Abstract
Since almost 4 decades, connexins have been discussed as important regulators of embryogenesis. Several different members of the gene family can be detected in the preimplantation embryo and during gastrulation. However, genetically engineered mice deficient for every connexin expressed during early development are available and even double-deficient mice were generated. Interestingly, all of these mice complete gastrulation without any abnormalities. This raises the question if the role of connexins has been overrated or if other gene family members compensate and mask their importance. To answer this question, embryos completely devoid of any gap junctional communication need to be investigated. This is challenging because a variety of connexin genes are co-expressed and some null mutations lead to a lethal phenotype. In addition, maternal connexin transcripts were described to persist until the blastocyst stage. In this review, we summarize the current knowledge about the role of connexins during preimplantation development and in embryonic stem cells. We propose that the use of pluripotent stem cells, trophoblast stem cells, as well as artificial embryo-like structures and organoid cultures in combination with multiplex CRISPR/Cas9-based genome editing provides a powerful platform to comprehensively readdress this issue and decipher the role of connexins during lineage decision, differentiation, and morphogenesis in a cell culture model for mouse and human development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Connexins: more than Gap junctions
The connexin gene family consists of 20 members in the mouse and 21 in the human genome (Sohl and Willecke 2004). Connexins are named by their theoretical molecular weight in kDa (e.g., Cx43 has a molecular weight of approx. 43 kDa). There is an alternative nomenclature for connexins in use: in this case the abbreviation Gj (for gap junction protein) is followed by the designation of the subgroup a–e and a number. For example, mouse Cx43 is also referred to as Gja1 (Sohl and Willecke 2003).
Connexins are the protein subunits of gap junctions. This statement is correct, but it is not the whole truth. Indeed, hexameric assemblies of connexins form a gap junction hemichannel termed connexon. Two connexons can dock to each other to build a full gap junctional intercellular conduit. Usually, up to hundreds of channels cluster within the plasma membrane to form a gap junction. Gap junctions allow the direct intercellular exchange of low molecular mass molecules (< 1.8 kDa) such as ions, second messengers, metabolites, and small peptides. By that, gap junctions mediate metabolic as well as electrical coupling between directly neighboring cells. Moreover, the transfer of siRNA and miRNA has been described in the literature (Valiunas et al. 2005; Wolvetang et al. 2007; Zong et al. 2016).
However, connexins are contributing not only to gap junctions but also to fulfill more unexpected roles. Similar to pannexins (Bruzzone et al. 2003; Bao et al. 2004), undocked gap junction hemichannels can release ATP into the interstitial space which contributes to the propagation of calcium signals (Stout et al. 2002). Interestingly, connexons can be also found at the mitochondrial membrane, where they contribute to mitochondrial K+ and Ca2+ homeostasis, release, and production of reactive oxygen species (ROS) as well as regulation of the respiratory complex I (Rodriguez-Sinovas et al. 2018).
Gap Junctions are present in the majority of tissues, e.g., skin, heart, bone, liver, blood vessels, nervous system, eye lens, and the inner ear. But besides in fully matured tissues, connexin expression is also found throughout embryonic development. Interestingly, most tissues, especially during embryonic development, simultaneously express several connexin genes, e.g., in cells of the inner cell mass (ICM) of a blastocyst at least Cx31, Cx43, and Cx45 are expressed as protein and several others only at the mRNA level (Davies et al. 1996). These connexins can form homomeric connexons consisting of the same connexin proteins as well as heteromeric connexons composed of different connexins (Fig. 1). For example, mouse ESCs show expression of the ICM connexins Cx31, Cx43, and Cx45 and these connexins assemble into heteromeric connexin hemichannels consisting of Cx31 and Cx43 or Cx43 and Cx45, whereas interaction of Cx31 and Cx45 within the same hemichannel could not be shown (Worsdorfer et al. 2008).
Connexins, connexons, and gap junctions. Six connexins form a hexameric hemichannel termed connexon. Two connexons of adjacent cells dock to each other to form a gap junction channel or remain as hemichannels within the plasma membrane mediating, e.g., ATP release into the interstitial space. Connexons consisting of the same connexins are termed homomeric; connexons formed by different connexins are referred to as heteromeric. Gap junction channels consisting of the same type of connexon are called homotypic; gap junctions formed by two different connexons are termed heterotypic
Are connexins important key players in embryonic development?
Since decades, connexins and gap junctions have been discussed as important regulators of embryogenesis. Gap junctional intercellular communication (GJIC) in the developing vertebrate embryo was first described in 1979 when Lo and Gilula demonstrated the spread of the low molecular weight (MW) tracer substances fluorescein (MW: 330) and Lucifer Yellow (MW: 457) among directly neighboring cells in the pre- as well as the post-implantation mouse embryo (Lo and Gilula 1979a, b). GJIC was detected from the eight-cell stage on and could be demonstrated among cells of the inner cell mass (ICM) as well as the trophoblast. Interestingly, inter-lineage dye transfer was observed between cells of the trophoblast and the ICM. After implantation ICM as well as trophoblast cells stay directly coupled to cells of the same lineage. However, inter-lineage GJIC is progressively limited as embryonic development proceeds. GJIC is also abundantly detectable among cells of the gastrulating embryo. During that phase, a formation of partially restricted communication compartments can be observed which appears to be associated with the delamination of the three germ layers (Kalimi and Lo 1988).
After the demonstration of GJIC by tracer injection into single cells at different early embryonic stages and the detection of embryonic Gap Junctions in electron microscopy, the question remained which of the 21 known connexin proteins are expressed during these developmental stages. A study by Davies and colleagues (Davies et al. 1996) addressed this question. They detected at least six different connexin transcripts (Cx30.3, Cx31, Cx31.1, Cx40, Cx43, and Cx45). Cx31, Cx43 (Fig. 3a–c), and Cx45 were expressed from the two-cell stage on, Cx30.3, Cx31.1, and Cx40 accumulated beginning in the eight-cell stage. A complementary study by Houghton et al. (Houghton et al. 2002) described that three additional connexin mRNAs (Cx30, Cx36, and Cx57) are expressed beginning in the two-cell stage. On the protein level, Cx31 (Armiger et al. 2018; Houghton et al. 2002), Cx31.1 (Davies et al. 1996; Houghton et al. 2002), Cx40 (Davies et al. 1996), Cx43 (Valdimarsson et al. 1991), and Cx45 (De Sousa et al. 1997) could be demonstrated in the preimplantation embryo. While all other connexin proteins were detectable in the cell membrane, Cx40 appeared to be localized in the cytoplasmic compartment (Davies et al. 1996). At the blastocyst stage, Cx31 and Cx43 proteins are expressed in the ICM as well as the trophoblast (Dahl et al. 1996) (Figs. 2, 3b). After implantation of the mouse embryo, Cx31 expression becomes restricted to the trophectodermal lineage, while Cx43 and Cx45 are found within the embryo proper (Dahl et al. 1996).
