Abstract
Gap junction proteins are essential for direct intercellular communication but also influence cellular differentiation and migration. The expression of various connexin gap junction proteins has been demonstrated in embryonic stem cells, with Cx43 being the most intensely studied. As Cx43 is the most prominent gap junction protein in the heart, cardiomyocyte-differentiated stem cells have been studied intensely. To date, however, little is known about the expression and the subcellular distribution of Cx43 in undifferentiated stem cells or about the structural arrangement of channels. We, therefore, here investigate expression of Cx43 in undifferentiated human cord-blood-derived induced pluripotent stem cells (hCBiPS2). For this purpose, we carried out quantitative real-time PCR and immunohistochemistry. For analysis of Cx43 ultrastructure and protein assembly, we performed freeze-fracture replica immunogold labeling (FRIL). Cx43 expression was detected at mRNA and protein level in hCBIPS2 cells. For the first time, ultrastructural data are presented on gap junction morphology in induced pluripotent stem (iPS) cells from cord blood: Our FRIL and electron microscopical analysis revealed the occurrence of gap junction plaques in undifferentiated iPS cells. In addition, these gap junctions were shown to contain the gap junction protein Cx43.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Communication between adjacent cells is an important factor during development and for the physiological functioning of almost all cells and organs. Connexins are a family of transmembrane proteins known to build gap junctions for direct intercellular communication and signaling between cells. Gap junctions are permeable for molecules with a molecular mass of up to 1 kDa. The connexin (Cx) protein family consists of 21 members in humans, which are named according to their approximate molecular weight (Sohl and Willecke 2004). Most connexin proteins show a distinct cell- and tissue-specific expression: Cx36 is a neuronal protein, Cx47 is expressed by oligodendrocytes, and Cx43 is found in astrocytes and cardiomyocytes, to name a few. Surprisingly, connexin proteins have also been detected in stem cells of various sources (Kar et al. 2012): Undifferentiated human embryonic stem cells (hESC) express mRNA for almost all known connexin subtypes and display intercellular dye transfer, which is characteristic of gap junctional communication (Huettner et al. 2006). Induced pluripotent stem cells (iPSC), obtained by the reprogramming of somatic fibroblast cells, were also shown to express most connexin subtypes as described for hESC (Oyamada et al. 2013). However, little is known about the subcellular distribution of connexin proteins in stem cells or the assembly of gap junctional plaques, and even less about their function.
Connexin proteins mostly differ in the length of their C terminus, but otherwise show a comparable structure: Four transmembrane domains are connected via two extracellular loops and one cytoplasmic loop with the C- and the N-termini being located within the cytoplasm. Six connexin proteins form the so-called connexon, a tubular transmembrane channel (Sosinsky and Nicholson 2005). Protein folding and assembly of connexons occur in the endoplasmic reticulum and the Golgi apparatus. After vesicular transport and insertion of a cluster of connexons into the plasma membrane, connexons can couple to another connexon from an adjacent cell to form a cell–cell-connecting gap junction channel or potentially function as hemichannels per se. Several channels usually unite to form gap junction plaques, which can differ in size from several channels up to thousands (Evans et al. 2006; Meier et al. 2004). The short half-life of connexin proteins ensures the high adaptability of gap junctional communication under pathologic circumstances, such as injuries and ischemia, or during different cellular responses, such as differentiation and proliferation. In view of this, the understanding of the mechanism underlying gap junction formation is an important issue. The life of gap junctions has been described as a process of generation and degeneration of gap junction plaques (Evans et al. 2006; Flores et al. 2012; Gaietta et al. 2002; Johnson et al. 2012). In light microscopy, several groups demonstrated the reformation of gap junction plaques after photobleaching, showing an assembly of channels along the outside edge of a gap junction plaque (Segretain and Falk 2004).
