Abstract
Uroplakin (Upk) 3 is one of the main structural components of the urothelium tissue. Although expression of UPK3B is seen in a wider variety of the tissues and organs than UPK3A, tissue-specific expression has not yet been analyzed. Here, we analyzed the Cre recombinase activity driven by the Upk3b promoter in transgenic mice and the endogenous localization of UPK3B. We generated Tg(Upk3b-Cre)/R26tdTomato mice by crossing ROSA26tm14(CAG-tdTomato) (R26tdTomato) mice with Tg(Upk3b-Cre) mice and investigated the spatiotemporal distribution of tdTomato in embryonic and adult mice. In embryos, we detected Cre recombinase activity in neural crest cells and the heart, liver, kidneys, and lungs. In adult mice, Cre recombinase activity was detected in male and female genital organs; however, the activity was absent in the bladder. Histological analyses revealed that both tdTomato and UPK3B were present in testicular and epididymal sperm; however, tdTomato was not present in the ductus epididymis, where the endogenous expression of UPK3B was detected. In female siblings, both tdTomato and UPK3B expressions were detected in the follicles of the ovary, whereas no tdTomato expression was found in the mucosal epithelium of the fallopian tubes, where the endogenous UPK3B was expressed. These data suggest that UPK3B may play a pivotal role in the maturation of gametes and gamete-delivery organs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Uroplakin proteins were purified and identified from rigid-looking plaques covering the apical surface of the urothelium, known as the asymmetric unit membrane (AUM) (Wu et al. 1990; Yu et al. 1990). Uroplakins comprise four different proteins that are tetraspanin-related molecules: uroplakin (UPK) UPK1A (27 kDa), UPK1B (28 kDa), and the single transmembrane proteins UPK2 (15 kDa) and UPK3 (47 kDa) (Wu et al. 1990; Yu et al. 1994). Important roles of uroplakins include permeability control of the apical urothelial membrane (Liang et al. 2001; Wu et al. 1994; Yu et al. 1994) and host cell receptor for bacteria causing urinary tract infection (Duncan et al. 2004; Wu et al. 1996; Zhou et al. 2001). UPK3A is a 47-kDa protein having a single transmembrane domain, a 20-kDa glycosylated luminal (long) domain, and a short 5-kDa cytoplasmic domain (Wu and Sun 1993). Genetic ablation of uroplakin3a (Upk3a) results in global anomalies of the urinary tract and causes primary vesicoureteral reflux (Hu et al. 2000). Upk3a knockout also up-regulates p35 protein, which is named as UPK3B (Deng et al. 2002). Upk3b CreERT2 knock-in mice have been generated; Upk3b CreERT2/CreERT2, the null mutant of UPK3B, does not show any change in urothelium structure (Rudat et al. 2014). Although Upk3a is only expressed in urothelial tissues, Upk3b is expressed in various tissues such as peritoneal, pleural, pericardial, and visceral mesothelia of the lung, heart, liver, spleen, intestine, and testis in adult mice (Kanamori-Katayama et al. 2011; Rudat et al. 2014). These data imply that UPK3B might function independently from UPK3A; however, the histological details of non-urothelial expression of UPK3B have not yet been analyzed.
Studies also show that UPK3 is a key player in frog embryo fertilization. UPK3 is expressed at the surface of the unfertilized egg of the African clawed frog, Xenopus laevis, and has been identified as a target of sperm-derived protease. This protease-mediated cleavage is required for transient increase in Ca2+ upon successful fertilization (Mahbub Hasan et al. 2005; Sakakibara et al. 2005). UPK3 serves as a phosphorylation substrate of the cytoplasmic tyrosine kinase Src (Sakakibara et al. 2005). UPK3-Src system occurs at lipid raft microdomains on unfertilized eggs (Mahbub Hasan et al. 2007), which is strictly regulated by sperm-derived protease (Mahbub Hasan et al. 2014). Despite the conservation of UPK3 within vertebrates, it is not yet clear whether the UPK-Src system is conserved for mammalian fertilization (Kurokawa et al. 2004).
Here, we have generated transgenic mice of harboring Cre recombinase driven by the Upk3b promoter. The Upk3b-Cre activity was monitored by mating with (ROSA)26Sortm14(CAG-tdTomato) mice (hereafter referred to as R26tdTomato mice; Madisen et al. 2010). The peritoneal, pleural, and mesentery mesothelial expressions of Upk3b mRNA have been previously analyzed by quantitative RT-PCR (Kanamori-Katayama et al. 2011). However, we were unsuccessful in generating Tg(Upk3b-Cre)/R26tdTomato mice with robust mesothelial expression of tdTomato; therefore, we investigated Cre recombinase activity in embryonic and adult tissues of Tg(Upk3b-Cre)/R26tdTomato mice. In this study, we have demonstrated that the embryonic expression of tdTomato is localized at the cranial and trunk neural crest cells, and the expression of UPK3B in different organs analyzed by immunohistochemistry in this study correlated well with the RT-qPCR-based expression analysis by Kanamori-Katayama et al. (2011), suggesting that Cre activity is dependent on the Upk3b promoter. Moreover, histological analysis revealed that both the endogenous UPK3B and Cre activities were observed in the adult testis and epididymis; in particular, genetic recombination exhibited robust efficiency in elongating spermatids. In addition, UPK3B expression was observed in the ovaries. The Cre activity and the endogenous UPK3B expression pattern were not identical in this study; however, the utility of the Cre line should be considered in terms of the intensity, uniformity, and specificity of reporter gene expression. Thus, we report that Tg(Upk3b-Cre) mice are a useful tool for labeling gametes in adult mice. Our data also suggest that UPK3B plays an important role in the maturation of haploid spermatids and follicular cells.
