Abstract
Hair shafts are one of the most common types of evidence at crime scenes, and mitochondrial DNA (mtDNA) has been analyzed as a valuable genetic marker for hair shafts in forensic casework. However, the mtDNA analysis strategy may vary according to the quantity and quality of DNA extracted from a forensic sample and the available massively parallel sequencing (MPS) platform in laboratories. Forensic practitioners often have to interpret mtDNA sequences exhibiting point heteroplasmy (PHP) that are analyzed using different analytical methods. In the present study, the whole mitochondrial genome (mtGenome) variants of hair shaft samples obtained from 20 donors, which were sampled in duplicate and stored at room temperature for > 1 year, were analyzed using the Precision ID mtDNA Whole Genome Panel and Ion S5 system. The whole mtGenome variants of 20 blood and 20 buccal swab samples (reference samples) from the hair shaft donors were analyzed using the Nextera XT DNA Library Prep Kit and MiSeq System. A total of 20 unique mtGenome haplotypes were observed, and 56 PHP variants were identified across the 4 sets of tissue. When the major nucleotide of PHP was considered, 16 of 20 haplotypes of the hair shaft samples matched those of the corresponding blood and buccal swab samples. In four donors, the major nucleotide of PHP was inverted at one nucleotide position between the hair shaft and reference samples. However, the data obtained on MPS, showing high PHP resolution, provided substantial information to avoid false exclusion when comparing two haplotypes containing PHP with inverted major nucleotides. In conclusion, the present study demonstrates the utility of MPS in forensic casework in the comparative analysis of mtGenome variants containing PHP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitochondrial DNA (mtDNA) testing is useful when analyzing poor-quality samples, particularly those in which the amount of nuclear DNA (nDNA) is extremely scarce to yield informative short tandem repeat profiles. The high copy number of mtDNA (approximately 103–104 copies/cell) [1] can provide clues in the analysis of forensic samples, such as highly degraded skeletal remains and hair shafts [2].
Numerous hair shafts are shed daily and can be useful as trace evidence at a crime scene. Therefore, many studies have identified mtDNA as a valuable genetic marker for hair shafts in forensic casework [3, 4]. Traditionally, most studies focused on the 1.1-kb mtDNA control region (CR), a noncoding region that contains 3 hypervariable regions, using Sanger sequencing [5, 6]. However, it is recognized that expanding the analysis range from the CR to whole mitochondrial genome (mtGenome) could increase the discrimination power [7]. Additionally, the application of massively parallel sequencing (MPS) has resulted in whole mtGenome sequencing becoming more feasible and sophisticated [8, 9].
Regarding the analysis of mtDNA variants by MPS, forensic practitioners generally consider the quantity and quality of DNA extracted from forensic samples and the available MPS platform in laboratories. For example, when dealing with forensic casework evidence such as aged hair shafts and skeletal remains, a short amplicon strategy is recommended because of the severely degraded condition of such samples [10, 11]. However, in the case of high-quality samples, such as blood and buccal swabs, a long-range (LR) amplification strategy is considered well-suited and efficient [8, 9, 12].
On the other hand, the proportion and distribution of point heteroplasmy (PHP) differ among tissues [13, 14], even if they are obtained from the same individual or maternal lineage. Therefore, PHP should be carefully considered when interpreting the mtDNA sequence match between questioned and known samples for their exclusion. Because MPS facilitates the detection of mtDNA PHP at a higher resolution than other methods [15], MPS data may reduce the possibility of false exclusion in cases wherein the major nucleotide of PHP differs among samples and/or the proportion of its minor nucleotide is very low. The latter case may cause confusion because it would be observed as homoplasmy when analyzed using a low-resolution method, particularly Sanger sequencing.