Connexin expression in blastocysts, embryonic stem cells (ESCs), and trophoblast stem cells (TSCs). Multiple connexins are co-expressed in blastocysts as well as embryonic stem cells. The published data show that the connexin expression pattern observed in the mouse ICM is also conserved in ESC in vitro. However, human ESCs display the expression of Cx40, which is not detectable in human blastocysts. This might be due to the epiblast stem cell-like identity of human ESCs. The scheme contains all connexin proteins which are published to be expressed in blastocysts, ESCs, or TSCs. More connexins were detected at mRNA level, and protein expression was not tested for all detected transcripts. Therefore, the list of expressed connexin proteins may be incomplete. (*) Detected in cytoplasm but not at the cell membrane; (**) Only persistent maternal protein detected. (***) Detected in EB3 ESCs but not in HM1 ESCs
Cx43 expression in the mouse blastocyst and in mouse ESCs. Cx43 is expressed in the Oct4-positive inner cell mass (ICM) as well as the trophoblast of mouse blastocysts (a, b). In c, a high magnification of the ICM is shown. Cx43 can be also detected in mouse ESCs (d). Using DAB staining for Cx43 in combination with transmission electron microscopy (TEM) we could detect signals at gap junctions between adjacent ESCs (e). Interestingly, signals were also frequently found at the mitochondrial membrane (f). In f, pictures of four different mitochondria are shown (a–d)
In addition, maternal Cx32 protein was described to be present as persistent oogenetic product through mouse preimplantation development until the blastocyst stage without being expressed by the embryo itself (Barron et al. 1989). Of note, additional connexin proteins (Cx37, Cx43, and Cx45) are also expressed in the ovarian follicle contributing to granulosa cell–oocyte coupling (Kidder and Mhawi 2002). Cx37 seems to be the main connexin expressed by the oocyte and maternal transcripts of this gene could similarly persist within the zygote, an aspect that has not been comprehensively analyzed yet.
The connexin expression pattern in the early rat embryo is quite similar. Cx31, Cx43, and Cx45 proteins were found at the blastocyst stage. In contrast to the mouse embryo, Cx26 is additionally expressed (Houghton et al. 2002). Cx30 was found as mRNA but no protein was detected. In the human blastocyst, Cx26, Cx31, Cx43, and Cx45 expressions were demonstrated on the protein level (Bloor et al. 2004; Hardy et al. 1996).
Taken together, the expression pattern of connexins during early embryogenesis is surprisingly complex and shows variations from species to species (e.g., Cx26 is expressed in rat but not mouse) and partially even between individual embryos (Bloor et al. 2004; Hardy et al. 1996). Cx31, Cx31.1, Cx43, and Cx45 seem to be the most important connexins expressed during mouse preimplantation development. Cx30.3 and Cx40 protein might be present as well and maternal Cx32 seems to persist until the blastocyst stage. However, it has to be mentioned that the analyses published so far are not complete as not all described connexins were tested at mRNA and protein level. It would be interesting to readdress this issue and comprehensively characterize all connexin transcripts and proteins being present from the oocyte to the late blastocyst. This analysis would help to better understand which connexin proteins might compensate in the knockout background.
The physiological necessity of gap junctional communication during early developmental processes such as compaction or early lineage segregation has been controversially discussed. In early studies, Lee et al. (Lee et al. 1987) raised polyclonal antibodies directed against the major 27/32 kDa protein eluted from rat liver gap junctions and injected such antibodies into cells of mouse eight-cell stage embryos, which strongly reduced electrical coupling and dye transfer. Interestingly, injected cells were excluded from embryo compaction. In a similar experiment, Bevilacqua and colleagues (Bevilacqua et al. 1989) injected antisense RNA targeted against the 27/32-kDa rat liver gap junction protein into the blastomeres of 2- and 4-cell zygotes which reduced the number of compacted embryos after 60 h of culture to 20% compared to 75% in embryos injected with an unrelated RNA. The conclusion that gap junctions are essentially required during preimplantation development was supported by Becker et al. (Becker et al. 1995). The authors generated several antibodies directed against the cytoplasmic loop of Cx43, which were injected into blastomeres of an eight-cell stage mouse embryo. Antibody injection blocked dye transfer (Lucifer Yellow, Cascade Blue) and led to the exclusion of the injected cell from compaction and further development. However, from today’s view, knowing that at least Cx31, Cx43, and Cx45 proteins are expressed in the two- to eight-cell stage mouse embryo and that Cx32 is not transcribed, it is clear that the injection of Cx32 antibodies or Cx32 antisense RNA, although it might strongly reduce GJIC, is not a specific approach. In contrast to these early experiments, two studies used the pharmacological gap junction inhibitor 18-alpha glycyrhetinic acid (18-AGA) which abolished GJIC among cells in the developing preimplantation embryo. This treatment had no impact on blastocyst formation, cell allocation to the trophectoderm, or inner cell mass (Vance and Wiley 1999) or apoptosis rates within the blastocyst (Houghton et al. 2002). However, it needs to be mentioned that not all connexins are sensitive to 18-AGA: for example, Cx43 is sensitive, while Cx31 and Cx45 are not (He et al. 2005; Worsdorfer et al. 2008). Between 1995 and 2007, mice deficient for each connexin expressed during preimplantation development were generated using homologous recombination in ESCs. Interestingly, none of these animals display a phenotype related to early development [Cx31 → (Plum et al. 2001); Cx43 → (Reaume et al. 1995); Cx45 → (Kruger et al. 2000; Kumai et al. 2000); Cx40 → (Simon et al. 1998); Cx30.3 → (Zheng-Fischhofer et al. 2007b); Cx31.1 → (Zheng-Fischhofer et al. 2007a)]. Even double-deficient animals for Cx43 and Cx31 (Kibschull et al. 2005) or Cx43 and Cx45 (Nishii et al. 2016) complete gastrulation in the expected mendelian ratio without any detected abnormalities. The triple deletion of Cx31, Cx43, and Cx45 would be of high interest as these are the main connexins expressed at the blastocyst stage. However, such mice could not be generated yet.
All these studies indicate either a minor contribution of connexins to preimplantation development or redundant functions of different connexin genes expressed in the early embryo and/or compensation by maternal transcripts. Moreover, it is possible that connexin expression is not essentially required under ideal conditions but protects the early embryo from stress such as delayed migration of embryos from the fallopian tube to the uterus, oxidative stress, environmental toxins, suboptimal maternal diet, or maternal disease. In addition, the preimplantation period is characterized by epigenetic reprogramming of embryonic cells, a procedure that might be affected by the loss of intercellular coupling. Gap junctions could be required to equalize levels of metabolites with epigenetic function (Etchegaray and Mostoslavsky 2016) such as alpha-ketogluterate (Carey et al. 2015) by allowing their intercellular diffusion among groups of cells. The resulting epigenetic effects could be mild and must not necessarily result in an obvious phenotype but might affect the developing embryo or even the resulting adult in a more subtile manner (e.g., decreased life-span or metabolic and mental disorders) as observed in cloned animals (Humpherys et al. 2001).
In summary, several connexins are expressed during early embryogenesis in the trophoblast and the inner cell mass. Expression continues during gastrulation and these connexins contribute to gap junctions and mediate functional GJIC. However, it has been controversially discussed if connexins are essential key regulators of development and what they are exactly required for. The use of injected antibodies, siRNAs, or pharmacological gap junction inhibitors as well as the analyses of connexin-deficient mouse lines led to contrasting results. Embryos or embryonic cells completely devoid of any connexin gene expressed during early development are essentially required to finally answer this question in a satisfying manner. Moreover, more subtile phenotypes such as altered epigenetic signatures should be considered.