Materials and methods
Isolation and subcloning of the uroplakin 3B promoter into the Cre recombinase vector
Genomic DNA was isolated from C57BL/6 N mouse. The 6.7-kbp region upstream of Upk3b gene was amplified by PCR, using the Tks Gflex DNA polymerase (Takara Bio, Shiga, Japan), and subcloned into the pCS2+-Cre recombinase vector. The primers for the amplification of the Upk3b promoter were 5′-TCATCGATGACCAGGCTTAGGGGCTGGTCC-3′ (forward) and 5′-ACCGTCTAGAAG GTGTGCCTTCGGTGAGA-3′ (reverse).
Generation of transgenic Tg(Upk3b-Cre)/ROSA26tm14(CAG-tdTomato) mice
The Upk3b-Cre vector DNA was linearized, and the purified DNA was microinjected into artificially fertilized C57BL/6 N mouse oocytes. Transgene expression was assessed in three mouse lines (two males and one female) among 82 mouse lines by Southern blot (data not shown). These mouse lines were designated as Tg(Upk3b-Cre) mice. The B6.Cg-Gt(ROSA)26Sor tm14(CAG-tdTomato)/Hze/J (R26tdTomato) mouse line (Stock No. 007914; Madisen et al. 2010) was obtained from Jackson Laboratory (CA, USA). Tg(Upk3b-Cre) and R26tdTomato mice were crossed and then genotyped by PCR. The primers for the amplification of wild-type ROSA26 allele were 5′-AAGGGAGCTGCAGTGGAGTA-3′ (forward) and 5′-CCGAAAATCTGTGGGA AGTC-3′ (reverse). The primers for the amplification of mutant R26tdTomato were 5′-GGCATTAAAGCAGCGTATCC-3′ (forward) and 5′-CTGTTCCTGTACGGCAT GG-3′ (reverse).
Quantitative genotyping of mice
The tail of Tg(Upk3b-Cre)/R26tdTomato homozygotes was biopsied. DNA was extracted with 0.25 N NaOH and neutralized with 40 mM Tris-HCl. The number of copies of Cre recombinase integrated into the genome was determined by quantitative PCR, which was performed using Fast Start Essential DNA Green Master and a LightCycler Nano real-time PCR system (Roche Life Science, Penzberg, Germany). The DNA amount for each mouse was estimated with the LightCycler Nano software (Roche). The primers for amplification of Foxn1 were 5′-CTTCCGCCTTTCTCCTTCAG-3′ (forward) and 5′-CC TCATGGAAGTGCCTCTTG-3′ (reverse). The primers for amplification of Cre were 5′-GCGGTCTGGCAGTAAAAACTATC-3′ (forward, olMR1084) and 5′-GTGAAACAGC ATTGCTGTCACTT-3′ (reverse, olMR1085; Jackson Laboratory).
Tissue processing, immunohistochemistry, and fluorescent immunostaining
The dissected tissues were rinsed in ice-cold phosphate-buffered saline (PBS) and then fixed in 4 % (w/v) paraformaldehyde for 16 h at 4 °C. For paraffin processing of tissues, the samples were dehydrated in ethanol, cleared in xylene, and embedded in paraffin. Paraffin blocks were sectioned into 3-µm-thick slices. For immunohistochemistry (IHC), the cross sections were treated with Target Retrieval solution, pH 9 (Dako, Glostrup, Denmark), blocked with normal donkey serum, and incubated with an anti-RFP polyclonal antibody, produced in rabbits (Cat. No. PM005; MBL, Nagoya, Japan). UPK3B was detected with an anti-UPK3B polyclonal antibody produced in rabbits (Cat. No. HPA010506; Sigma-Aldrich, MO, USA). The HRP-conjugated secondary antibody and the reagents for coloring reaction of HRP were provided in the ChemMate ENVISION kit (Dako, Glostrup, Denmark).