In this study, we analyzed the whole mtGenome sequences of hair shafts, blood, and buccal swab samples obtained from 20 donors using different MPS methods. Hair shaft samples had been stored at room temperature for > 1 year and were analyzed using the Precision ID mtDNA Whole Genome Panel (Thermo Fisher Scientific, Waltham, MA, USA) on the Ion S5 system (Thermo Fisher Scientific). Blood and buccal swab samples were analyzed using the Nextera XT DNA Library Prep Kit on the MiSeq system (Illumina, San Diego, CA, USA). Here, we have demonstrated a method to analyze whole mtGenome variants with the adopted MPS platform by considering the tissue type. In addition, we have demonstrated the utility of MPS when interpreting PHP in the evaluation of mtGenome variants in forensic casework.
Materials and methods
Sample preparation and DNA extraction from hair shaft samples
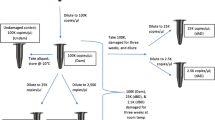
Hair shaft samples were collected from 20 unrelated Korean males after obtaining informed consent. More than 5 hair shafts were collected from each donor, 2 of which were randomly selected for analysis. The samples had been stored at room temperature for > 1 year prior to DNA extraction to simulate the uncollected hair shafts from crime scenes often encountered by forensic practitioners. The hair shaft samples were analyzed independently and consistently throughout all experimental processes. The first proximal 2-cm fragment of each hair shaft sample, including the root, was removed to exclude nDNA from mtDNA analysis. The second proximal 2-cm fragment was prepared in duplicate as follows: 1 ml of 5% UV-irradiated TergAZyme (Alconox, White Plains, NY, USA) was dispensed into a tube and a hair shaft section was inserted. After vortexing for 1 min, the samples were sonicated at room temperature for 15 min. A filter cup was placed on a new tube and prewetted with dH2O; then, the sonicated hair shaft was placed in the filter cup using forceps and sequentially washed with 1 ml of UV-irradiated dH2O, 0.85% saline, and 100% ethanol. Next, the filter cup containing the hair shaft sample was separated from the tube, inverted, and air dried. Then, the desiccated hair was transferred to a clean 1.5-ml tube for DNA extraction using the QIAamp DNA Investigator Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
mtDNA amplification and MPS library preparation of hair shaft samples
Extracted DNA (3 μl) from each hair shaft sample was subjected to PCR amplification of the whole mtGenome using the Precision ID mtDNA Whole Genome Panel with 22 thermal cycles on the Veriti 96-Well Thermal Cycler (Thermo Fisher Scientific). Barcoded MPS libraries were prepared using the 2-in-1 method with the Precision ID Library Kit (Thermo Fisher Scientific) and constantly purified with 1.5× Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). The MPS library was quantified using the Ion Library TaqMan Quantitation Kit (Thermo Fisher Scientific) on the AB 7500 Real-Time PCR System (Thermo Fisher Scientific) and normalized to 30 pM. Finally, pooled libraries composed of 8 MPS libraries with the same quantity were enriched on the Ion Chef system (Thermo Fisher Scientific) and sequenced on the Ion S5 system (Thermo Fisher Scientific) with the Ion 520 Chip (Thermo Fisher Scientific). All procedures for MPS library preparation and sequencing were performed according to the application guide for the Precision ID mtDNA Whole Genome Panel.
Sample collection and DNA extraction from blood and buccal swab samples
Peripheral blood and buccal swab samples were collected from each Korean male who donated hair shafts. In brief, approximately 2 ml of blood was collected from each donor and immediately dispensed into a blood collection tube containing EDTA solution. Buccal swab samples were obtained by rubbing the inside of the donors’ mouths with a sterile cotton swab 2–4 times. All samples were stored at 4 °C until genomic DNA extraction using the Qiagen QIAamp DNA Mini Kit according to the manufacturer’s protocol. The extracted DNA was quantified using the Quantifiler Trio DNA Quantification Kit (Thermo Fisher Scientific) on the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s protocol; then, it was diluted to a concentration of 1.0 ng/μl for downstream analyses. This study was approved by the Institutional Review Board of Severance Hospital, Yonsei University, Seoul, Korea.