Embryonic stem cells and trophoblast stem cells as in vitro models
A cell culture platform that guarantees unlimited access to early embryonic cells and allows for their easy manipulation is the use of embryonic stem cells (ESCs) or trophoblast stem cells (TSCs). These cells are an interesting in vitro model to study the role of gap junctions during early lineage decision and pluripotency maintenance.
Embryonic stem cells (ESCs) are the in vitro equivalent of ICM cells. ESCs are isolated from the ICM of blastocysts. They can expand virtually unlimited in cell culture under appropriate conditions and retain their pluripotent differentiation potential. Mouse ESC cultures were first established in 1981 (Evans and Kaufman 1981; Martin 1981), and human ESCs in 1998 (Thomson 1998). Human and mouse ESCs display a similar connexin expression pattern as observed in the embryo. Mouse ESCs express Cx31, Cx43 (Fig. 2d, e), and Cx45 protein (Worsdorfer et al. 2008). Cx30.3 protein was also described but might not be expressed in every mESC line (Saito et al. 2017; Worsdorfer et al. 2008), probably due to a different genetic background (Sharova et al. 2007). Interestingly, Cx30.3 expression seems to be regulated by LIF/STAT signaling (Saito et al. 2017) and also Cx31 mRNA expression is strongly downregulated within 24 h of LIF starvation and was, therefore, suggested as “Pluri” gene (Trouillas et al. 2009). In contrast, Cx31.1 and Cx45 mRNA expressions were shown to increase upon siRNA induced Oct4 silencing (Trouillas et al. 2009).
Human ESCs express Cx43, Cx45, and Cx40 protein (Huettner et al. 2006; Wong et al. 2004). Unlike in ICM cells in vivo, Cx40 was detected as protein in the membrane of hESCs. This might be explained by the fact that human ESCs and human iPSCs resemble more primed epiblast stem cells than naive ICM cells. Cx26 was found at the mRNA level, but protein expression was not tested (Huettner et al. 2006). Cx43 and Cx45 can be found on a “consensus hESC gene list” published by Assou and colleagues which analyzed datasets from 38 microarray analyses performed on human pluripotent stem cells. The list contains genes which expression was detected in hESCs by at least three independent studies (Assou et al. 2007).
In 2008, a study by Todorova and co-workers reported that the reduction of GJIC using the pharmacological inhibitor 18-AGA or siRNAs targeted against Cx43 mRNA leads to reduced proliferation and increased apoptosis in mouse ESCs (Todorova et al. 2008). Moreover, such ESCs were no longer capable of forming embryoid bodies in suspension culture. A study on human ESCs came to a similar result reporting increased apoptosis and reduced colony growth under serum-free conditions (Wong et al. 2006). However, these results are in contrast to studies from several other groups that showed that a single ablation of either Cx43 or Cx45 in mESCs has no obvious effect on their proliferation rates or in vitro differentiation potential (Egashira et al. 2004; Gutstein et al. 2001; Reaume et al. 1995; Worsdorfer et al. 2008, 2017). Interestingly, Cx45-deficient ESCs fail to integrate into blastocysts of WT mice, unlike Cx43-deficient ESCs, which form viable chimeras (Egashira et al. 2004; Gutstein et al. 2001). Cx43/Cx45 double-deficient ESCs show unaltered proliferation, apoptosis rates as well as pluripotency marker expression in maintenance culture. However, when differentiated in 3D culture as embryoid bodies, Cx43/45-deficient cells fail to differentiate into cells of the primitive endoderm and the cell aggregates contain a high number of undifferentiated Nanog-positive cells even after 6 days of differentiation (Worsdorfer et al. 2017). Prolonged culture of aggregates for more than 20 days results in differentiation mostly to the neural lineage (unpublished data from our lab). Similar observations were made by Sheardown and Hooper in 1992 who investigated the differentiation potential of metabolic coupling-deficient embryonic stem cells generated by general mutagenesis and “kiss of death” selection (Sheardown and Hooper 1992). These results stand in contrast to the situation observed in Cx43/Cx45 double-deficient mice, which develop normally until embryonic day 9.5, when the Cx45 mutation proves lethal due to disturbed cardiac development (Kumai et al. 2000). A possible explanation for the differences observed between the in vivo vs. in vitro situation could be that compensating maternal connexins—such as Cx32- which are found in the preimplantation embryo (Barron et al. 1989), are no longer present in long-term cultured ESCs.
Trophoblast stem cells (TSCs) are the in vitro equivalent of cells found within the blastocysts polar trophectoderm. Mouse TSCs were first derived in 1998 (Tanaka et al. 1998), and human trophoblast stem cells were recently described by Okae et al. (Okae et al. 2018). TSCs can be permanently maintained in culture and differentiate into trophoblast subtypes in vitro and at least mouse TSCs do also contribute to the trophoblast lineage in chimeras in vivo. Mouse TSCs exclusively express Cx31 protein and Cx31.1 transcripts and upon differentiation Cx26 and Cx43 become induced. Connexin expression in human TSCs has not been investigated yet. TSCs derived from Cx31-deficient mice reveal an accelerated differentiation toward giant cells compared to controls (Kibschull et al. 2004). In vivo, Cx31 is expressed in the spongiotrophoblast and Cx31-deficient mice display transient placental dysmorphogenesis with an increased proportion of giant cells that leads to a loss of 60% of the embryos between embryonic day 10.5 and 13.5 (Plum et al. 2001). Cx31.1 deficiency in mice leads to a loss of 30% of the embryos between embryonic day 10.5 and 13.5, and a deficiency in Cx31.1, in contrast to Cx31, delays TSC differentiation in vitro (Kibschull et al. 2014).
Recapitulating development in the culture dish: pluripotent stem cells, genome editing, and artificial tissues
In 2006, Takahashi and Yamanaka first demonstrated that pluripotency can be induced in somatic cells (Takahashi and Yamanaka 2006). The initial experiments were performed converting mouse embryonic fibroblasts and 1 year later the first human-induced pluripotent stem cells (iPSCs) from adult dermal fibroblasts were reported by two independent groups (Takahashi et al. 2007; Yu et al. 2007). iPSCs are almost identical to ESCs derived from an embryo (Choi et al. 2015) and new transgene free or non-integrative reprogramming methods allow for the generation of iPSCs devoid of any “footprints” caused by the reprogramming method such as stably integrated retroviruses or loxP sites (Worsdorfer et al. 2013). Human iPSCs avoid the ethical dilemma associated with the derivation of ESCs from human embryos and make such cells easily available to every lab. Induced trophoblast stem cells (iTSCs) from murine fibroblasts were generated as well (Benchetrit et al. 2015; Kubaczka et al. 2015). Human iTSCs are not described yet.
Although, connexin protein expression in iPSCs has not been comprehensively investigated, Cx43 and Cx45 expressions were demonstrated in human iPSCs (Beckmann et al. 2016; Ke et al. 2013, 2017) and expression of Cx43 as well as Cx45 was shown to enhance the generation of human iPSCs from dermal fibroblasts (Ke et al. 2013, 2017).