For fluorescent immunostaining of tissues, samples were equilibrated with 7 % (w/v) sucrose (in PBS) overnight at 4 °C and treated with 14 % (w/v) calf bone gelatin with 7 % (w/v) sucrose (in PBS) for 30 min at 42 °C. The gelatin blocks were solidified by cooling, and the samples were excised. The gelatin blocks were embedded with optimum cutting temperature compound (Sakura Finetek, Tokyo, Japan), and the frozen samples were sectioned into 10- to 12-µm-thick slices using a Leica CM3050-S research cryostat (Leica Biosystems, Nussloch, Germany). The sections were mounted on MAS-coated slide glasses (Matsunami Glass Ind. Ltd., Osaka, Japan), blocked with 5 % donkey serum, immunoreacted with anti-Sox9 antibody (Abcam, Cambridge, UK, ab3697), and visualized with AlexaFluor 488 anti-Rabbit IgG antibody (Thermo Scientific, Waltham, MA, USA, ab150073). For the identification of the stages of spermatogenesis, we used FITC-labeled peanut agglutinin (PNA) (Vector, Burlingame, CA, USA, FL-1071). For SLA (TRA54) monoclonal antibody (Abnova, Taipei, Taiwan, MAB7952), we used AlexaFluor 488 or 647 anti-Rat IgG antibody (Thermo Scientific).
Results
Generation of transgenic mice expressing Cre recombinase driven by the Upk3b promoter
To generate Cre recombinase transgenic mice driven under the Upk3b promoter (Tg(Upk3b-Cre) mice), we cloned −6.7 kbp upstream of the mouse Upk3b gene. Two males (#6, #32) and one female (#73) transgenic mice were generated and identified (data not shown), and we repeatedly crossed the mice derived from #6 and #32 to increase the activity of Cre recombinases. Tg(Upk3b-Cre) mice were crossed with Gt(ROSA)26Sortm14(CAG-tdTomato) mice, which have a loxP-flanked STOP cassette upstream of the ROSA26/tandem dimer (td) Tomato (modified red fluorescent protein; Madisen et al. 2010). Cre recombinase activity was assessed by fluorescence or IHC. Although tdTomato expression was detected in the mesentery of a small number of transgenic mice (data not shown), a homozygous mouse line with robust Cre recombinase activity in several tissues and cells was identified, indicating that Cre recombinase activity is regulated by the Upk3b gene promoter. In embryonic stages, Cre recombinase activity was detected in cranial and trunk neural crest cells (Fig. 1a, b). The tdTomato expression overlapped with that of Sox9, a known neural crest marker (Cheung and Briscoe 2003; Fig. 1c–e). In some of sibling mice, tdTomato expression was weak in the mesenchyme of the embryo, which may be due to Cre recombination in neural crest cells, also neural-crest-specific Wnt1-Cre mice display similar mesenchymal expression of reporter (Chai et al. 2000; Jiang et al. 2002) (data not shown). Robust Cre recombinase activity was detected in the liver (Fig. 1g). The endogenous UPK3B was observed in heart, liver, kidney, and neural crest-derived cells in each somite (Fig. 1h–n). UPK3B expression was also detected weakly in the lungs (Fig. 1k, arrows), confirming a previously published study (Kanamori-Katayama et al. 2011; Rudat et al. 2014). The expression of UPK3B was also detected in the developing gonad cells (Fig. 1m); therefore, we looked at the further development of gonad.
Cre recombinase activity and UPK3B expression in embryonic mice. a, b Cre recombinase activity was monitored by floxed-tdTomato (red) expression in embryonic mice. TdTomato expression around the brain (Br) of E11 mice (a) and the neural tube (NT) of E13 mice (b). Each scale bar in a, b is 100 µm. c–e Localization of tdTomato-positive cells (red) and Sox9-positive cells (green) in Tg (Upk3b-Cre)/R26tdTomato embryonic mice. Each scale bar is 20 µm. The localization of tdTomato (d) overlapped with that of Sox9 (e). Arrowheads indicate Sox9+ neural crest cells. f Schematic drawing of the cryosectioned region in (g). g Localization of RFP in the liver (Lv), heart (Ht), and intestine (Int). h–n Each scale bar is 250 µm. h HE staining of heart (Ht) and liver (Lv) on the sagittal section of E13.5 embryo. i Localization of UPK3B in the similar section to (h). j HE staining of lung region (Lu) on the sagittal section of E13.5 embryo. k Localization of UPK3B in the similar section to (j) (Arrows). l HE staining of kidney (Kd) and gonad (Go) region. m Localization of UPK3B in the similar section to (l). n Cryosection of somites (So) and kidney (Kd) region. The neural crest cells migrate along somites
Assessment of Cre recombinase activity in adult organs
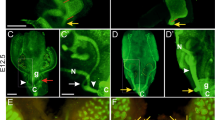