MPS library construction for blood and buccal swab samples
The LR-PCR mixture was prepared to a final volume of 20 μl that contained 2.0 μl of 10× LA PCR Buffer II with Mg2+, 2.5 mM of each dNTP, 1.5 U LA Taq DNA Polymerase Hot Start Version (all Takara Bio, Inc., Kusatsu, Shiga, Japan), 0.5 μM of the primer set validated by Fendt et al. [16], and 1 ng of genomic DNA extracted from the blood and buccal swab samples. The PCR mixture was subjected to an initial denaturation at 94 °C for 1 min, followed by 26 cycles of denaturation at 94 °C for 20 s, annealing at 60 °C for 30 s, and extension at 68 °C for 9 min as well as a final extension step at 72 °C for 10 min on the Veriti 96-Well Thermal Cycler. The amplicon sizes and quantities were estimated using the Agilent DNA 12000 Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A total of 250 pg of PCR product containing 125 pg of each LR amplicon per sample was run. MPS libraries were constructed using the Nextera XT DNA Library Prep Kit with 250 pg of PCR products. The size range and concentration of the constructed library were assessed using the Agilent High Sensitivity DNA Kit on the Agilent 2100 Bioanalyzer. Subsequent PCR cleanup and size selection were performed with 0.6× Agencourt AMPure XP beads, and the libraries were quantified using the KAPA Library Quantification Kit for Illumina platforms (KAPA Biosystems, Wilmington, MA, USA) on the AB 7500 Real-Time PCR System. Finally, each library was normalized to 10 nM and pooled by volume, and the pooled libraries were sequenced on the MiSeq system with the MiSeq Reagent Kit v3 (2 × 300 cycles; Illumina). All procedures for MPS library preparation and sequencing were performed according to the reference guide of the Nextera XT DNA Library Prep Kit.
MPS data analysis
For each blood and buccal swab sample, we mapped the reads on the revised Cambridge Reference Sequence (rCRS) [17] and rotated rCRS with the BWA-mem [18] after trimming the adapter sequences using the Cutadapt v1.18 software [19]. The general approach for mapping reads to rCRS is problematic because it misses reads on CR, which is the break point to construct a linear reference from the circular DNA that it is located in. To overcome the relatively lower coverage of such regions including the break point, we aligned and called variants separately for the coding regions with rCRS and CR with rotated rCRS. We used the GATK Base Recalibration module to recalibrate the base quality score considering sequencing errors for each aligned file [20]. Variations were called using GATK MuTect2 with the mitochondrial mutation calling mode. The uncertain variations caused by technical artifacts and sequencing errors were filtered out using the GATK FilterMutectCalls module. The separately obtained variants were merged after removing the leftovers for aligned reads on rotated rCRS.
For hair shaft samples, MPS data generated using the Ion S5 system were transferred to the Converge 2.1 plug-in and analyzed using HID Genotyper 2.1. (Thermo Fisher Scientific). Data were aligned to rCRS + 80 bp as a reference genome to adjust for the overlapping designs of tiled amplicon multiplexes [17, 21].
All variants were called only at positions with a read depth of > 100×. Called variants were confirmed according to the International Society for Forensic Genetics (ISFG) guidelines [22]. Homoplasmic indels were confirmed with the Integrative Genomics Viewer (IGV) [23] and notated using the EMPOP mtDNA database v4 (http://empop.online) [24]. For hair shaft samples, the variants were automatically called using the Converge 2.1 software following ISFG guidelines. PHP was reported using more stringent thresholds to exclude the effects of system noise in both MPS data analyses. PHP was called when the total coverage on a nucleotide reached a read depth of > 400×, and the minor nucleotides of PHP were observed in > 5% of the total coverage. Variants in length heteroplasmy containing a poly-C stretch were not considered in this study.