The possibility to generate human iPSCs from skin biopsies or peripheral blood samples of every individual allows the generation of patient-specific iPSCs. Several genetic human disorders are associated with mutations in connexin genes (Srinivas et al. 2018). A good example is the rare autosomal dominant disorder Oculodentodigital dysplasia (ODDD) linked to a variety of different point mutations in the Cx43 gene (Paznekas et al. 2003). iPSCs derived from ODDD patients are an interesting model system to understand the developmental aspects of the disease. A first iPSC line from an ODDD patient has been recently described and first analyses reveal a negative effect of the mutated Cx43 on osteoblast and chondrocyte differentiation (Esseltine et al. 2017).
The CRISPR/Cas9 technology is a new and powerful tool for the deletion of genes in mouse and human ESCs (Cho et al. 2013; Jinek et al. 2013; Mali et al. 2013). Multiplex CRISPR/Cas9-based genome editing allows for the simultaneous homozygous deletion of several target genes (Cong et al. 2013). By that, ESC or iPSC lines deficient of three or even more different connexin genes can be generated and different combinations of connexin deficiency can be explored. Moreover, specific mutations linked to genetic disorders can be introduced or individual domains of connexin proteins can be deleted/modified.
In 2014, a fascinating culture system for the assessment of the intrinsic tendency of pluripotent stem cells to create patterns in a very controlled and reproducible fashion was described. The authors show that geometric confinement and BMP4 stimulation triggers human pluripotent stem cells to form a tissue structure with an outer ring of trophectoderm, an inner ectodermal cell population, and a ring of mesendoderm expressing primitive streak markers in between (Warmflash et al. 2014). The publication stimulated an important and interesting discussion on ethical issues raised by synthetic human entities with embryo-like features (SHEEFS) (Aach et al. 2017) (Fig. 4). By combining mouse ESCs and trophoblast stem cells (TSCs) in a 3D-scaffold, Harrison et al. generated structures, so-called ETS-embryos, whose morphogenesis is remarkably similar to natural embryos and that develop till mesoderm-specification in vitro (Harrison et al. 2017) (Fig. 4). A very recent publication describes the generation of “blastoids”, blastocyst-like structures generated by the co-culture of mouse ESCs and TSCs in a micro-well array (Rivron et al. 2018). These artificial embryonic structures closely resemble natural blastocysts, display the formation of primitive endoderm, and robustly implant and trigger decidualization in utero (Fig. 4). Artificial embryo-like structures generated from CRISPR/Cas9 engineered ESCs or iPSCs could be an interesting experimental platform to investigate the influence of connexins on early embryonic lineage decisions and patterning in vitro.
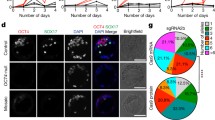
Cell culture models for embryonic development. Pluripotent stem cells (PSCs) such as ESCs and iPSCs as well as TSCs and iTSCs can be isolated directly from blastocyst stage embryos or generated artificially by cellular reprogramming or direct conversion from adult somatic cells (e.g., fibroblasts or peripheral blood cells). (i) PSCs and (i) TSCs can be genetically modified using CRISPR/Cas9 technology. The example shows an experiment from our lab where the Cx43 gene was mutated in hIPSCs. Unmodified or genetically altered PSCs or TSCs can serve as basis for the generation of in vitro models of early lineage decision and morphogenesis. Examples are so-called “gastruloids”, “blastoids” or “ETS-embryos”. Moreover 3D-organoid cultures can be also generated from PSCs modeling more mature tissue development, e.g., brain development in cerebral organoids
Within the last few years, ESC- and iPSC-derived organoid cultures were developed representing simplified mini-versions of organs or tissues such as gut, brain, eye, and kidney (Clevers 2016). Organoid models are promising tools to investigate developmental processes and to study the differentiation of human pluripotent stem cells with different combinations of ablated connexins in a tissue-like context. For example, neural connexins (Cina et al. 2007) such as Cx36 (Condorelli et al. 1998; Sohl et al. 1998) and Cx43 (Duval et al. 2002; Elias et al. 2007), which essentially contribute to neuronal differentiation and migration, can be genetically ablated in pluripotent cells, either individually or in different combinations, and the consequences could be studied in human cerebral organoids. These self-organizing artificial tissues closely recapitulate several key aspects of embryonic neurogenesis such as neural tube formation, cortical development and neuronal migration in an in vitro setup (Lancaster et al. 2013).
In summary, the absence of maternal transcripts, the variety of well-established differentiation protocols, the possibility to study the situation in humans, and finally the easy manipulation via CRISPR/Cas9 technology make ESCs and especially human iPSCs a very attractive model system to investigate the role of connexins during early developmental lineage decisions. In addition, blastoids (Rivron et al. 2018), gastruloids (Warmflash et al. 2014), and organoids (Clevers 2016) as 3D in vitro models for tissue development open new possibilities to gain insight into the elementary processes controlling the early steps of embryonic development.
How can connexins modulate cellular differentiation?
The fundamental question regarding the role of connexins in embryonic development and stem cell differentiation is, by which pathways connexons or gap junctions might modulate proliferation, cellular differentiation and apoptosis. Different mechanisms have been proposed in the literature. In the following part of the review, we will focus on two hypothetic mechanisms that could mediate the effect of connexins in pluripotent stem cells.
IP3, Ca2+, and NFAT-signaling
After translation at the ER, connexins oligomerize and assemble into hexameric structures, usually within the trans-Golgi network, to form hemichannels, so-called connexons. These connexons traffic to the cell membrane and dock to their counterparts on directly neighboring cells to form a full intercellular conduit. Usually, a few to hundreds of such tightly packed channels cluster at certain membrane domains and contribute to gap junctional plaques (Laird 2006). Gap junctions assure electrical coupling and allow the diffusion of low molecular weight molecules such as ions, metabolites, or second messengers between adjoining cells. The interesting question is: which of these molecules have the ability to influence gene expression and trigger, synchronize, or modulate cellular differentiation? IP3 and Ca2+ are candidate molecules which can affect cellular differentiation and their intercellular diffusion via gap junctions (Niessen et al. 2000) could synchronize gene expression among a group of neighboring cells. IP3 can release calcium from intracellular stores. Ca2+ itself activates Calcineurin/NFAT-signaling (Crabtree and Olson 2002) (Fig. 5), a signaling pathway, which directly modulates gene expression and was described to critically regulate early lineage specification in mouse embryonic stem cells and embryos (Li et al. 2011). A link between connexin expression and synchronized NFATc3 activation has been previously described in the developing heart (Kumai et al. 2000) as well as in the lymphatic system (Sabine et al. 2012) and could also be a reason why Cx43/Cx45 double-deficient ESCs fail to differentiate into primitive endoderm in 3D embryoid body cultures (Worsdorfer et al. 2017).
