Abstract
Y-chromosomal short tandem repeats (Y-STRs) have proven to be very useful in investigating sexual assault cases and in paternity lineage differentiation. However, currently available commercial Y-STR multiplex amplification systems bear the limitations in the identification of related males from the same paternal lineage due to there being an insufficient number of loci in any single amplification kit. The aim of this study was to establish and validate a novel 6-dye, 36-plex Y-STR multiplex amplification system that incorporated all of the loci present in the Yfiler™ Plus kit (DYS19, DYS385a/b, DYF387S1, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS449, DYS456, DYS458, DYS460, DYS481, DYS518, DYS533, DYS570, DYS576, DYS627, DYS635, Y_GATA_H4) as well as a further nine highly polymorphic Y-STR loci (DYS388, DYS444, DYS447, DYS522, DYS527a/b, DYS549, DYS596, DYS643). The novel system was optimized and validated by a series of studies that tested the effect of different PCR-based conditions as well as the species specificity, sensitivity, stability, stutter precision, suitability for use on DNA mixtures, reproducibility, and parallel testing of the system, as well as its performance on casework samples and population analysis, according to the SWGDAM developmental validation guidelines. A total of 246 haplotypes were found for the 36 Y-STRs among 247 Guangdong Han unrelated males. Collectively, the results demonstrate that the developed 36-plex Y-STR system is sensitive, robust, reliable, and highly informative for use in forensic genetics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Y-chromosome short tandem repeats (Y-STRs) are inherited with the characteristics of the non-coding region no switching and non-recombination in the genetic inheritance from father to son [1, 2]. Because of this, they have unique application value in forensic science, particularly in paternal lineage differentiation, male identification in sexual assault cases, male component detection in male-male or male-female DNA mixtures, and in DNA genealogy construction [3,4,5]. In the past few years, several commercial Y-STR testing kits have been developed, for example PowerPlex® Y23 system, AGCU Y24 kit, and Goldeneye 20Y kit [6,7,8]. However, these kits are limited in their ability to differentiate male lineages in isolated and inbred populations with insufficient discrimination power [9, 10]. The Yfiler® Plus kit was recently developed in part to resolve this problem [11, 12]. It has a higher discrimination ability which is to a large part due to being based on the identification of seven rapidly mutating Y-STR loci (DYS387S1, DYS449, DYS518, DYS570, DYS576, DYS627) [13, 14].
This study aimed to develop a novel, feasible, amplification system with an increased number of Y-STR loci, high levels of haplotype diversity, and discriminatory capacity. The minimal haplotype (MHT) with nine Y-STRs [15] and the haplotype of the Scientific Working Group for DNA Analysis Methods (SWGDAM) with 11 Y-STRs [16] are still included in the commonly used Y-STR kits in current forensic applications. Thus, all the loci of the Yfiler® Plus kit were selected for inclusion in the novel amplification system. In addition, nine Y-STR loci that exhibited a high degree of polymorphism were also selected for inclusion (DYS388, DYS444, DYS447, DYS522, DYS527a/b, DYS549, DYS596, DYS643). With the exception of DYS596, these additional Y-STR loci were contained in either the AGCU Y24 kit or the PowerPlex® Y23 kit [17, 18]. Thus, we designed the new amplification system based on 36 Y-STR loci in a 6-dye configuration and named it the “36-plex Y-STR multiplex amplification system”, or “36-plex Y-STR system” for short. To evaluate the efficiency of the system, we tested a variety of PCR conditions, including the amount of added Taq polymerase and primers, PCR reaction volume, and annealing temperature, and performed a series of validation studies. This developmental validation study was based on the Scientific Working Group on DNA Analysis Methods (SWGDAM) developmental validation guidelines [16, 19].
Materials and methods
DNA samples
Blood card samples from 247 unrelated males were obtained from the Guangzhou Forensic Science Institute, Guangzhou, China. Nonhuman samples from cattle, sheep, pigs, cats, dogs, chickens, rats, fish, and Escherichia coli were obtained from AGCU ScienTech Incorporation, Wuxi, China and were selected for species specificity studies. The blood stain samples were punched to 1.2 mm in diameter using a BSD 600-DUET stiletto instrument (BSD, Australia) and placed into 96-well plates for direct PCR amplification. The biological material that could not be amplified directly was extracted using the Chelex-100 method for PCR amplification [20]. The project was approved by the Ethics Committee of Southern Medical University, China before the study, and signed informed consent was obtained from all participants prior to sample collection.
Primer design
The 36 selected Y-STR loci were divided into five groups based upon the needs of multiplex amplification. Information about the STR loci is presented in Table 1. Primers were developed for each locus based on unified parameters using Oligo 6.0 software (Premier Biosoft International, Palo Alto CA, USA). Preliminarily amplification specificity of the primers was evaluated using the BLAST function of GeneBank on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The resulting primer for each locus was optimized and redesigned to meet the needs of multiplex amplification.