Sanger sequencing for verifying inverted major nucleotides
To confirm the inverted major nucleotide observed in the MPS data, we performed Sanger sequencing. The target mtDNA region was amplified in a PCR mixture prepared with 2.0 μl of Gold ST★R 10× Buffer (Promega Corp., Madison, WI, USA), 0.8 μM of the primer set shown in Supplementary Table S1, 1.5 U of AmpliTaq Gold DNA Polymerase (Thermo Fisher Scientific), and DNA template (3 μl of hair shaft extracts and 1 ng of blood or buccal swab extracts, respectively). Thermal cycling was performed using the Veriti 96-Well Thermal Cycler under the following conditions: 95 °C for 11 min; 37 cycles for hair shaft samples and 33 cycles for blood and buccal swab samples at 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 7 min. The PCR products were purified with 2 μl of ExoSAP-IT™ PCR Product Cleanup Reagent (Thermo Fisher Scientific) and incubated at 37 °C for 45 min followed by at 80 °C for 15 min. Sanger sequencing was performed at Macrogen, Inc. (Seoul, Republic of Korea) using a universal primer (Supplementary Table S1), and the results were analyzed using the SeqSacpe 2.6 software (Thermo Fisher Scientific).
Results
MPS coverage of the whole mtGenome
A total read count of 23,666,165 was obtained by combining hair shaft sets #1 and #2. For hair shaft set #1 (n = 20), 12,152,521 reads were obtained and the average read count was 607,626 (mean depth: 4316×). Sample 004 showed the highest coverage (average: 878,577 reads, mean depth: 6262×), whereas Sample 001 showed the lowest coverage (average: 456,159 reads, mean depth: 3227×). Hair shaft set #2 (n = 20) had a total of 11,513,644 reads and an average read count of 575,682 (mean depth: 4108×). Sample 012 showed the highest coverage (average: 733,106 reads, mean depth: 5239×), whereas Sample 001 showed the lowest coverage (average: 346,456 reads, mean depth: 2467×).
MtDNA extracted from the 20 blood and 20 buccal swab samples were sequenced on the MiSeq system, yielding a total of 23,745,480 (average: 1,187,274 reads, mean depth: 15,979×) and 23,833,444 (average: 1,191,672 reads, mean depth: 11,840×) reads, respectively. Among the blood samples, Sample 014 showed the highest coverage (average: 1,879,932, mean depth: 27,655×), whereas Sample 001 showed the lowest coverage (average: 832,178, mean depth: 11,216×). Among the buccal swab samples, Sample 010 showed the highest coverage (average: 1,876,768 reads, mean depth: 17,370×), whereas Sample 004 showed the lowest coverage (average: 784,444 reads, mean depth: 7794×).
Although the coverage observed for the hair shaft samples was less than that observed for the blood and buccal swab samples, their coverage per sample was sufficient according to the Precision ID mtDNA Whole Genome Panel User Manual [21].
Whole mtGenome haplotype and haplogroup determination
Whole mtGenome variants were obtained from hair shaft samples as well as blood and buccal swab samples using the respective MPS platforms without any missing areas. In particular, although the hair shaft samples had been stored at room temperature for > 1 year, the variants across their whole mtGenome were successfully analyzed. The sequence variations and haplogroup of each sample were confirmed using the EMPOP database v4 [24]. We identified 20 unique mtDNA haplotypes and haplogroups. Further, the mtDNA haplogroups of all samples had origins belonging to East Asians: Y1, G3a2+152, B4h1, N9a1, D4g2b1, G2a5+152, B4d3a1, D4b2b1, A5a1a, D4h1c1, B5b3a, C4a1b, M10a1b, D4a1a1, B4b1a1, N9a7, B4a1b1, A15c, Z3, and D5a2a1b [25]. The representative whole mtGenome variants analyzed from the blood sample of each donor are presented in Supplementary Table S2.
Observed PHP of whole mtGenomes
With a 5% PHP detection threshold, we identified a total of 56 PHP variants among 15 donors across the 4 sets of tissue: 23 PHP variants in hair shaft set #1, 17 in hair shaft set #2, 8 in blood samples, and 8 in buccal swab samples (Table 1). The PHP variants were distributed more in the coding regions (n = 32) than in CRs (n = 24). Regarding the number of PHP variants per donor, up to 5 variants were observed in a hair shaft set #1 sample, 3 in a hair shaft set #2 sample, 2 in a blood sample, and 2 in a buccal swab sample. The proportion of PHP variants in the 4 sets of tissue varied: 5.4–45.6% in hair shaft set #1, 5.8–45.2% in hair shaft set #2, 6.4–28.3% in blood samples, and 5.8–49.0% in buccal swab samples.