How can connexins modulate differentiation? Connexons form gap junctions between neighboring cells (1), are present as undocked hemichannels in the cell membrane (2), and can be found as connexons within the mitochondrial membrane (3). (1) Gap junctions allow the diffusional exchange of ions, metabolites, and second messenger molecules. Among these, IP3 is an interesting candidate which triggers the release of calcium from intracellular stores and might regulate gene expression in adjacent cells via the calcineurin/NFAT signaling pathway. Calcineurin/NFAT signaling plays an important role in stem cell differentiation. (2) Undocked hemichannels can open under pathological and maybe also physiological conditions and release, e.g., ATP into the interstitial space. Extracellular ATP acts in an autocrine and paracrine manner and binds to purinergic receptors (P2Y, P2X) which activate signaling pathways such as phospholipase C (PLC) signaling. Activated PLC produces the second messenger IP3 which can result in elevated intracellular calcium levels and subsequent activation of the calcineurin/NFAT signaling pathway. (3) Mitochondrial connexons have been shown to modulate, among others, the production and release of reactive oxygen species (ROS). ROS production and mitochondrial dynamics play an important role in modulating stem cell differentiation
Another way how connexins could activate Calcium signaling in adjacent cells is the release of ATP into the interstitial space. Connexons do not necessarily dock to each other to form intercellular conduits. They can also function at the membrane as hemichannels which open under certain conditions and release ATP into the interstitium (Stout et al. 2002). ATP activates purinergic receptors on the surface of ESCs (Heo and Han 2006) in an autocrine or paracrine fashion which leads to the mobilization of intracellular calcium and activation of signaling pathways such as protein kinase C (Heo and Han 2006a) and calcineurin/NFAT-signaling (Kawano et al. 2006). These pathways were described to have an impact on ESC proliferation (Heo and Han 2006) and differentiation (Li et al. 2011) (Fig. 5). Functional connexin hemichannels were demonstrated in human embryonic stem cells by dye uptake studies and recordings from solitary human ESCs (Huettner et al. 2006).
In this context, it is important to mention that ATP release is also mediated by another distinct channel family build up by a class of proteins termed pannexins (Panx) (Bao et al. 2004; Bruzzone et al. 2003). The pannexin gene family consists of three members (Panx1–3). Pannexins are expressed, e.g., in neural stem cells (NSCs) and essentially contribute to NSC proliferation and maintenance (Wicki-Stordeur et al. 2012, 2016). We could show that Panx1 mRNA can be detected in mouse ESCs while Panx2 and Panx3 seem to be absent. However, we could not detect any Panx1 protein by immunoblot analyses (Worsdorfer et al. 2008). Therefore, we conclude that pannexin channels are not involved in ATP release in pluripotent stem cells. A recently published study investigated the expression of pannexins in human ESCs and iPSCs and comes to a similar conclusion. The authors detected all pannexin mRNAs (Panx1–3) with Panx1 showing the highest expression level. However, immunoblot analyses could not demonstrate pannexin protein expression in human pluripotent stem cells (Hainz et al. 2018).
Connexins at mitochondria: modulators of mitochondrial activity?
Connexins are well known as building blocks of gap junctions, directly connecting the cytoplasm of two neighboring cells. Moreover, connexon hemichannels mediate the release of ATP into the intercellular space as described above. A third and less recognized aspect is the localization of connexons at the mitochondrial membrane. Mitochondrial Cx43 was first described in cardiomyocytes (Boengler et al. 2005; Rodriguez-Sinovas et al. 2006) where it is localized to the inner mitochondrial membrane and essentially contributes to the cardioprotective effect of ischemic preconditioning. This effect appears to be related to reactive oxygen species (ROS) signaling which was found to be attenuated in Cx43-deficient mice (Heinzel et al. 2005) (for review see Rodriguez-Sinovas et al. 2018). Additional studies indicate the involvement of mitochondrial Cx43 in the modulation of K+ influx, mitochondrial respiration and apoptosis (Boengler et al. 2012; Goubaeva et al. 2007; Miro-Casas et al. 2009).
Interestingly, although mitochondrial connexons are described since more than 10 years, most studies still focus almost exclusively on Cx43 in cardiomyocytes. However, given the assumption that mitochondrial connexons are important regulators of mitochondrial function, it is very likely that they are also present in other cell types including stem cells. Only few studies addressed this issue so far. However, it could be shown that mitochondrial Cx43 is present in brown adipose tissue (Kim et al. 2017), mesenchymal stem cells (Lu et al. 2009), and human vascular endothelial cells (Li et al. 2002; Trudeau et al. 2012). Moreover, if Cx43 is localized at the mitochondrial membrane, it is likely that other connexin proteins are found there as well. In this context, Cx40 was detected at mitochondria of coronary endothelial cells (Guo et al. 2017) and Cx32 at mitochondria of hepatocytes (Fowler et al. 2013). However, a comprehensive analysis investigating all cell types showing mitochondrial connexins and all connexin proteins involved is still missing and would be of high interest.
Within the last years, mitochondrial activity represented by, e.g., cellular metabolism, ATP content and especially ROS production is getting more and more recognized as important trigger and regulator of stem cell differentiation (Khacho et al. 2016) (for comprehensive review see Lisowski et al. 2018). While undifferentiated embryonic stem cells rely mainly on glycolysis and display small mitochondria with few cristae, they switch to oxidative phosphorylation accompanied by mitochondrial fusion and increased morphological complexity. Moreover, a transient increase of ROS production can be observed during the transition from the pluripotent toward a differentiated state (Lisowski et al. 2018). Preliminary data from our lab demonstrates Cx43 localization at mitochondria of mouse ESCs (Fig. 3f). Similarly to the situation in cardiomyocytes (Heinzel et al. 2005), mitochondrial Cx43 might be involved in regulating ROS production in pluripotent stem cells, an important regulator of cellular differentiation (Khacho et al. 2016; Schmelter et al. 2006). Therefore, mitochondrial connexons could be one important mechanistic link how connexins regulate stem cell maintenance and differentiation in vitro as well as in the embryo in vivo (Fig. 5).
Conclusions
The fact that connexins are expressed in the early embryo is known since almost 4 decades and although most people would agree that connexins play an important role during development, the exact mechanisms are not well understood and it is still unclear if connexin expression is necessarily required during preimplantation development. Although, the early antibody injection experiments suggested an essential requirement of connexins during development, genetic mouse models could not proof that hypothesis. This might be due to the presence of maternal transcripts, the redundant function of several simultaneously expressed connexins as well as other compensatory mechanisms in the complex environment of the embryo. It would be highly interesting to investigate if embryos devoid of all gap junction proteins can still develop beyond gastrulation. However, as null mutations in some connexins expressed at the blastocyst stage display an embryonically lethal phenotype (Cx26, Cx43, Cx45) it will be difficult to create mice deficient in all relevant connexins by natural cross-breeding.
Pluripotent stem cells and more complex cell culture models containing different cell types and mimicking some aspects of embryonic development (blastoids, gastruloids, and organoids) in combination with multiplex CRISPR/Cas9-based genome editing might serve as suitable tools to create tissue structures devoid of any embryonic connexins and could present an experimental platform to decipher the role of connexins in embryogenesis. Moreover, induced pluripotent stem cells enable the investigation of connexins in the context of human development and even in cells directly derived from patients affected by mutated connexin genes.
Finally, changes in cellular metabolism and ROS production are more and more recognized as key drivers of cellular differentiation. In this context, it will be important to investigate mitochondrial connexins in PSCs and their role in controlling cellular metabolism and thereby epigenetics, lineage decision and cellular differentiation.