Although CreERT expression of Upk3b CreERT/wt mice has already been analyzed in a previous report (Rudat et al. 2014), the histological details of the Cre activity or the endogenous expression of UPK3B protein in adult mice have not yet been analyzed. Although weak endogenous p35 bands of UPK3B were observed in each lane, tdTomato was only observed in the Cre recombinase-positive testis (Fig. 2a). To confirm the specificity of UPK3B antibody, we examined the absorption of the antibody using HaloTag fusion protein of UPK3B peptide (Fig. S1A). We performed the IHC on the similar section to Fig. 1i with the pre-absorbed UPK3B antibodies, and the result confirmed that the organ expressions were UPK3B-specific (Fig. S1B). Next, in order to obtain robust and uniform expression of tdTomato, we crossed two lines of Tg(Upk3b-Cre) mice, leading to amplification of randomly integrated Upk3b-Cre copies. To estimate the copy number of Cre recombinase in mice, we performed quantitative genotyping. Foxn1 amplification served as an internal control for crude genomic DNA from the tail, and the Cre amount was normalized to that of Foxn1 and compared within the same siblings (Fig. 2b). Cre was not amplified from mouse #3, whereas the Cre level in mouse #2 was approximately 4 times higher than that in the parent mouse (Fig. 2b). We then compared tdTomato expression of these mice; the testis tissue of mouse#2 fluoresced bright red, indicative of tdTomato expression (Fig. 2c), whereas fluorescence was absent in#3 (Fig. 2c’). Although UPK expression correlates with urothelial development, there was no tdTomato expression in the bladder of Cre-positive (#2) and Cre-negative (#3) mice (Fig. 2d, e), similar to a previous report of Upk3b knock-in mice (Rudat et al. 2014). TdTomato expression was also detected in the ovary of the Cre-positive female sibling (Fig. 2g). No abnormalities were apparent in the mice, even those harboring high Cre recombinase copy number.
Cre recombinase activity and quantitative genotyping of adult mice. a Testis (Te), lung (Lu), kidney (Kd), bladder (Bd), muscle (Ms), and trachea with thyroid gland (Tr) lysates were electrophoresed, and immunoblots were probed with anti-RFP (tdTomato), α-tubulin, and UPK3B antibodies. Te+, Upk3b-Cre-positive mouse testis; Te-, Upk3b-Cre-negative mouse testis. Intense bands are seen in (Lu) and (Bd); however, they are too short to be p35 (UPK3B) protein. b Quantitative genotyping of mice by PCR. The number of copies of Cre recombinase integrated into the genome was estimated as a ratio of Cre from Foxn1-control amplification. c, c’ tdTomato expression (red) in the testis of Cre-positive (c) and Cre-negative (c’) mice. d tdTomato expression in the bladder and testis of Cre-positive mice. Bladder was tdTomato-negative. e Absence of tdTomato expression in the bladder and testis of Cre-negative mice. Red autofluorescence was observed. f tdTomato expression in the ovary and uterus of Cre-positive mice. g tdTomato expression in the ovary (Ov) of Cre-positive mice. TdTomato expression in the fallopian tubes (FT) and uterus was insignificant
Histological analysis of Cre-positive/Cre-negative testis and epididymis
To determine whether tdTomato expression is dependent on Upk3b expression, IHC was performed on the testis and epididymis of Cre-positive and Cre-negative mice (Fig. 3a–d). Although the endogenous UPK3B protein was detected in a few of the seminiferous tubules (SeT) (Fig. 3a), genetic recombination was robust in the testis of Cre-positive mice (Fig. 3b), resulting in bright tdTomato fluorescence (Fig. 2c). Endogenous UPK3B expression was also detected in the ductus epididymis (DE) (Fig. 3a, c); however, genetic recombination did not occur even in Cre-positive DE (Fig. 3b). TdTomato was not detected in the testis and epididymis of Cre-negative mice (Fig. 3d). The distribution of the endogenous UPK3B in SeT (Fig. 3e) was similar to that of tdTomato (Fig. 3f), except that fewer SeTs were positive for UPK3B (20.4 ± 6.88 %) compared to tdTomato (96.7 ± 0.66 %). To see whether the copy number of Cre recombinase affects on the expression of tdTomato, we compared the tdTomato-positive ratio between relatively high and low copy Cre mice (Fig. S2). We counted tdTomato-positive or tdTomato-negative SeT and statistically analyzed. Each mouse had a specific value of tdTomato-positive ratio (p = 0.041 < 0.05, one-way ANOVA); however, F value was 2.73 from one-way ANOVA analysis. We could say that the tdTomato-positive ratio of each mouse was not significantly different, and there was no correlation among the copy number of Cre, tdTomato protein level, and the ratio of tdTomato-positive SeT (see Fig. S2), indicating that Upk3b-Cre activity in the testis does not exhibit a mosaic-like pattern. To identify tdTomato-positive cells in the testis, fluorescent immunostaining was performed (Fig. 3g–k). Pachytene spermatocytes at stages VII–VIII (Fig. 3g) and elongating (step 2–12) spermatids at stages XI (Fig. 3h) were immunoreactive for SLA, a spermatocyte and spermatid marker (Pereira et al. 1998), indicating that genetic recombination occurs in haploid spermatids. Furthermore, we stained the acrosomes of spermatids with the lectin peanut agglutinin (PNA)/DAPI double staining (Nakata et al. 2014; Fig. 3i), or PNA/SLA/DAPI triple staining to identify accurate stages of spermatogenesis (Fig. 3j, k). As the results, UPK3B-Cre activities were well correlated to S10-S16 spermatids (data not shown).