In this study, we analyzed the clone number two of human cord-blood-derived induced pluripotent stem cells (HCBiPS2), which are known to express Cx43 after differentiation into cardiomyocytes (Haase et al. 2009). As it is not known whether undifferentiated hCBiPS2 cells show gap junction formation or even express connexins, the aim of the study was to investigate the expression of Cx43 and to analyze the extent of Cx43 gap junction formation. For monitoring Cx43 mRNA and protein expression, quantitative real-time polymerase chain reaction and immunofluorescence analysis were applied. The morphology of Cx43 gap junction plaques in hCBiPS2 cells was investigated at the ultrastructural level. Freeze-fracture replica immunogold labeling (FRIL) is an outstanding method for studying the assembling of gap junctions at the subcellular level as it combines the freeze-fracture method for high-resolution electron microscopy with the advantages of protein detection by means of immunogold-labeled antibodies (Fujimoto 1995; Rash et al. 2000; Robenek and Severs 2008). Here, we identify gap junctions ultrastructurally in undifferentiated cord-blood-derived induced pluripotent stem cells and show that these gap junctions were shown to contain Cx43.
Materials and methods
Cell culture
hiPSC lines were generated by lentiviral transduction of cord-blood-derived endothelial cells (hCBiPS2) as previously described (Haase et al. 2009). The hCBiPS2 cells were cultured on irradiated mouse embryonic fibroblasts (MEFs) in knockout Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20 % knockout serum replacement, 1 mM l-glutamine, 0.1 mM β-mercaptoethanol, 1 % non-essential amino acid stock (all from life technologies, Darmstadt, Germany), and 10 ng/ml basic fibroblast growth factor (bFGF; supplied by the Institute for Technical Chemistry, Leibniz University, Hannover, Germany) (Chen et al. 2012). For FRIL and electron microscopy, the cells were expanded as monolayer cultures on Geltrex® (life technologies) for 4 days as previously described (Burridge et al. 2011). Under these culture conditions, cells maintained pluripotency and did not differentiate (Burridge et al. 2011; Haase et al. 2009; Xu et al. 2001).
The human embryonic stem cell line HES-3 was cultured and expanded under standard hESC culture conditions (Haase et al. 2009).
The astrocytoma cell line U87-MG was a gift from Prof. Gerald Thiel, University of Saarland, Department of Medical Biochemistry and Molecular Biology. Cells were cultivated in high-glucose DMEM supplemented with 10 % fetal calf serum, glutamine and penicillin/streptomycin (all from Life Technologies) at 37 °C in a 5 % CO2 atmosphere.
RNA isolation, cDNA synthesis, standard PCR, and quantitative real-time PCR
Total RNA was isolated from hCBiPS2 and HES-3 cells frozen in TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. After thawing, 0.2 ml chloroform was added to 1 ml TRIzol reagent/cell sample and mixed vigorously for 15 s. After incubation for 2–3 min, samples were centrifuged, and thereafter, the upper aqueous phase containing RNA was transferred to a new tube. RNA was precipitated by adding 100 % isopropanol to the aqueous phase with subsequent incubation for a period of 10 min. Following centrifugation, the supernatant was discarded and the RNA pellet was purified by adding 1 ml 75 % ethanol. Finally, the mixture was centrifuged and the RNA pellet was dried after removal of the supernatant. The RNA pellet was resuspended in 60 µl RNase-free water. Afterward, genomic DNA was eliminated by DNase digestion using RNAse-free DNase I set (Qiagen, Hilden, Germany). DNase I in RDD buffer was added to the RNA solution with subsequent incubation for 10 min. After incubation, the RNA solution was mixed with RLT buffer and 100 % ethanol and given onto an RNeasy Mini spin column (Qiagen, Hilden, Germany). After several washing steps, the RNA was eluted from the RNeasy Mini spin column membrane with RNase-free water. Subsequently, the RNA was quantified using the spectrophotometer Nano Drop ND-1000 (Thermo Fisher Scientific, Heidelberg, Germany).
Based on 500 ng RNA per sample, cDNA was synthesized using M-MuLV reverse transcriptase with oligo(dt) primer, dNTP mix, RNase inhibitor and RT buffer (all reagents New England BioLabs, Frankfurt/Main, Germany). To exclude incomplete genomic DNA elimination, we performed controls without MuLV reverse transcriptase, and to test the purity of the reagent/chemicals used, we performed a control transcription without RNA.