References
Aach J, Lunshof J, Iyer E, Church GM (2017) Addressing the ethical issues raised by synthetic human entities with embryo-like features. Elife. https://doi.org/10.7554/eLife.20674
Armiger TJ, Lampi MC, Reinhart-King CA, Dahl KN (2018) Determining mechanical features of modulated epithelial monolayers using subnuclear particle tracking. J Cell Sci. https://doi.org/10.1242/jcs.216010
Assou S et al (2007) A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells 25:961–973. https://doi.org/10.1634/stemcells.2006-0352
Bao L, Locovei S, Dahl G (2004) Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett 572:65–68. https://doi.org/10.1016/j.febslet.2004.07.009
Barron DJ, Valdimarsson G, Paul DL, Kidder GM (1989) Connexin32, a gap junction protein, is a persistent oogenetic product through preimplantation development of the mouse. Dev Genet 10:318–323. https://doi.org/10.1002/dvg.1020100407
Becker DL, Evans WH, Green CR, Warner A (1995) Functional-analysis of amino-acid-sequences in connexin43 involved in intercellular communication through gap-junctions. J Cell Sci 108:1455–1467
Beckmann A, Schubert M, Hainz N, Haase A, Martin U, Tschernig T, Meier C (2016) Ultrastructural demonstration of Cx43 gap junctions in induced pluripotent stem cells from human cord blood. Histochem Cell Biol 146:529–537. https://doi.org/10.1007/s00418-016-1469-9
Benchetrit H et al (2015) Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell 17:543–556. https://doi.org/10.1016/j.stem.2015.08.006
Bevilacqua A, Loch-Caruso R, Erickson RP (1989) Abnormal development and dye coupling produced by antisense RNA to gap junction protein in mouse preimplantation embryos. Proc Natl Acad Sci USA 86:5444–5448
Bloor DJ, Wilson Y, Kibschull M, Traub O, Leese HJ, Winterhager E, Kimber SJ (2004) Expression of connexins in human preimplantation embryos in vitro. Reprod Biol Endocrinol 2:25. https://doi.org/10.1186/1477-7827-2-25
Boengler K et al (2005) Connexin 43 in cardiomyocyte mitochondria and its increase by ischemic preconditioning. Cardiovasc Res 67:234–244. https://doi.org/10.1016/j.cardiores.2005.04.014
Boengler K et al (2012) Mitochondrial connexin 43 impacts on respiratory complex I activity and mitochondrial oxygen consumption. J Cell Mol Med 16:1649–1655. https://doi.org/10.1111/j.1582-4934.2011.01516.x
Bruzzone R, Hormuzdi SG, Barbe MT, Herb A, Monyer H (2003) Pannexins, a family of gap junction proteins expressed in brain. Proc Natl Acad Sci USA 100:13644–13649. https://doi.org/10.1073/pnas.2233464100
Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB (2015) Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518:413–416. https://doi.org/10.1038/nature13981
Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. https://doi.org/10.1038/nbt.2507
Choi J et al (2015) A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 33:1173–1181. https://doi.org/10.1038/nbt.3388
Cina C, Bechberger JF, Ozog MA, Naus CCG (2007) Expression of connexins in embryonic mouse neocortical development. J Comp Neurol 504:298–313. https://doi.org/10.1002/cne.21426
Clevers H (2016) Modeling development and disease with organoids. Cell 165:1586–1597. https://doi.org/10.1016/j.cell.2016.05.082
Condorelli DF, Parenti R, Spinella F, Trovato-Salinaro A, Belluardo N, Cicirata VC F (1998) Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur J Neurosci 10:1202–1208. https://doi.org/10.1046/j.1460-9568.1998.00163.x
Cong L et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Crabtree GR, Olson EN (2002) NFAT signaling: choreographing the social lives of cells. Cell 109:S67–S79. https://doi.org/10.1016/S0092-8674(02)00699-2
Dahl E, Winterhager E, Reuss B, Traub O, Butterweck A, Willecke K (1996) Expression of the gap junction proteins connexin31 and connexin43 correlates with communication compartments in extraembryonic tissues and in the gastrulating mouse embryo, respectively. J Cell Sci 109(Pt 1):191–197
Davies TC, Barr KJ, Jones DH, Zhu D, Kidder GM (1996) Multiple members of the connexin gene family participate in preimplantation development of the mouse. Dev Genet 18:234–243. https://doi.org/10.1002/(SICI)1520-6408(1996)18:3<234::AID-DVG4>3.0.CO;2-A
De Sousa PA et al (1997) Normal development of preimplantation mouse embryos deficient in gap junctional coupling. J Cell Sci 110(Pt 15):1751–1758
Duval N, Gomes D, Calaora V, Calabrese A, Meda P, Bruzzone R (2002) Cell coupling and Cx43 expression in embryonic mouse neural progenitor cells. J Cell Sci 115:3241–3251
Egashira K, Nishii K, Nakamura KI, Kumai M, Morimoto S, Shibata Y (2004) Conduction abnormality in gap junction protein connexin45-deficient embryonic stem cell-derived cardiac myocytes. Anat Rec Part A 280a:973–979. https://doi.org/10.1002/ar.a.20110
Elias LAB, Wang DD, Kriegstein AR (2007) Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448:901–903. https://doi.org/10.1038/nature06063
Esseltine JL, Shao Q, Brooks C, Sampson J, Betts DH, Seguin CA, Laird DW (2017) Connexin43 mutant patient-derived induced pluripotent stem cells exhibit altered differentiation potential. J Bone Miner Res 32:1368–1385. https://doi.org/10.1002/jbmr.3098
Etchegaray JP, Mostoslavsky R (2016) Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell 62:695–711. https://doi.org/10.1016/j.molcel.2016.05.029
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156. https://doi.org/10.1038/292154a0
Fowler SL, Akins M, Zhou H, Figeys D, Bennett SA (2013) The Liver connexin32 interactome is a novel plasma membrane-mitochondrial signaling nexus. J Proteome Res 12:2597–2610. https://doi.org/10.1021/pr301166p
Goubaeva F, Mikami M, Giardina S, Ding B, Abe J, Yang J (2007) Cardiac mitochondrial connexin 43 regulates apoptosis. Biochem Bioph Res Commun 352:97–103. https://doi.org/10.1016/j.bbrc.2006.10.177
Guo R, Si R, Scott BT, Makino A (2017) Mitochondrial connexin40 regulates mitochondrial calcium uptake in coronary endothelial cells. Am J Physiol-Cell Ph 312:C398–C406. https://doi.org/10.1152/ajpcell.00283.2016
Gutstein DE, Morley GE, Vaidya D, Liu FY, Chen FL, Stuhlmann H, Fishman GI (2001) Heterogeneous expression of gap junction channels in the heart leads to conduction defects and ventricular dysfunction. Circulation 104:1194–1199. https://doi.org/10.1161/hc3601.093990
Hainz N, Beckmann A, Schubert M, Haase A, Martin U, Tschernig T, Meier C (2018) Human stem cells express pannexins. BMC Res Notes 11:54. https://doi.org/10.1186/s13104-018-3125-z
Hardy K, Warner A, Winston RM, Becker DL (1996) Expression of intercellular junctions during preimplantation development of the human embryo. Mol Hum Reprod 2:621–632
Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M (2017) Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. https://doi.org/10.1126/science.aal1810
He LQ et al (2005) Cx31 is assembled and trafficked to cell surface by ER-Golgi pathway and degraded by proteasomal or lysosomal pathways. Cell Res 15:455–464. https://doi.org/10.1038/sj.cr.7290314