Cre recombinase activity in spermatids and spermatozoa. a–d) UPK3B and tdTomato expression in the testis (Te) and epididymis (Edd) of Cre-positive and Cre--negative mice by immunohistochemistry. Each scale bar is 200 µm. a UPK3B expression was observed in 20.4 ± 2.88 % of seminiferous tubules (SeT) in the testis of Cre-positive mice. The ductus epididymis (DE) was positive for UPK3B. b tdTomato expression was observed in 96.4 ± 0.66 % of SeT in the testis of Cre-positive mice. c UPK3B expression in the testis and epididymis of Cre-negative mice. d Absence of tdTomato expression in the testis and epididymis of Cre-negative mice. e, f UPK3B and tdTomato expression in SeT from the testis of Cre-positive mice. Each scale bar is 50 µm. g–k Alexa488/647 fluorescent immunostaining of anti-SLA (TRA54, haploid-specific antigen), a spermatocyte and spermatid marker, in SeT from the testis of Cre-positive mouse with tdTomato fluorescence and DAPI nucleus staining. Roman numerals correspond to the stages of the seminiferous epithelial cycle. Each scale bar in (g, h) is 20 µm. i FITC-conjugated lectin peanut agglutinin (PNA) staining with DAPI and tdTomato fluorescences. The stage of spermatids is S13. The scale bar indicates 10 µm. j, k 4-color images: PNA (FITC: cyan), SLA (Alexa647: yellow), tdTomato (magenta), and DAPI (gray). Each scale bar is 10 µm. l, m UPK3B and tdTomato expression in the epididymis of Cre-positive mice. Each scale bar is 50 µm. n The endogenous UPK3B localization in the isolated sperm in the epididymis lumen. DAB signal is localized at the middle of the sperm tail (arrowhead). The scale car indicates 12.5 µm. SP sperm, P pachytene spermatocytes, Prl preleptotene spermatocytes, Z zygotene spermatocytes, Se Sertoli cells, S11–S16 elongating spermatids, DE ductus epididymis, Nuc the head nuclei of sperm
Epididymal expression pattern of the endogenous UPK3B differs from the observed Upk3b-Cre activity
Even though the endogenous expression of UPK3B in ductal epithelium was relatively high (Fig. 3l), the Cre activity in DE was probably not enough to cleave the floxed genome from ROSA alleles (Fig. 3m). A high cellular turnover rate in rapidly proliferating tissue (Akagi et al. 1997) could also be a reason for loss of the tdTomato signal, as cells positive for recombination would have died prior to our observation. To compare the IHC intensity of the endogenous UPK3B and promoter-derived tdTomato expressions in the sperm of epididymal lumen, we estimated the cell numbers from red (DAB, IHC signals) and blue (nucleus, HE staining) image analysis by using ImageJ. When we count the nucleus and UPK3B-positive particles separately, the ratio of UPK3B-positive sperm was approximately 99.95 % (Nucleus: 1036.68 ± 1.29, UPK3B-positive particles: 1036.15 ± 1.72); however, the co-localization ratio of UPK3B on nucleus was 44.19 ± 2.7 % (Fig. 3l). On the other hand, tdTomato-positive ratio was approximately 87.4 % calculated from the estimated cell numbers (tdTomato: 2106.31 ± 2.25, nucleus: 2408.74 ± 1.94; Fig. 3m). When we looked at the isolated sperm in the epididymal lumen, we found that DAB signal of UPK3B was specifically localized at the middle of the sperm tail (Fig. 3n). Therefore, the DAB staining and the nucleus should not be overlapped. Unfortunately, UPK3B antibody does not suit for immunofluorescence; thus, further analysis will be needed for clarifying the positive ratio of UPK3B in the isolated sperm. Because of these observed differences between sperm and sperm-delivery organs, we analyzed tdTomato expression in female genitals.