Standard PCR was performed with the synthesized cDNA and with all controls in a volume of 25 μl containing dNTP mix (Thermo Fisher Scientific), 10 × Thermo Polymerase Buffer, Taq polymerase (both New England BioLabs) and specific forward and reverse primers (Table 1). Amplification was performed in a LifeTouch Thermal Cycler (Biozym, Hess. Oldendorf, Germany) under the following conditions: Each cycle was performed with an initial denaturation at 95 °C for 30 s, annealing at primer-specific temperature (Table 1) for 30 s and extension at 68 °C for 2 min. After primer-specific cycles, final extension was performed at 68 °C for 10 min. The PCR products were separated by electrophoresis on a 2 % agarose gel containing ethidium bromide for visualization of the PCR product.
Quantitative real-time PCR was performed using Takyon Rox SYBR MasterMix dTTP Blue (Eurogentec, Köln, Germany) according to the manufacturer’s instructions on a StepOnePlus Real-Time PCR System (Applied Biosystems/Life Technologies, Darmstadt, Germany). In brief, cDNA was diluted 1:3 with water and then mixed with Takyon Mastermix and specific primers (Table 1). All probes were measured in triplicate. The relative expression was calculated using the ΔΔCt method according to Pfaffl (Pfaffl 2001). The mean of all triplets from genes of interest were normalized to the mean of 18S rRNA as reference.
Immunohistochemistry
For immunohistochemical staining, hCBiPS2 or U87-MG cells were fixed with 100 % ethanol for 20 min at RT. Blocking was performed with incubation in 10 % normal goat serum (NGS) and 0.1 % triton in PBS for 1 h at RT. Primary antibodies against Cx43 (ab11370, Abcam, Cambridge, UK) were diluted 1:100 in primary blocking solution for 2 h at RT. After several rinses with PBS, incubation in secondary blocking solution 0.2 % bovine serum albumin (BSA) in PBS was performed for 30 min at RT. Anti-rabbit Alexa Fluor®488 diluted 1:3000 in secondary blocking solution was used as secondary antibodies. Incubation was performed for 1 h at RT. After several rinses with PBS, the cells were mounted in fluoromount with DAPI. Analysis was performed with a fluorescent microscope with respective filter sets and digital camera (Zeiss, Jena, Germany).
Freeze-fracture and immunogold labeling (FRIL)
For FRIL analysis, hCBiPS2 or U87-MG cells were fixed and processed as described previously. Freeze fracturing of double replicas was achieved by cracking the double carrier open and was performed at −162 °C and 8 × 10−8 mbar (EMBAF060, Leica, Wetzlar, Germany). Using a quartz crystal thickness monitor to determine coating thickness, fractured samples were coated with a 1-nm pre-carbon coat applied at a shadowing angle of 90°, a 1-nm platinum–carbon coat was applied at 60° and a second carbon coat of 20 nm was applied at 90°. The frozen replicas were stabilized on a gold index grid (Plano, Wetzlar, Germany) using a drop of 1 % Lexan polycarbonate plastic dissolved in dichloroethane (DCE; Acros, Geel, Belgium) (Rash et al. 1997). The package of carrier-sample-replica-Lexan-grid was incubated at −20 °C for 16 h to evaporate the DCE. Then the samples were thawed at room temperature and the carriers removed. Digestion of the sample tissue was performed in SDS-digestion buffer (2.5 % SDS, 10 mM Tris–HCl, pH 8.9) at 60 °C with agitation for a total of 27 h. The replica was then digested in 2 % collagenase D (Roche, Mannheim, Germany) in SPB at 37 °C with agitation for 90 min. Blocking of nonspecific binding sites was achieved by incubation in labeling blocking buffer (LBB) (10 % goat serum, 1.5 % teleost gelatin in 0.15 M SPB) at 25 °C for 90 min. Primary antibodies against Cx43 (ab11370, Abcam) were diluted 1:10 in LBB and incubation was carried out overnight at 25 °C. Incubation with 12 and 18 nm anti-rabbit colloid gold-conjugated antibodies, diluted 1:20 and 1:10, respectively, in LBB (Jackson ImmunoResearch, Newmarket, UK), was performed at 25 °C under gentle agitation for 22 h. Thereafter, the replicas were washed in SPB and water and dried prior to application of a 20-nm carbon backing coat. Finally, the Lexan was removed from the replica by dipping the replica in DCE at a temperature of 60°.