We also observed 4 cases in which the major nucleotide of PHP was inverted according to the tissue type. For example, in Sample 001, T was dominant in hair shaft set #2 and blood samples at nucleotide position (np) 152; however, C was dominant in hair shaft set #1 and buccal swab samples at the same position. In Sample 004, G was the most prominent nucleotide in all tissue types at np 709, except in hair shaft set #1. At np 15034 in Sample 015, A was the major nucleotide in hair shaft set #2, blood, and buccal samples, whereas G was the major nucleotide in hair shaft set #1. Finally, at np 16103 in Sample 017, A was dominant in hair shaft sets #1 and #2 samples, whereas G was dominant in blood and buccal swab samples. Based on the tissue type, the major nucleotide of PHP was inverted in three cases of hair shaft sets—at np 152 in Sample 001, np 709 in Sample 004, and np 15034 in Sample 015. Remarkably, PHP state differed even between reference samples. As previously described, at np 152 in Sample 001, T was dominant in the blood sample, whereas C was dominant in the buccal swab sample. In addition, one type of reference sample showed PHP, whereas the other exhibited homoplasmy in two cases—at np 16150 in Sample 007 and np 8517 in Sample 009 (Table 1).
Sanger sequencing results of inverted major nucleotide
Sanger sequencing was performed at the target region of the 4 samples that contained inverted major nucleotides as per the MPS data (i.e., np 152 in Sample 001, np 709 in Sample 004, np 15034 in Sample 015, and np 16103 in Sample 017). For all 4 samples, the major nucleotide depending on the tissue types as per the MPS data correlated with the Sanger sequencing results. Moreover, the proportion of PHP variants in the MPS data was correlated with the peak height ratio observed on the capillary electrophoresis (CE) (Fig. 1, Supplementary Fig S1a–c). For example, A was the major nucleotide in all tissue types at np 15034, except in hair shaft set #1, according to the MPS data (Fig. 1). This is consistent with the findings of Sanger sequencing electropherogram. Moreover, regarding the MPS coverage ratio of G, it increased in the order of buccal swab, blood, hair shaft set #2, and hair shaft set #1, consistent with the results of the contributing sequence variants obtained on Sanger sequencing electropherogram. Notably, although PHP in hair shaft set #1 could be identified using MPS at a low level (5.9%), it was difficult to observe them with Sanger sequencing electropherogram, and it was probably omitted due to the limited Sanger sequencing resolution.
The Sanger sequencing results for Sample 015, which showed an inverted major nucleotide according to tissue type in the MPS data. In all tissue types, the major nucleotide was identical between the Sanger sequencing and MPS data. Furthermore, the proportion of PHP relatively corresponded between them
Discussion
In forensic casework, because most mtDNA analyses demand the application of a PCR strategy for degraded samples such as hair shafts, a method that generates short amplicons has been adopted [10, 11]. Parson et al. analyzed the whole mtGenome extracted from hair shafts by an MPS method designed to yield short amplicons (range: 300–500 bp) using 62 primer sets [26]. Recently, several studies have shown that the Precision ID mtDNA Whole Genome Panel, which produces much shorter amplicons (average: 163 bp), could be useful for analyzing whole mtGenome variants extracted from degraded samples, including hair shaft samples [27]. In this study, although the hair shafts used had been stored at room temperature for > 1 year, MPS libraries were successfully generated with 22 PCR cycles of 3 μl of DNA extracted from hair shaft samples using the 2-in-1 method. Each of 8 libraries were loaded on the Ion 520 Chip and resulted in sufficient coverage (average: 578,937 reads/chip). Furthermore, analysis using the Converge 2.1 software supported user-friendly data processing for MPS, including automatic haplotype designation following the forensic nomenclature ISFG guidelines [22].