Histological analyses of the ovary, uterus, and fallopian tubes of Tg(Upk3b-Cre) mice
Upk3b expression in the ovary, uterus, and fallopian tubes of Cre-positive female siblings was studied. The endogenous UPK3B was detected in ovary, infundibulum, and fallopian tubes (Figs. 2g, 4a, b) while tdTomato expression was detected in the ovary but not in the uterus and fallopian tubes (Figs. 2g, 4c). The primary follicles were not UPK3B-positive, although tdTomato was weakly expressed (Fig. 4d–f). UPK3B and tdTomato expressions were gradually increased from secondary follicles to vesicular follicles (Fig. 4g–l). Notably, tdTomato expressions were also detected in corpus luteum (Fig. 4o) and degenerating corpus luteum (data not shown); therefore, tdTomato fluorescence looks ubiquitous from whole mount sample (Fig. 2g). Although tdTomato-IHC was very weak in fallopian tubes (Fig. 4r), the endogenous expression of UPK3B was observed in the mucosal epithelium (MEp) of the infundibulum and fallopian tubes (Fig. 4q). Interestingly, the endogenous UPK3B expression was seen both in sperm and DE of males and in the follicles and MEp of females, whereas Cre activity was absent from the epithelium of gamete-delivery organs. It is tempting to consider a similar role of UPK3B for male and female gamete maturation; however, further research is warranted for this conclusion.
Cre recombinase activity in the ovary. a Cross section of the ovary (Ov), fallopian tubes (FT), and infundibulum (In) stained with hematoxylin and eosin. b, c Localization of UPK3B and tdTomato in these tissues by immunohistochemistry. Scale bars in (a–c) are 0.5 mm. Each scale bar in (d–r) is 50 µm. d–f A primary follicle. g–i A secondary follicle. j–l A vesicular follicle. m–o A corpus luteum. p–r The section of fallopian tubes. d, g, j, m, p HE staining. e, h, k, n, q UPK3B-IHC. f, i, l, o, r tdTomato expression. Both UPK3b and tdTomato were observed everywhere in ovary. Especially in the secondary or the vesicular follicle, the signals in the granulosa cells of these follicles are relatively strong. p In the fallopian tubes, the mucosal epithelium (MEp) was observed with a lot of cytosolic granules. q UPK3B expression was not overlapped with the granules. r tdTomato-staining was very faint
In conclusion, histological analysis revealed that UPK3B protein is localized to the elongating spermatids and the ductal epithelium of epididymis in male mice genital organs and ubiquitously expressed in the ovary, especially in the follicles and the mucosal epithelium of the fallopian tubes of female mice. In the Upk3b-Cre mouse line, the flanked stop codons are cleaved from the genome in developing sperm and follicles. Thus, the Upk3b-Cre mouse is a useful tool to label gametes in both sexes and distinguish them from the surrounding gamete-delivery tissues.
Discussion
To our knowledge, it is the first study to analyze UPK3B expression in the mature testis. Histological analysis demonstrated that UPK3B is specifically activated at the maturation stage of spermatid. We also observed endogenous expression of UPK3B in sperm; therefore, future studies on its role in sperm function could be anticipated. Our Tg(Upk3b-Cre) mice have robust expression in the testis and ovary, which can be differentiated from the gonad. Thus, the −6.7-kb Upk3b promoter drives Cre recombinase independent of sex determination. The epididymis differentiates from Wolffian duct, which regresses in females (Blecher and Erickson 2007). On the other hand, the uterine tubes differentiate from the Mullerian duct, which regresses in males (Blecher and Erickson 2007). Although the epididymis and the uterine tubes are mutually exclusive as they are sex-dependent, endogenous UPK3B was strongly detected in both the epididymis and uterine tubes (Figs. 3, 4). Therefore, the endogenous UPK3B expression in gamete-delivery organs could also be sex-independent. Possibly, UPK3B expression is controlled as per structural requirements. In the urothelium, UPK3A plays an essential role as a structural component; therefore, knockout of UPK3A causes defects in urothelium function (Hu et al. 2000). However, the expression level of UPK3B is usually 10 % of that of UPK3A in the bladder (Rudat et al. 2014) and is activated as compensation for ablation of UPK3A (Deng et al. 2002). Therefore, we propose that UPK3B could be the major structural protein of mucosal epithelium of the gamete-delivery organs. Since there is no study on the phenotypic effects of Upk3b knockout on fertility, further investigation of the effect of conditional double mutants of UPK3A and 3B on delivery organs and gametes is required.