Electron microscopy
Labeled replicas were analyzed in a FEI Technai G2 transmission electron microscope (FEI, Hillsboro, Oregon, USA) at 100 kV. Pictures were taken with an 8-bit camera with an image size of 1.42 Megapixel (Olympus MegaView III; Olympus, Hamburg, Germany). To distinguish gold on the tissue side from nonspecific labeling on the replica side, photographs were taken as stereo pairs at an 8° angle and viewed stereoscopically (Rash and Yasumura 1999). The international standard nomenclature for freeze fracture was used to describe the FRIL data (Branton et al. 1975). For antibodies directed against the membrane protein of interest itself, a gold bead-to-particle distance <30 nm, corresponding to the length of two antibodies, was considered particle-associated labeling (Fujimoto 1997; Rash et al. 1998, 2004).
Results
Transcription of Cx30.2 and Cx43 in undifferentiated human induced pluripotent stem cells from cord blood
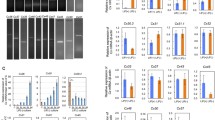
As it was unknown whether undifferentiated hCBiPS2 cells transcribe connexin mRNA, we initially performed cDNA synthesis plus standard PCR with specific primers (Table 1). We chose Cx30.2 as a connexin known from myelinated cells and expressed at low levels in stem cells. The second connexin analyzed was Cx43, which is expressed in most tissues, and was shown to play a role in the reprogramming of somatic cells during iPS generation (Ke et al. 2013). ES cells were used for comparison, as they are known to express both types of connexins (Huettner et al. 2006). cDNA of HES-3 and hCBiPS2 cells as well as appropriate controls served as templates. Amplification of 18S rRNA served as reference gene. PCR results showed that Cx30.2 and Cx43 both were expressed in HES-3 and hCBiPS2 cells (Fig. 1a).
Detection of Cx30.2 and Cx43 mRNAs in HES-3 and hCBiPS2 cells. a RNA of HES-3 and hCBiPS2 cells was isolated and cDNAs transcribed via reverse transcription. Cx30.2 and Cx43 were amplified with specific primers. PCR-probes were separated via agarose gel electrophoresis. For both connexins and 18S, gels showed a band of the expected height, b–d quantitative real-time PCR of Cx30.2 and Cx43 in stem cells. qPCR data for Cx30.2 (b) or Cx43 (c) of hCBiPS2 cells compared to HES-3 cells, d relative RNA expression of Cx43 in hCBiPS2 cells compared to Cx30.2
Next, the RNA levels were determined at a quantitative level (Fig. 1b–d). We performed cDNA synthesis plus quantitative real-time PCR with connexin-specific primers (Table 1). Quantification revealed that Cx30.2 is highly expressed in hCBiPS2 cells, 56-fold higher than in HES-3 cells (Fig. 1b). For Cx43, hCBiPS2 cells showed a 1.4-fold higher relative expression rate than HES-3 cells (Fig. 1c). Comparing the expression of both connexins in hCBiPS2 cells, Cx43 expression was more than eightfold higher than that of Cx30.2 (Fig. 1d).
Expression of Cx43 protein in human induced pluripotent stem cells from cord blood
Due to the high mRNA level of Cx43, we focused on the analysis of Cx43 protein expression in hCBiPS2 cells. For this purpose, hCBiPS2 cells were stained with specific antibodies against Cx43. In hCBiPS2 cells, a characteristic punctate immunofluorescent signal of Cx43 gap junction plaques was detected in the plasma membrane (Fig. 2a–c). In comparison, the human glioblastoma–astrocytoma cell lineage U87-MG was used as a positive control for astrocytic Cx43 expression and showed a similar but noticeably less intense pattern of immunolabeling at sites of cell contact (Fig. 2d, e).