On the other hand, most blood and buccal swab samples are often collected directly from a suspect; thus, they have a low possibility of undergoing DNA degradation. In such cases, it is relatively easy to amplify the whole mtGenome with only 2 fragments. Systematic studies have revealed that the use of LR amplification and library preparation with the Nextera XT Kit is suitable for high-quality samples and enables a rapid workflow to produce high-throughput results [8, 9, 12]. While > 35 thermal cycles were applied to amplify the whole mtGenome in previous reports [8, 9, 12, 16], we could obtain sufficient PCR yield with 1 ng of genomic DNA using only 26 thermal cycles.
Among the 56 PHP variants observed in 15 donors across the 4 sets of tissue, mtDNA PHP was present more abundantly in hair shaft samples (n = 40) than in blood and buccal swab samples (n = 16), consistent with the results of previous reports [14, 28]. The narrow bottlenecks during hair follicle development and high-energy requirements for keratinizing hair shafts could be conducive to the presence of higher amount of PHP in hair shaft samples than in blood and buccal swabs samples [29]. Most major nucleotides of mtDNA PHP were consistent with those observed in other tissues. However, the inverted major nucleotide, also known as heteroplasmic variant drift, was observed in only 4 samples (Samples 001, 004, 015, and 017) (Table 1). There are some possible explanations for the presence of inverted major nucleotides within these tissue types. In the case of inter-hairs, different populations of heteroplasmic variants are likely to be present on an individual’s scalp because mtDNA molecules in the supplied cells are limited or cloned due to bottlenecks [29, 30]. Furthermore, variant drift in hairs could occur more easily because each hair strand originates from clusters or follicles with independent growth cycles. In the case of inter-tissue types, each tissue originates from a different germ layer during the early stage of histogenesis [28], which possibly caused the variant drift observed among tissues. Moreover, there are some hot spots that are already known to have high mutation rates, and one such hot spot is np 152 [31], which could explain the result obtained for Sample 001.
Meanwhile, when comparing mtGenome variants based on the major nucleotide of PHP, the haplotypes of 80% (16/20) donors matched across the whole mtGenome in the 4 sets of tissue. The inverted major nucleotides were confirmed using Sanger sequencing. In all 4 cases, the major nucleotide according to tissue type was identical with to those obtained on MPS, and the visual peak height ratio between the major and minor nucleotides on electropherograms corresponded to their read depth ratio obtained in MPS data. This finding implies that inverted major nucleotides across all tissue types were not a result of a difference in the MPS platform but was a unique characteristic that existed inherently in individuals. It also has been suggested that MPS of the whole mtGenome would produce consistent and robust results although each forensic sample are sequenced and analyzed on different MPS platforms.
From the perspective of interpretation, the inverted major nucleotide observed in MPS data does not indicate that the questioned and known samples were excluded as deriving from the same individual or maternal lineage. For example, in Fig. 1, let us suppose that Sample 015 from hair shaft set #1 was found as evidence at a crime scene. The major nucleotide at np 15034 was different from those obtained for other known samples (blood and buccal swabs) and another hair shaft sample (hair shaft set #2); the major nucleotide was G in hair shaft set #1 and A in the other sets of tissue. However, A was dominant at np 15034 in hair shaft set #1 that showed low-level heteroplasmy (5.9%), which did not provide adequate evidence for excluding hair shaft set #1 and other samples obtained from the same donor. However, if the results were analyzed by CE based on Sanger sequencing, the interpretation could be confused. Regarding hair shaft set #1, np 15034 in Sample 015 would have only the homoplasmic variant G if the normal CE threshold (approximately 20%) is applied. It would even seem that there were no minor nucleotides at np 15034 in blood and buccal swab samples. In this context, the CE method could result in imprecise interpretation because of the low resolution and/or current chemistry of dye terminator sequencing [22]. On the other hand, MPS increased the detection resolution of PHP to 5% [15] or even finer [32], and it could provide more accurate PHP information. Therefore, the use of higher resolution MPS data for mtDNA PHP can aid forensic practitioners to conduct more informed investigation, particularly in the interpretation of mtDNA variants extracted from various tissue types. In this study, a limited amount of mtGenome data demonstrated the potential utility of MPS based-mtGenome PHP analysis. However, if a larger amount of data for mtGenome PHP were accumulated across various tissue types with the MPS method, it would increase the value of mtDNA in forensic practice.