Comparison between Tg(Upk3b-Cre) mice and previously reported spermatid-specific Cre transgenic mice
Several spermatid-specific Cre recombinant lines have been reported; however, histological analysis was not available for them. We hereby address the similarity of our study to others, as well as the enhanced information our Upk3b-Cre transgenic line provides. Tspy-Cre and Z/EG double-transgenic mice were generated, and Cre activity was observed in spermatids and parts of the brain (Kido and Lau 2005). Although UPK3B was not seen in spermatogonia, endogenous TSPY expression was observed in several germ cell populations, including spermatogonia (Kido and Lau 2005). In a previous report, AQP2-Cre (aquaporin2) expression was analyzed in the kidney and testis (Nelson et al. 1998). The endogenous AQP2 is localized in spermatids; however, the localization of Cre protein driven by the AQP2 promoter was unclear (Nelson et al. 1998). Cre activity monitored by the reporter gene in the testis has been reviewed by Smith (2011); however, the expression in the kidney and epididymis was not known. Protamine1 (Prm1)-Cre mice have also been generated, in which Cre activity was observed in haploid spermatids (O’Gorman et al. 1997), and probably most popular testis-specific Cre mice. Notably, the Prm1-Cre gene was segregated by meiosis; therefore, the ratio of Cre-positive spermatids should be 50 %. However, the actual percentage increased to 92 % because the gene products were shared between germ cells via cytoplasmic bridges (O’Gorman et al. 1997; Smith 2011). It could be similar for Upk3b-Cre mice, where 96.7 % of spermatids are reporter-positive, whereas the endogenous UPK3B-positive cells were only 20.4 % (Fig. 3). For Prm1-Cre/R26GFP mice, GFP expression was observed in the center of the SeT (Bell et al. 2014). The endogenous protamin-1 (PRM1) distribution has been analyzed as its expression was observed in elongating spermatids but was absent in round spermatids (Fukuda et al. 2004). Thus, the expression patterns of Upk3B-Cre and Prm1-Cre were quite similar.
Conservation of spermatid-specific UPK3 expression among vertebrates
In spite of the large number of studies describing the role of UPK3 in urothelium, there is a lack of understanding of the role of UPK3 in mammalian fertilization. UPK3 is expressed on the surface of Xenopus eggs and is cleaved by sperm-specific protease during fertilization (Mahbub Hasan et al. 2005). The spatiotemporal and subcellular localizations of Src phosphorylation are dependent on UPK3, and the Src activation is precisely regulated by sperm protease (Mahbub Hasan et al. 2007, 2014). Therefore, the cleavage of UPK3 on the egg surface is essential for fertilization in Xenopus. In mammalian fertilization, the Src activation is not essential for triggering the calcium wave (Kurokawa et al. 2004). Thus, the role of UPK3 in mammalian fertilization does not seem to be conserved. In this study, we demonstrate that UPK3B-Cre activity existed in both sperm and follicles; moreover, endogenous UPK3B protein was observed in gamete-delivery organs of both sexes. Therefore, we speculate the absence of sexually biased expression of UPK3B on the egg surface. Since this is the first report of UPK3B expression in tissues of reproductive organs, further research is required to address its precise role in these organs.
References
Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A (1997) Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res 25:1766–1773
Bell EL, Nagamori I, Williams EO, Del Rosario AM, Bryson BD, Watson N, White FM, Sassone-Corsi P, Guarente L (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141:3495–3504
Blecher SR, Erickson RP (2007) Genetics of sexual development: a new paradigm. Am J Med Genet A 143A:3054–3068
Chai Y, Jiang X, Ito Y, Bringas P Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM (2000) Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127:1671–1679
Cheung M, Briscoe J (2003) Neural crest development is regulated by the transcription factor Sox9. Development 130:5681–5693
Deng FM, Liang FX, Tu L, Resing KA, Hu P, Supino M, Hu CC, Zhou G, Ding M, Kreibich G, Sun TT (2002) Uroplakin IIIb, a urothelial differentiation marker, dimerizes with uroplakin Ib as an early step of urothelial plaque assembly. J Cell Biol 159:685–694
Duncan MJ, Li G, Shin JS, Carson JL, Abraham SN (2004) Bacterial penetration of bladder epithelium through lipid rafts. J Biol Chem 279:18944–18951
Fukuda N, Yomogida K, Okabe M, Touhara K (2004) Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci 117:5835–5845
Hu P, Deng FM, Liang FX, Hu CM, Auerbach AB, Shapiro E, Wu XR, Kachar B, Sun TT (2000) Ablation of uroplakin III gene results in small urothelial plaques, urothelial leakage, and vesicoureteral reflux. J Cell Biol 151:961–972
Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM (2002) Tissue origins and interactions in the mammalian skull vault. Dev Biol 241:106–116
Kanamori-Katayama M, Kaiho A, Ishizu Y, Okamura-Oho Y, Hino O, Abe M, Kishimoto T, Sekihara H, Nakamura Y, Suzuki H, Forrest AR, Hayashizaki Y (2011) LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS ONE 6:e25391