Undifferentiated hCBiPS2 cells express Cx43. Immunohistochemical staining for Cx43; nuclei were stained with DAPI (blue). a–c hCBiPS2 cells were immunohistochemically stained with antibodies against Cx43. a Phase contrast, b Cx43 immunofluorescence signal (green), c Cx43 immunofluorescence signal (green) plus DAPI staining of nuclei (blue), d–f in comparison, U87-MG cells were stained with antibodies against Cx43, d differential interference contrast (DIC) light microscopy, e Cx43 immunofluorescence signal (green), f Cx43 immunofluorescence signal (green) plus DAPI staining of nuclei (blue). Scale bars 100 µm
Cx43 gap junction plaques in human induced pluripotent stem cells from cord blood
To investigate the ultrastructure and assembly of Cx43 protein in membranes of hCBiPS2 cells, freeze-fracture replica immunogold labeling was performed. As known in freeze fracturing, protoplasmic (P-face) and extraplasmic (E-face) leaflets of fractured membranes can be observed (Branton et al. 1975). The process visualized several gap junction plaques labeled for Cx43 (Fig. 3a, b), which displayed the typical assembly of connexon particles with characteristic particle size of approximately 10 nm diameter and periodicity within the gap junction plaque. In some gap junctions, the fracture plane ran through a gap junction plaque, thereby separating P- and E-faces of a membrane. Thus, P-face gap junction particles were adjacent to arrayed pits on adjoining E-faces (Fig. 3c), with distinct narrowing of the extracellular space within the margins of the gap junctions. These neighboring P- and E-faces of connexons and narrowing of the extracellular space confirm that intercellular gap junctions have in fact been formed. A schematic illustration of the resulting replica labeled for Cx43 is presented in Fig. 3d, e.
Undifferentiated hCBiPS2 cells show gap junction plaques in the plasma membrane, which consist of Cx43. The cells were vitrified, freeze-fractured under high vacuum and replicated with carbon and platinum. The replica was incubated with primary antibodies against Cx43 and colloidal-gold-coupled secondary antibodies (12 and 18 nm diameter). a Overview of the fractured and replicated membrane, showing two gap junctions in the upper right. b Magnification of the membrane area boxed in a, showing compact gap junction plaques (areas colored in green). Membrane faces are labeled by E (E-face) or P (P-face), c stereo pair at higher magnification of boxed area in b. Photographs were taken at an 8° angle to allow stereoscopic view and assessment of three-dimensional structure. Membrane fracture divides the gap junction plaque in an E- and a P-face half, d schematic illustration of a mature gap junction plaque between adjacent cells coupled via Cx43 proteins (green) assembled to hexameric connexons with identical spacing The fracture plane is illustrated by a red line, e the illustration shows the fracture plane (red) dividing membranes in an E- and a P-face, the platinum coat (violet) after freeze-fracture and possible binding of antibodies. Arrow indicates the direction of platinum shadowing; labeling is illustrated by primary antibodies against Cx43 (blue), secondary antibodies (green) and conjugated gold particles (black circles). Note that “cryptic labeling” is closely associated with the margins of the gap junctions, potentially indicating insertion of clusters of connexons in the apposed cell’s unfractured membrane. Scale bars in a and b are 0.2 µm, in c 0.1 µm. Schematic illustrations are not to scale
Several aspects illustrate the specificity of immunolabeling. P-face particles and E-face pits are considered labeled if they are located within a radius of <30 nm of a gold bead (Fujimoto 1997; Rash et al. 1998, 2004). With a signal-to-noise ratio of approximately 370:1 (determined in one of the replicas; signal = gold on morphologically identified gap junctions; noise = gold on membrane areas distant from gap junctions) (Beckmann et al. 2016; Rash and Yasumura 1999), background labeling was very low. Interestingly, in areas close to a gap junction—and only there (Fig. 3a)—many more gold beads were visible, located primarily beneath E-faces (Fig. 3a–c). In our interpretation, these are likely to arise from cryptic labeling (Rash and Yasumura 1999), particularly as both sizes of gold are located in identical regions. However, we cannot exclude nonspecific binding of antibodies at these sites nor can we exclude cryptic labeling of unfractured gap junctions in incompletely digested gap junctions in overlapping cell appositions beneath the replicated freeze-fractured membrane (see Fig. 2 for evidence of overlapping cell margins). The examination of stereo pairs showed that almost all gold beads were located on the tissue side of the replica. The epitope of the antibody used was on the cytoplasmic C terminus of Cx43 (Beckmann et al. 2016; Johnson et al. 2012). Labeling for Cx43 was performed using two different secondary antibodies conjugated to colloidal gold of different size, and both displayed an overlapping labeling for Cx43 (Fig. 3a–c).