Heinzel FR et al (2005) Impairment of diazoxide-induced formation of reactive oxygen species and loss of cardioprotection in Connexin 43 deficient mice. Circ Res 97:583–586. https://doi.org/10.1161/01.Res.0000181171.65293.65
Heo JS, Han HJ (2006) ATP stimulates mouse embryonic stem cell proliferation via protein kinase C, phosphatidylinositol 3-kinase/Akt, and mitogen-activated protein kinase signaling pathways. Stem Cells 24:2637–2648. https://doi.org/10.1634/stemcells.2005-0588
Houghton FD et al (2002) Functional significance of gap junctional coupling in preimplantation development. Biol Reprod 66:1403–1412. https://doi.org/10.1095/biolreprod66.5.1403
Huettner JE, Lu AW, Qu Y, Wu YJ, Kim M, McDonald JW (2006) Gap junctions and connexon hemichannels in human embryonic stem cells. Stem Cells 24:1654–1667. https://doi.org/10.1634/stemcells.2005-0003
Humpherys D et al (2001) Epigenetic instability in ES cells and cloned mice. Science 293:95–97. https://doi.org/10.1126/science.1061402
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. Elife 2:e00471. https://doi.org/10.7554/eLife.00471
Kalimi GH, Lo CW (1988) Communication compartments in the gastrulating mouse embryo. J Cell Biol 107:241–255
Kawano S et al (2006) ATP autocrine/paracrine signaling induces calcium oscillations and NFAT activation in human mesenchymal stem cells. Cell Calcium 39:313–324. https://doi.org/10.1016/j.ceca.2005.11.008
Ke Q et al (2013) Connexin 43 is involved in the generation of human-induced pluripotent stem cells. Hum Mol Genet 22:2221–2233. https://doi.org/10.1093/hmg/ddt074
Ke Q et al (2017) Enhanced generation of human induced pluripotent stem cells by ectopic expression of Connexin 45. Sci Rep 7:458. https://doi.org/10.1038/s41598-017-00523-y
Khacho M et al (2016) Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell 19:232–247. https://doi.org/10.1016/j.stem.2016.04.015
Kibschull M et al (2004) Connexin31-deficient trophoblast stem cells: a model to analyze the role of gap junction communication in mouse placental development. Dev Biol 273:63–75. https://doi.org/10.1016/j.ydbio.2004.04.037
Kibschull M, Magin TM, Traub O, Winterhager E (2005) Cx31 and Cx43 double-deficient mice reveal independent functions in murine placental and skin development. Dev Dyn 233:853–863. https://doi.org/10.1002/dvdy.20424
Kibschull M, Colaco K, Matysiak-Zablocki E, Winterhager E, Lye SJ (2014) Connexin31.1 (Gjb5) deficiency blocks trophoblast stem cell differentiation and delays placental development. Stem Cells Dev 23:2649–2660. https://doi.org/10.1089/scd.2014.0013
Kidder GM, Mhawi AA (2002) Gap junctions and ovarian folliculogenesis. Reproduction 123:613–620. https://doi.org/10.1530/rep.0.1230613
Kim SN et al (2017) Connexin 43 is required for the maintenance of mitochondrial integrity in brown adipose tissue. Sci Rep UK 7:7159. https://doi.org/10.1038/s41598-017-07658-y
Kruger O et al (2000) Defective vascular development in connexin 45-deficient mice. Development 127:4179–4193
Kubaczka C et al (2015) Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell 17:557–568. https://doi.org/10.1016/j.stem.2015.08.005
Kumai M, Nishii K, Nakamura K, Takeda N, Suzuki M, Shibata Y (2000) Loss of connexin45 causes a cushion defect in early cardiogenesis. Development 127:3501–3512
Laird DW (2006) Life cycle of connexins in health and disease. Biochem J 394:527–543. https://doi.org/10.1042/Bj20051922
Lancaster MA et al (2013) Cerebral organoids model human brain development and microcephaly. Nature 501:373. https://doi.org/10.1038/nature12517
Lee S, Gilula NB, Warner AE (1987) Gap junctional communication and compaction during preimplantation stages of mouse development. Cell 51:851–860
Li H et al (2002) Paradoxical overexpression and translocation of connexin43 in homocysteine-treated endothelial cells. Am J Physiol Heart C 282:H2124–H2133. https://doi.org/10.1152/ajpheart.01028.2001
Li XA et al (2011) Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell 8:46–58. https://doi.org/10.1016/j.stem.2010.11.027
Lisowski P, Kannan P, Mlody B, Prigione A (2018) Mitochondria and the dynamic control of stem cell homeostasis. Embo Rep 19:e45432. https://doi.org/10.15252/embr.201745432
Lo CW, Gilula NB (1979a) Gap junctional communication in the post-implantation mouse embryo. Cell 18:411–422
Lo CW, Gilula NB (1979b) Gap junctional communication in the preimplantation mouse embryo. Cell 18:399–409
Lu G, Haider HK, Jiang SJ, Ashraf M (2009) Sca-1(+) stem cell survival and engraftment in the infarcted heart dual role for preconditioning-induced Connexin-43. Circulation 119:2587–2107. https://doi.org/10.1161/Circulationaha.108.827691
Mali P et al (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. https://doi.org/10.1126/science.1232033
Martin GR (1981) Isolation of a pluripotent cell-line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem-cells. Proc Natl Acad Sci Biol 78:7634–7638. https://doi.org/10.1073/pnas.78.12.7634
Miro-Casas E et al (2009) Connexin43 in cardiomyocyte mitochondria contributes to mitochondrial potassium uptake. Cardiovasc Res 83:747–756. https://doi.org/10.1093/cvr/cvp157
Niessen H, Harz H, Bedner P, Kramer K, Willecke K (2000) Selective permeability of different connexin channels to the second messenger inositol 1,4,5-trisphosphate. J Cell Sci 113(Pt 8):1365–1372
Nishii K, Kobayashi Y, Shibata Y (2016) Absence of connexin43 and connexin45 does not disturb pre- and peri-implantation development. Zygote 24:457–464. https://doi.org/10.1017/S0967199415000386
Okae H et al (2018) Derivation of human trophoblast stem cells. Cell Stem Cell 22:50. https://doi.org/10.1016/j.stem.2017.11.004
Paznekas WA et al (2003) Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet 72:408–418. https://doi.org/10.1086/346090
Plum A et al (2001) Connexin31-deficiency in mice causes transient placental dysmorphogenesis but does not impair hearing and skin differentiation. Dev Biol 231:334–347. https://doi.org/10.1006/dbio.2000.0148
Reaume AG et al (1995) Cardiac malformation in neonatal mice lacking connexin43. Science 267:1831–1834
Rivron NC et al (2018) Blastocyst-like structures generated solely from stem cells. Nature 557:106. https://doi.org/10.1038/s41586-018-0051-0
Rodriguez-Sinovas A et al (2006) Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ Res 99:93–101. https://doi.org/10.1161/01.RES.0000230315.56904.de
Rodriguez-Sinovas A, Ruiz-Meana M, Denuc A, Garcia-Dorado D (2018) Mitochondrial Cx43, an important component of cardiac preconditioning. BBA Biomembr 1860:174–181. https://doi.org/10.1016/j.bbamem.2017.06.011
Sabine A et al (2012) Mechanotransduction, PROX1, and FOXC2 cooperate to control Connexin37 and Calcineurin during lymphatic-valve formation. Dev Cell 22:430–445. https://doi.org/10.1016/j.devcel.2011.12.020
Saito M, Asai Y, Imai K, Hiratoko S, Tanaka K (2017) Connexin30.3 is expressed in mouse embryonic stem cells and is responsive to leukemia inhibitory factor. Sci Rep UK 7:42403. https://doi.org/10.1038/srep42403