Conclusion
In this study, we analyzed the whole mtGenome variants of 4 sets of tissue (duplicated hair shaft, blood, and buccal swab samples) obtained from 20 donors using suitable MPS platforms by considering the characteristics of forensic samples. Although hair shafts had been stored at room temperature for > 1 year, their whole mtGenome variants were successfully obtained using the Precision ID mtDNA Whole Genome Panel, which was validated for degraded DNA analysis with the Ion S5 system. The whole mtGenome variants of blood and buccal swabs with 2 LR-PCR products were analyzed using the Nextera XT DNA Library Prep Kit on the MiSeq system. Although 56 PHP variants were identified across the 4 sets of tissue, the whole mtGenome haplotypes were not excluded according to each tissue type. In 4 samples, the major nucleotide of PHP was inverted at one nucleotide position in the hair shaft and reference samples. However, the MPS data, which provide a high resolution for PHP, did not result in exclusion of that both haplotypes were derived from the same donor. Accumulation of mtGenome PHP data will facilitate the application of MPS in the comparative analysis of mtGenome variants containing PHP to forensic casework.
References
Robin ED, Wong R (1988) Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol 136:507–513
Budowle B, Allard MW, Wilson MR, Chakraborty R (2003) Forensics and mitochondrial DNA: applications, debates, and foundations. Annu Rev Genomics Hum Genet 4:119–141
Allen M, Engstrom AS, Meyers S et al (1998) Mitochondrial DNA sequencing of shed hairs and saliva on robbery caps: sensitivity and matching probabilities. J Forensic Sci 43:453–464
Melton T, Dimick G, Higgins B, Lindstrom L, Nelson K (2005) Forensic mitochondrial DNA analysis of 691 casework hairs. J Forensic Sci 50:73–80
Irwin JA, Saunier JL, Strouss KM et al (2007) Development and expansion of high-quality control region databases to improve forensic mtDNA evidence interpretation. Forensic Sci Int Genet 1:154–157
Chaitanya L, Van Oven M, Brauer S et al (2016) High-quality mtDNA control region sequences from 680 individuals sampled across the Netherlands to establish a national forensic mtDNA reference database. Forensic Sci Int Genet 21:158–167
Just RS, Scheible MK, Fast SA, Sturk-Andreaggi K, Röck AW, Bush JM, Higginbotham JL, Peck MA, Ring JD, Huber GE, Xavier C, Strobl C, Lyons EA, Diegoli TM, Bodner M, Fendt L, Kralj P, Nagl S, Niederwieser D, Zimmermann B, Parson W, Irwin JA (2015) Full mtGenome reference data: Development and characterization of 588 forensic-quality haplotypes representing three U.S. populations. Forensic Sci Int Genet 14:141–155
McElhoe JA, Holland MM, Makova KD, Su MS, Paul IM, Baker CH, Faith SA, Young B (2014) Development and assessment of an optimized next-generation DNA sequencing approach for the mtgenome using the Illumina MiSeq. Forensic Sci Int Genet 13:20–29
King JL, LaRue BL, Novroski NM, Stoljarova M, Seo SB, Zeng X, Warshauer DH, Davis CP, Parson W, Sajantila A, Budowle B (2014) High-quality and high-throughput massively parallel sequencing of the human mitochondrial genome using the Illumina MiSeq. Forensic Sci Int Genet 12:128–135
Gabriel MN, Huffine EF, Ryan JH, Holland MM, Parsons TJ (2001) Improved MtDNA sequence analysis of forensic remains using a “mini-primer set” amplification strategy. J Forensic Sci 46:247–253
Eichmann C, Parson W (2008) “Mitominis”: multiplex PCR analysis of reduced size amplicons for compound sequence analysis of the entire mtDNA control region in highly degraded samples. Int J Legal Med 122:385–388
Peck MA, Brandhagen MD, Marshall C, Diegoli TM, Irwin JA, Sturk-Andreaggi K (2016) Concordance and reproducibility of a next generation mtGenome sequencing method for high-quality samples using the Illumina MiSeq. Forensic Sci Int Genet 24:103–111