Assembly of Cx43 connexons in human induced pluripotent stem cells from cord blood
In addition to the compact assembly of gap junction channels, areas with a less ordered assembly of particles were observed. Figure 4 shows an example of such an assembly. The particles had an average diameter of 9.41 nm (±1.11), as determined by the measurement of 130 randomly selected particles. This diameter corresponds to the typical size of replicated connexons (Fujimoto 1995). The particular assembly shown in Fig. 4 consists of 531 particles and has a size of 235.8 nm2. Accumulation of particles was observed to occur in chains, in dense arrangements or in a less ordered, loose pattern. In areas with highly regular assembly of particles, 9700 particles per µm2 were counted. In all other areas, particle density was reduced: In less dense areas, 4322.6 particles per µm2 were counted. In areas with unorganized particle arrangements, the ratio was 2251.8 particles per µm2 (in contrast, the surrounding displayed 1847.8 particles of different diameter per µm2). E-faces, like those seen in Cx43 gap junctions of U87-MG cells (Fig. 4b), always displayed a compact and regular assembly of connexons (compare Fig. 3). Figure 4c shows a schematic illustration of the fractured and replicated assembly of P-face particles, including areas devoid of connexons.
Gap junction formation in undifferentiated hCBiPS2 cells. a Colored area shows a less compact form of gap junction plaque labeled for Cx43 (gold particles, 12 and 18 nm). Several particles aggregate in loose chains (arrow heads) with particle free areas (light green) within the gap junction plaque, b the insert in a shows the E-face of a typical compact gap junction labeled for Cx43 (gold particles, 12 and 18 nm) in U87-MG cells, c schematic illustration of P-face connexons in a single cell membrane after freeze-fracture and shadowing. Arrow indicates the direction of platinum shadowing (violet); labeling is illustrated showing primary antibodies against Cx43 (blue), secondary antibodies (green) and conjugated gold particles (black circles). The green arrow points to particle free areas within the gap junction, which were observed within gap junctions (a; light green area). Scale bar (a, b) 0.2 µm. Schematic illustrations are not to scale
Discussion
Up to now, Cx43 expression in stem cells has been mainly discussed with a view to cardiomyocyte development and differentiation. Here, we present data on a differentiation-independent expression of Cx43 in undifferentiated iPS cells from human cord blood, pointing to a function of Cx43 in these cells:
-
1.
We have demonstrated ultrastructurally that gap junctions occur in near confluent cultures of iPS cells from human cord blood.
-
2.
We show by FRIL analysis that those gap junctions contain Cx43.
Cx43 was identified to be a major protein component of these gap junction plaques. We identified Cx43-immunolabeled particles in compact gap junction assemblies with identical distances between individual connexons. In those gap junction plaques connecting two cells, the fracture plane might travel through each of the adjacent cell membranes. The resulting replica contains an E- and a P-face as for instance shown in Fig. 3. This substantiates the hypothesis of cell–cell communication and the transfer of molecules smaller than 1 kDa via the docked connexons of two adjacent cells. Note that the extracellular space is narrowed within the margins of the immunogold-labeled, ultrastructurally identified gap junctions. Cx43 is a remarkable protein with several putative roles. First of all, Cx43 is abundantly expressed in most tissues and cell types. Cx43 is the gap junction protein of cardiomyocytes, expressed at intercalated disks and maintaining correct rhythm of the heartbeat, thus, preventing arrhythmias (Beyer et al. 1987). In the brain, Cx43 is the most abundant connexin, expressed for instance in astrocytes. In these cells, Cx43 contributes to the maintenance of homoeostasis, the initiation of calcium waves, etc. (Naus et al. 2016). Secondly, Cx43 exhibits an important function in the migration of cells as studies on neural progenitors (Elias et al. 2007; Naus et al. 2016) and cardiac neural crest (Huang et al. 1998) have shown. A third important function of Cx43 was described in 2013, when Ke and colleagues described a crucial role of Cx43 during the reprogramming of somatic cells (Ke et al. 2013) and therefore identified Cx43 as a suitable marker for pluripotency (Oyamada et al. 2013).