Kido T, Lau YF (2005) A Cre gene directed by a human TSPY promoter is specific for germ cells and neurons. Genesis 42:263–275
Kurokawa M, Sato K, Smyth J, Wu H, Fukami K, Takenawa T, Fissore RA (2004) Evidence that activation of Src family kinase is not required for fertilization-associated [Ca2 +]i oscillations in mouse eggs. Reproduction 127:441–454
Liang FX, Riedel I, Deng FM, Zhou G, Xu C, Wu XR, Kong XP, Moll R, Sun TT (2001) Organization of uroplakin subunits: transmembrane topology, pair formation and plaque composition. Biochem J 355:13–18
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140
Mahbub Hasan AK, Sato K, Sakakibara K, Ou Z, Iwasaki T, Ueda Y, Fukami Y (2005) Uroplakin III, a novel Src substrate in Xenopus egg rafts, is a target for sperm protease essential for fertilization. Dev Biol 286:483–492
Mahbub Hasan AK, Ou Z, Sakakibara K, Hirahara S, Iwasaki T, Sato K, Fukami Y (2007) Characterization of Xenopus egg membrane microdomains containing uroplakin Ib/III complex: roles of their molecular interactions for subcellular localization and signal transduction. Genes Cells 12:251–267
Mahbub Hasan AK, Hashimoto A, Maekawa Y, Matsumoto T, Kushima S, Ijiri TW, Fukami Y, Sato K (2014) The egg membrane microdomain-associated uroplakin III-Src system becomes functional during oocyte maturation and is required for bidirectional gamete signaling at fertilization in Xenopus laevis. Development 141:1705–1714
Nakata H, Wakayama T, Takai Y, Iseki S (2014) Quantitative analysis of the cellular composition in seminiferous tubules in normal and genetically modified infertile mice. J Histochem Cytochem 63:99–113
Nelson RD, Stricklett P, Gustafson C, Stevens A, Ausiello D, Brown D, Kohan DE (1998) Expression of an AQP2 Cre recombinase transgene in kidney and male reproductive system of transgenic mice. Am J Physiol 275:C216–C226
O’Gorman S, Dagenais NA, Qian M, Marchuk Y (1997) Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A 94:14602–14607
Pereira LA, Tanaka H, Nagata Y, Sawada K, Mori H, Chimelli LM, Nishimune Y (1998) Characterization and expression of a stage specific antigen by monoclonal antibody TRA 54 in testicular germ cells. Int J Androl 21:34–40
Rudat C, Grieskamp T, Rohr C, Airik R, Wrede C, Hegermann J, Herrmann BG, Schuster-Gossler K, Kispert A (2014) Upk3b is dispensable for development and integrity of urothelium and mesothelium. PLoS ONE 9:e112112
Sakakibara K, Sato K, Yoshino K, Oshiro N, Hirahara S, Mahbub Hasan AK, Iwasaki T, Ueda Y, Iwao Y, Yonezawa K, Fukami Y (2005) Molecular identification and characterization of Xenopus egg uroplakin III, an egg raft-associated transmembrane protein that is tyrosine-phosphorylated upon fertilization. J Biol Chem 280:15029–15037
Smith L (2011) Good planning and serendipity: exploiting the Cre/Lox system in the testis. Reproduction 141:151–161
Wu XR, Sun TT (1993) Molecular cloning of a 47 kDa tissue-specific and differentiation-dependent urothelial cell surface glycoprotein. J Cell Sci 106(Pt 1):31–43
Wu XR, Manabe M, Yu J, Sun TT (1990) Large scale purification and immunolocalization of bovine uroplakins I, II, and III. Molecular markers of urothelial differentiation. J Biol Chem 265:19170–19179
Wu XR, Lin JH, Walz T, Haner M, Yu J, Aebi U, Sun TT (1994) Mammalian uroplakins. A group of highly conserved urothelial differentiation-related membrane proteins. J Biol Chem 269:13716–13724
Wu XR, Sun TT, Medina JJ (1996) In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc Natl Acad Sci U S A 93:9630–9635
Yu J, Manabe M, Wu XR, Xu C, Surya B, Sun TT (1990) Uroplakin I: a 27-kD protein associated with the asymmetric unit membrane of mammalian urothelium. J Cell Biol 111:1207–1216
Yu J, Lin JH, Wu XR, Sun TT (1994) Uroplakins Ia and Ib, two major differentiation products of bladder epithelium, belong to a family of four transmembrane domain (4TM) proteins. J Cell Biol 125:171–182
Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP (2001) Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J Cell Sci 114:4095–4103
Acknowledgments
The authors thank Dr. Ryouma Haraguchi (Univ. Ehime, Ehime, JAPAN) for his comments, Dr. Daniel Buchholz (Univ. Cincinnati, OH, USA) for providing the pCS2 + -Cre vector, and Dr. Naoki Takeda (CARD-Kumamoto University, Kumamoto, Japan) for microinjection of the mouse oocytes. Technical support was provided by the Scientific Support Programs for Cancer Research Grant-in-Aid for Scientific Research on Innovative Areas, Ministry of Education, Culture, Sports, Science and Technology (MEXT). SK was supported by MEXT KAKENHI 25111702 and JSPS KAKENHI 15K06822. MT was supported by JSPS KAKENHI 25290042 and 26640068.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
All animal experiments were approved by, and conducted in compliance with the protocol which was reviewed by Animal Health Committee in Akita University.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kuriyama, S., Tamiya, Y. & Tanaka, M. Spatiotemporal expression of UPK3B and its promoter activity during embryogenesis and spermatogenesis. Histochem Cell Biol 147, 17–26 (2017). https://doi.org/10.1007/s00418-016-1486-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-016-1486-8