaudhary SC, Kuzynski M, Bottini M et al (2016) Phosphate induces formation of matrix vesicles during odontoblast-initiated mineralization in vitro. Matrix Biol 52:284–300. https://doi.org/10.1016/j.matbio.2016.02.003
Chen L, Feng Z, Yue H et al (2018) Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat Commun 9:4585. https://doi.org/10.1038/s41467-018-07006-2
Gaete M, Lobos N, Torres-Quintana MA (2004) Mouse tooth development time sequence determination for the ICR/Jcl strain. J Oral Sci 46:135–141. https://doi.org/10.2334/josnusd.46.135
Garant PR, Szabo G, Nalbandian J (1968) The fine structure of the mouse odontoblast. Arch Oral Biol 13:857–876. https://doi.org/10.1016/0003-9969(68)90002-2
Goldberg M, Kulkarni AB, Young M, Boskey A (2011) Dentin: structure, composition and mineralization: the role of dentin ECM in dentin formation and mineralization. Front Biosci (Elit Ed) 3:711–735. https://doi.org/10.2741/e281
Gori A, Romanato A, Greta B et al (2020) Membrane-binding peptides for extracellular vesicles on-chip analysis. J Extracell Vesicles 9:1751428. https://doi.org/10.1080/20013078.2020.1751428
Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30:3481–3500. https://doi.org/10.1080/20013078.2020.1751428
Joyner CJ, Bennett A, Triffitt JT (1997) Identification and enrichment of human osteoprogenitor cells by using differentiation stage-specific monoclonal antibodies. Bone 21:1–6. https://doi.org/10.1016/S8756-3282(97)00074-4
Mahdee A, Eastham J, Whitworth JM, Gillespie JI (2018) Evidence for programmed odontoblast process retraction after dentine exposure in the rat incisor. Arch Oral Biol 85:130–141. https://doi.org/10.1016/j.archoralbio.2017.10.001
Massa LF, Ramachandran A, George A, Arana-Chavez VE (2005) Developmental appearance of dentin matrix protein 1 during the early dentinogenesis in rat molars as identified by high-resolution immunocytochemistry. Histochem Cell Biol 124:197–205. https://doi.org/10.1007/s00418-005-0009-9
Menck K, Sönmezer C, Worst TS et al (2017) Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J Extracell Vesicles 6:1378056. https://doi.org/10.1080/20013078.2017.1378056
Nanci A, Fortin M, Ghitescu L (1996) Endocytotic functions of ameloblasts and odontoblasts: immunocytochemical and tracer studies on the uptake of plasma proteins. Anat Rec 245:219–234. https://doi.org/10.1002/(SICI)1097-0185(199606)245:2%3c219::AID-AR9%3e3.0.CO;2-R
Nieuwenhuis HK, Van Oosterhout JJG, Rozemuller E et al (1987) Studies with a monoclonal antibody against activated platelets: evidence that a secreted 53,000-molecular weight lysosome-like granule protein is exposed on the surface of activated platelets in the circulation. Blood 70:838–845. https://doi.org/10.1182/blood.v70.3.838.838
Nydegger S, Khurana S, Krementsov DN et al (2006) Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol 173:795–807. https://doi.org/10.1083/jcb.200508165
Ohki R, Matsuki-Fukushima M, Fujikawa K et al (2021) In the absence of a basal lamina, ameloblasts absorb enamel in a serumless and chemically defined organ culture system. J Oral Biosci 63:66–73. https://doi.org/10.1016/j.job.2020.12.004
Orsini G, Ruggeri A, Mazzoni A et al (2007) Immunohistochemical identification of decorin and biglycan in human dentin: a correlative field emission scanning electron microscopy/transmission electron microscopy study. Calcif Tissue Int 81:39–45. https://doi.org/10.1007/s00223-007-9027-z
Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I et al (2013) The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem 288:11649–11661. https://doi.org/10.1074/jbc.M112.445304
Sasaki T, Garant PR (1996) Structure and organization of odontoblasts. Anat Rec 245:235–249. https://doi.org/10.1002/(SICI)1097-0185(199606)245:2%3c235::AID-AR10%3e3.0.CO;2-Q
Sasaki R, Aoki S, Yamato M et al (2008) Neurosphere generation from dental pulp of adult rat incisor. Eur J Neurosci 27:538–548. https://doi.org/10.1111/j.1460-9568.2008.06026.x
Schröder J, Lüllmann-Rauch R, Himmerkus N et al (2009) Deficiency of the tetraspanin CD63 associated with kidney pathology but normal lysosomal function. Mol Cell Biol 29:1083–1094. https://doi.org/10.1128/mcb.01163-08
Schulze U, Brast S, Grabner A et al (2017) Tetraspanin CD63 controls basolateral sorting of organic cation transporter 2 in renal proximal tubules. FASEB J 31:1421–1433. https://doi.org/10.1096/fj.201600901R
Shapiro JL, Wen X, Okamoto CT et al (2007) Cellular uptake of amelogenin, and its localization to CD63, and lamp1-positive vesicles. Cell Mol Life Sci 64:244–256. https://doi.org/10.1007/s00018-006-6429-4
Stratmann U, Wen X, Schaarschmidt K et al (1997) The mineralization of mantle dentine and of circumpulpal dentine in the rat: an ultrastructural and element-analytical study. Anat Embryol 195:289–297. https://doi.org/10.1007/s004290050048
Swanson WB, Gong T, Zhang Z et al (2020) Controlled release of odontogenic exosomes from a biodegradable vehicle mediates dentinogenesis as a novel biomimetic pulp capping therapy. J Control Release 324:679–694. https://doi.org/10.1016/j.jconrel.2020.06.006
Tominaga N, Hagiwara K, Kosaka N et al (2014) RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol Cancer 13:1–11. https://doi.org/10.1186/1476-4598-13-134
Veziroglu EM, Mias GI (2020) Characterizing extracellular vesicles and their diverse RNA contents. Front Genet 11:1–30. https://doi.org/10.3389/fgene.2020.00700
Wang HS, Yang FH, Wang YJ et al (2019) Odontoblastic exosomes attenuate apoptosis in neighboring cells. J Dent Res 98:1271–1278. https://doi.org/10.1177/0022034519869580
Yamakoshi Y, Hu JCC, Fukae M et al (2005) Dentin glycoprotein: the protein in the middle of the dentin sialophosphoprotein chimera. J Biol Chem 280:17472–17479. https://doi.org/10.1074/jbc.M413220200
Funding
This work was supported by an intramural grant of Showa University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest.
Ethical approval
Approval for this study was obtained from the ethics committee of the Showa University School of Dentistry (approval number 19006 and 12018 for ICR mice, 13034 for C57BL/6 mice).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsuki-Fukushima, M., Fujikawa, K., Inoue, S. et al. Expression and localization of CD63 in the intracellular vesicles of odontoblasts. Histochem Cell Biol 157, 611–622 (2022). https://doi.org/10.1007/s00418-022-02072-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-022-02072-6