Ke et al. associated Cx43 expression with the reprogramming progress (Ke et al. 2013) and demonstrated that the presence of Cx43 enhanced the reprogramming efficiency. However, the application of pharmacological inhibitors of gap junctional communication did not inhibit this effect, pointing to a function of Cx43, which is independent of intercellular communication (Ke et al. 2013). Worsdorfer et al. demonstrated an effect of Cx43 on embryonic stem cell survival. Interestingly, they observed that pharmacologic inhibition of Cx43-induced apoptotic cells death, whereas the deletion of Cx43 in transgenic stem cells did not influence their survival (Worsdorfer et al. 2008). The importance of Cx43 for multi- or pluripotency seems a more generalized phenomenon, as it has also been described for skin-derived stem cells that their multipotency was dependent on Cx43 (Dyce et al. 2014).
In summary, we here identified gap junction channels in undifferentiated iPS cells from human cord blood ultrastructurally and demonstrated that these channels were composed of the gap junction protein Cx43. In view of the multiple cellular functions of Cx43, these data provide the basis for ultrastructural analysis and association of Cx43 assembly with distinct functional stem cell properties.
Abbreviations
- iPSC:
-
Induced pluripotent stem cells
- Cx43:
-
Connexin 43
- FRIL:
-
Freeze-fracture replica immunogold labeling
- hCBiPS2:
-
Human cord-blood-derived induced pluripotent stem cell clone 2
- hESC:
-
Human embryonic stem cells
References
Beckmann A, Grissmer A, Krause E, Tschernig T, Meier C (2016) Pannexin-1 channels show distinct morphology and no gap junction characteristics in mammalian cells. Cell Tiss Res 363:751–763. doi:10.1007/s00441-015-2281-x
Beyer EC, Paul DL, Goodenough DA (1987) Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol 105:2621–2629
Branton D et al (1975) Freeze-etching nomenclature. Science 190:54–56
Burridge PW et al (2011) A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS ONE 6:e18293. doi:10.1371/journal.pone.0018293
Chen R et al (2012) Cytokine production using membrane adsorbers: human basic fibroblast growth factor produced by Escherichia coli. Eng Life Sci 12:29–38. doi:10.1002/elsc.201100045
Dyce PW, Li D, Barr KJ, Kidder GM (2014) Connexin43 is required for the maintenance of multipotency in skin-derived stem cells. Stem Cells Dev 23:1636–1646. doi:10.1089/scd.2013.0459
Elias LA, Wang DD, Kriegstein AR (2007) Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448:901–907. doi:10.1038/nature06063
Evans WH, De Vuyst E, Leybaert L (2006) The gap junction cellular internet: connexin hemichannels enter the signalling limelight. Biochem J 397:1–14. doi:10.1042/BJ20060175
Flores CE, Nannapaneni S, Davidson KG, Yasumura T, Bennett MV, Rash JE, Pereda AE (2012) Trafficking of gap junction channels at a vertebrate electrical synapse in vivo. Proc Natl Acad Sci USA 109:E573–E582. doi:10.1073/pnas.1121557109
Fujimoto K (1995) Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci 108(Pt 11):3443–3449
Fujimoto K (1997) SDS-digested freeze-fracture replica labeling electron microscopy to study the two-dimensional distribution of integral membrane proteins and phospholipids in biomembranes: practical procedure, interpretation and application. Histochem Cell Biol 107:87–96
Gaietta G et al (2002) Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503–507. doi:10.1126/science.1068793
Haase A et al (2009) Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5:434–441. doi:10.1016/j.stem.2009.08.021
Huang GY, Cooper ES, Waldo K, Kirby ML, Gilula NB, Lo CW (1998) Gap junction-mediated cell-cell communication modulates mouse neural crest migration. J Cell Biol 143:1725–1734
Huettner JE, Lu A, Qu Y, Wu Y, Kim M, McDonald JW (2006) Gap junctions and connexon hemichannels in human embryonic stem cells. Stem Cells 24:1654–1667. doi:10.1634/stemcells.2005-0003
Johnson RG et al (2012) Gap junction assembly: roles for the formation plaque and regulation by the C-terminus of connexin43. Mol Biol Cell 23:71–86. doi:10.1091/mbc.E11-02-0141