Calloway CD, Reynolds RL, Herrin GL, Anderson WW (2000) The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am J Hum Genet 66:1384–1397
Naue J, Hörer S, Sänger T, Strobl C, Hatzer-Grubwieser P, Parson W, Lutz-Bonengel S (2015) Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion 20:82–94
Just RS, Irwin JA, Parson W (2015) Mitochondrial DNA heteroplasmy in the emerging field of massively parallel sequencing. Forensic Sci Int Genet 18:131–139
Fendt L, Zimmermann B, Daniaux M, Parson W (2009) Sequencing strategy for the whole mitochondrial genome resulting in high quality sequences. BMC Genomics 10:139
Andrews RM, Kubacka I, Chinnery PF et al (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147
Li H, Durbin R (2009) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12
Van der Auwera GA, Carneiro MO, Hartl C et al (2014) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 11:1–42
Thermo Fisher Scientific (2016) Precision ID mtDNA panels with the HID Ion S5TM/HID Ion GeneStudioTM S5 System
Parson W, Gusmão L, Hares DR, Irwin JA, Mayr WR, Morling N, Pokorak E, Prinz M, Salas A, Schneider PM, Parsons TJ, DNA Commission of the International Society for Forensic Genetics (2014) DNA Commission of the International Society for Forensic Genetics: revised and extended guidelines for mitochondrial DNA typing. Forensic Sci Int Genet 13:134–142
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2012) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192
Parson W, Dür A (2007) EMPOP-a forensic mtDNA database. Forensic Sci Int Genet 1:88–92
Lee HY, Yoo JE, Park MJ, Chung U, Kim CY, Shin KJ (2006) East Asian mtDNA haplogroup determination in Koreans: haplogroup-level control region SNP analysis and subhaplogroup-level control region sequence analysis. Electrophoresis 27:4408–4418
Parson W, Huber G, Moreno L, Madel MB, Brandhagen MD, Nagl S, Xavier C, Eduardoff M, Callaghan TC, Irwin JA (2015) Massively parallel sequencing of complete mitochondrial genomes from hair shaft samples. Forensic Sci Int Genet 15:8–15
Strobl C, Eduardoff M, Bus MM, Allen M, Parson W (2018) Evaluation of the precision ID whole MtDNA genome panel for forensic analyses. Forensic Sci Int Genet 35:21–25
Gallimore JM, McElhoe JA, Holland MM (2018) Assessing heteroplasmic variant drift in the mtDNA control region of human hairs using an MPS approach. Forensic Sci Int Genet 32:7–17
Linch CA, Whiting DA, Holland MM (2001) Human hair histogenesis for the mitochondrial DNA forensic scientist. J Forensic Sci 46:844–853
Desmyter S, Bodner M, Huber G, Dognaux S, Berger C, Noël F, Parson W (2016) Hairy matters: MtDNA quantity and sequence variation along and among human head hairs. Forensic Sci Int Genet 25:1–9
Irwin JA, Saunier JL, Niederstätter H, Strouss KM, Sturk KA, Diegoli TM, Brandstätter A, Parson W, Parsons TJ (2009) Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples. J Mol Evol 68:516–527
Li M, Schröder R, Ni S et al (2015) Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc Natl Acad Sci 112:2491–2496
Acknowledgments
The authors would like to thank P.H. Kim and C.H. Lee from Thermo Fisher Scientific Korea for their generous support for this research.
Funding
This study was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2014M3A9E1069989).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board of Severance Hospital, Yonsei University, Seoul, Korea (4-2015-1068) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, B.M., Hong, S.R., Chun, H. et al. Comparison of whole mitochondrial genome variants between hair shafts and reference samples using massively parallel sequencing. Int J Legal Med 134, 853–861 (2020). https://doi.org/10.1007/s00414-019-02205-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-019-02205-y