Schmelter M, Ateghang B, Helmig S, Wartenberg M, Sauer H (2006) Embryonic stem cells utilize reactive oxygen species as transducers of mechanical strain-induced cardiovascular differentiation. FASEB J 20:1182. https://doi.org/10.1096/fj.05-4723fje
Sharova LV et al (2007) Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol 307:446–459. https://doi.org/10.1016/j.ydbio.2007.05.004
Sheardown SA, Hooper ML (1992) A relationship between gap junction-mediated intercellular communication and the invitro developmental capacity of murine embryonic stem-cells. Exp Cell Res 198:276–282. https://doi.org/10.1016/0014-4827(92)90380-Q
Simon AM, Goodenough DA, Paul DL (1998) Mice lacking connexin40 have cardiac conduction abnormalities characteristic of atrioventricular block and bundle branch block. Curr Biol 8:295–298
Sohl G, Willecke K (2003) An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes 10:173–180
Sohl G, Willecke K (2004) Gap junctions and the connexin protein family. Cardiovasc Res 62:228–232. https://doi.org/10.1016/j.cardiores.2003.11.013
Sohl G, Degen J, Teubner B, Willecke K (1998) The murine gap junction gene connexin36 is highly expressed in mouse retina and regulated during brain development. Febs Lett 428:27–31. https://doi.org/10.1016/S0014-5793(98)00479-7
Srinivas M, Verselis VK, White TW (2018) Human diseases associated with connexin mutations. Biochim Biophys Acta 1860:192–201. https://doi.org/10.1016/j.bbamem.2017.04.024
Stout CE, Costantin JL, Naus CCG, Charles AC (2002) Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. J Biol Chem 277:10482–10488. https://doi.org/10.1074/jbc.M109902200
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. https://doi.org/10.1016/j.cell.2006.07.024
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi.org/10.1016/j.cell.2007.11.019
Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J (1998) Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075. https://doi.org/10.1126/science.282.5396.2072
Thomson JA (1998) Embryonic stem cell lines derived from human blastocysts. Science 282(1147):1827–1827
Todorova MG, Soria B, Quesada I (2008) Gap junctional intercellular communication is required to maintain embryonic stem cells in a non-differentiated and proliferative state. J Cell Physiol 214:354–362. https://doi.org/10.1002/jcp.21203
Trouillas M et al (2009) Three LIF-dependent signatures and gene clusters with atypical expression profiles, identified by transcriptome studies in mouse ES cells and early derivatives. BMC Genomics 10:73. https://doi.org/10.1186/1471-2164-10-73
Trudeau K, Muto T, Roy S (2012) Downregulation of mitochondrial connexin 43 by high glucose triggers mitochondrial shape change and cytochrome c Release in retinal endothelial cells. Invest Ophth Vis Sci 53:6675–6681. https://doi.org/10.1167/iovs.12-9895
Valdimarsson G, Desousa PA, Beyer EC, Paul DL, Kidder GM (1991) Zygotic expression of the Connexin43 gene supplies subunits for gap junction assembly during mouse preimplantation development. Mol Reprod Dev 30:18–26. https://doi.org/10.1002/mrd.1080300103
Valiunas V et al (2005) Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J Physiol Lond 568:459–468. https://doi.org/10.1113/jphysiol.2005.090985
Vance MM, Wiley LM (1999) Gap junction intercellular communication mediates the competitive cell proliferation disadvantage of irradiated mouse preimplantation embryos in aggregation chimeras. Radiat Res 152:544–551
Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH (2014) A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 11:847–854. https://doi.org/10.1038/Nmeth.3016
Wicki-Stordeur LE, Dzugalo AD, Swansburg RM, Suits JM, Swayne LA (2012) Pannexin 1 regulates postnatal neural stem and progenitor cell proliferation. Neural Dev 7:11. https://doi.org/10.1186/1749-8104-7-11
Wicki-Stordeur LE, Sanchez-Arias JC, Dhaliwal J, Carmona-Wagner EO, Shestopalov VI, Lagace DC, Swayne LA (2016) Pannexin 1 differentially affects neural precursor cell maintenance in the ventricular zone and peri-infarct cortex. J Neurosci 36:1203–1210. https://doi.org/10.1523/JNEUROSCI.0436-15.2016
Wolvetang EJ, Pera MF, Zuckerman KS (2007) Gap junction mediated transport of shRNA between human embryonic stem cells. Biochem Biophys Res Commun 363:610–615. https://doi.org/10.1016/j.bbrc.2007.09.035
Wong RCB, Pebay A, Nguyen LTV, Koh KLL, Pera MF (2004) Presence of functional gap junctions in human embryonic stem cells. Stem Cells 22:883–889. https://doi.org/10.1634/stemcells.22-6-883
Wong RC, Dottori M, Koh KL, Nguyen LT, Pera MF, Pebay A (2006) Gap junctions modulate apoptosis and colony growth of human embryonic stem cells maintained in a serum-free system. Biochem Biophys Res Commun 344:181–188. https://doi.org/10.1016/j.bbrc.2006.03.127
Worsdorfer P et al (2008) Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells 26:431–439. https://doi.org/10.1634/stemcells.2007-0482
Worsdorfer P, Thier M, Kadari A, Edenhofer F (2013) Roadmap to cellular reprogramming—manipulating transcriptional networks with DNA, RNA, proteins and small molecules. Curr Mol Med 13:868–878. https://doi.org/10.2174/1566524011313050017
Worsdorfer P et al (2017) Abrogation of gap junctional communication in ES cells results in a disruption of primitive endoderm formation in embryoid bodies. Stem Cells 35:859–871. https://doi.org/10.1002/stem.2545
Yu J et al (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920. https://doi.org/10.1126/science.1151526
Zheng-Fischhofer Q et al (2007a) Characterization of connexin31.1-deficient mice reveals impaired placental development. Dev Biol 312:258–271. https://doi.org/10.1016/j.ydbio.2007.09.025
Zheng-Fischhofer Q et al (2007b) Characterization of connexin30.3-deficient mice suggests a possible role of connexin30.3 in olfaction. Eur J Cell Biol 86:683–700. https://doi.org/10.1016/j.ejcb.2007.01.005
Zong L, Zhu Y, Liang RQ, Zhao HB (2016) Gap junction mediated miRNA intercellular transfer and gene regulation: a novel mechanism for intercellular genetic communication. Sci Rep UK 6:19884. https://doi.org/10.1038/srep19884
Acknowledgements
We are grateful to Helena Dambacher for providing immunofluorescence pictures of Cx43-deficient hIPSCs generated by CRISPR/Cas9 technology. Figures 1, 2, 4, and 5 were produced using graphical elements taken from the image bank of Servier Medical Art (http://www.servier.com) licensed under a Creative Commons Attribution 3.0 Unported License.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wörsdörfer, P., Wagner, N. & Ergün, S. The role of connexins during early embryonic development: pluripotent stem cells, gene editing, and artificial embryonic tissues as tools to close the knowledge gap. Histochem Cell Biol 150, 327–339 (2018). https://doi.org/10.1007/s00418-018-1697-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-018-1697-2