Kar R, Batra N, Riquelme MA, Jiang JX (2012) Biological role of connexin intercellular channels and hemichannels. Arch Biochem Biophys 524:2–15. doi:10.1016/j.abb.2012.03.008
Ke Q et al (2013) Connexin 43 is involved in the generation of human-induced pluripotent stem cells. Hum Mol Genet 22:2221–2233. doi:10.1093/hmg/ddt074
Meier C, Dermietzel R, Davidson KG, Yasumura T, Rash JE (2004) Connexin32-containing gap junctions in Schwann cells at the internodal zone of partial myelin compaction and in Schmidt-Lanterman incisures. J Neurosci 24:3186–3198. doi:10.1523/JNEUROSCI.5146-03.2004
Naus CC, Aftab Q, Sin WC (2016) Common mechanisms linking connexin43 to neural progenitor cell migration and glioma invasion. Semin Cell Dev Biol 50:59–66. doi:10.1016/j.semcdb.2015.12.008
Oyamada M, Takebe K, Endo A, Hara S, Oyamada Y (2013) Connexin expression and gap-junctional intercellular communication in ES cells and iPS cells. Front Pharmacol 4:85. doi:10.3389/fphar.2013.00085
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acid Res 29:e45
Rash JE, Yasumura T (1999) Direct immunogold labeling of connexins and aquaporin-4 in freeze-fracture replicas of liver, brain, and spinal cord: factors limiting quantitative analysis. Cell Tiss Res 296:307–321
Rash JE, Duffy HS, Dudek FE, Bilhartz BL, Whalen LR, Yasumura T (1997) Grid-mapped freeze-fracture analysis of gap junctions in gray and white matter of adult rat central nervous system, with evidence for a “panglial syncytium” that is not coupled to neurons. J Comp Neurol 388:265–292
Rash JE, Yasumura T, Hudson CS, Agre P, Nielsen S (1998) Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc Natl Acad Sci USA 95:11981–11986
Rash JE, Staines WA, Yasumura T, Patel D, Furman CS, Stelmack GL, Nagy JI (2000) Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin-36 but not connexin-32 or connexin-43. Proc Natl Acad Sci USA 97:7573–7578
Rash JE, Davidson KG, Yasumura T, Furman CS (2004) Freeze-fracture and immunogold analysis of aquaporin-4 (AQP4) square arrays, with models of AQP4 lattice assembly. Neuroscience 129:915–934. doi:10.1016/j.neuroscience.2004.06.076
Robenek H, Severs NJ (2008) Recent advances in freeze-fracture electron microscopy: the replica immunolabeling technique. Biol Proced Onl 10:9–19. doi:10.1251/bpo138
Segretain D, Falk MM (2004) Regulation of connexin biosynthesis, assembly, gap junction formation, and removal. Biochim Biophys Acta 1662:3–21. doi:10.1016/j.bbamem.2004.01.007
Sohl G, Willecke K (2004) Gap junctions and the connexin protein family. Cardiovasc Res 62:228–232. doi:10.1016/j.cardiores.2003.11.013
Sosinsky GE, Nicholson BJ (2005) Structural organization of gap junction channels. Biochim Biophys Acta 1711:99–125. doi:10.1016/j.bbamem.2005.04.001
Worsdorfer P et al (2008) Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cell 26:431–439. doi:10.1634/stemcells.2007-0482
Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK (2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol 19:971–974. doi:10.1038/nbt1001-971
Acknowledgments
The authors wish to thank Andrea Rabung and Alexander Grissmer for excellent technical assistance. They also thank Ann Soether for linguistic editing and Alina Mattheis for the schematic illustrations. The authors are very grateful to Prof. Dr. John E. Rash for his continuous support and expert advice on the manuscript. The authors acknowledge financial support by the German Research Foundation and the Saarland, who funded the freeze-fracture unit. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
All authors declare that research was done without any potential conflict of interest. No competing financial interests exist.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beckmann, A., Schubert, M., Hainz, N. et al. Ultrastructural demonstration of Cx43 gap junctions in induced pluripotent stem cells from human cord blood. Histochem Cell Biol 146, 529–537 (2016). https://doi.org/10.1007/s00418-016-1469-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-016-1469-9