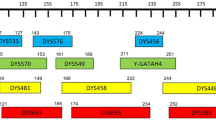
Primers were labeled by fluorescent dyes: primers of DYS392, DYS389I/II, DYS447, DYS438, DYS527 a/b, DYS522, and DYS596 were labeled with 6-FAM fluorescent dye (blue); primers of DYS391, DYS456, DYS19, DYS388, DYS448, DYS385a/b, and DYS549 were labeled with HEX fluorescent dye (green); primers of DYS437, DYS481, DYS533, DYS390, DYS627, DYS458, and DYS460 were labeled with TAMRA fluorescent dye (yellow); primers of DYS393, Y_GATA_H4, DYS439, DYS635, DYS444, and DYS643 were labeled with ROX fluorescent dye (red); and primers of DYS576, DYS570, DYF387S1, DYS449, and DYS518 were labeled with VIG fluorescent dye (purple). Fragments included in the internal lane standard were detected in the orange channel and labeled with SIZ-500. A schematic diagram of the fluorescence detection method is shown in Fig. 1. All primers were synthesized by Shanghai Sangon Biological Engineering Technology & Services Company, Shanghai, China.
PCR amplification
Unless otherwise mentioned, PCR amplification was performed on a GeneAmp® PCR System 9700 thermal cycler (Life Technologies, CA, USA). Each amplification reaction contained 10.0 μL PCR buffer mixture containing 125 mM Tris-HCl buffer, 125 mM KCl, 7.5 mM dNTPs, and 5.0 mM MgCl2, as well as 5.0 μL primer set (concentration 0.045–0.360 μM, 1 μL heat activated Taq polymerase (5 U/μl), 0.3–1 ng of template DNA, or a 1.2 mm punch of the blood card sample and ddH2O, to obtain a final reaction volume of 25 μL.
Standard thermal cycling conditions included an initial denaturation step at 95 °C for 2 min, followed by subsequent cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 1 min, elongation at 72 °C for 1 min repeated 30 times, and followed by a final extension step at 60 °C for 30 min. The DNA sample was amplified under standard conditions, and both positive and negative controls were included in all experiments.
Sample electrophoresis and data analysis
Electrophoresis was performed by Applied Biosystems 3500xl Genetic Analyzer using 36-cm capillary arrays with POP-4® Polymer (Life Technologies, CA, USA). Spectral calibration was performed using the J6 Dyeset with the 6 Dye Matrix Standards. Capillary Electrophoresis (CE) was performed by adding 1.0 μL of the PCR product, or 36 Y-STR allelic ladders, to 10.0 μL of formamide/SIZ-500 mixture (comprising 9.5 μL of deionized Hi-Di™ Formamide and 0.5 μL of Marker SIZ-500 size standard). The samples were denatured for 3 min at 95°C then chilled on ice prior to immediately performing electrophoresis. The PCR products for CE were analyzed on an Applied Biosystems 3500xl Genetic Analyzer with a 3 kV, 10 s injection, and electrophoresis at 15 kV for 1500 s at 60 °C. Fragment sizes and genotyping were determined by GeneMapper ID-X software (Life Technologies, USA) using a peak amplitude of 150 RFU as the peak detection threshold for allele calls.
Species specificity
Cattle, sheep, pigs, cats, dogs, chickens, rats, fish, and E. coli samples were used for the species specificity studies. For each species, a 10 ng of purified DNA was amplified by the 36-plex Y-STR system following the standard PCR protocol.
Sensitivity
Sensitivity studies were performed with control DNA 9948 (Ori-Gene company, USA), and total DNA inputs were prepared in a serial dilution to 1 μL with the following template amounts: 1.0, 0.5, 0.25, 0.125, 0.0625, and 0.03125 ng.
DNA mixtures
A total of 1 ng of human genomic DNA was prepared in a reaction volume of 25 μL for the mixture samples: Male-male mixtures were prepared using 9948 and 007 human genomic DNA, with mixture ratios of 1:1, 1:2, 1:4, 1:8, and 1:15, respectively. Each mixture was tested in triplicate to reduce the accidental error and ensure the accuracy of the results analysis.
Inhibitors study
To validate the anti-interference capability of the 36-plex Y-STR system, six common forensic inhibitors including hematin, indigo, humic acid, calcium ion, EDTA, and hemoglobin were tested. The quantity of control DNA 9948 was held constant at 0.5 ng with the inhibitors at the following concentrations: 25, 50, 75, 100, or 125 μmol/L of hematin; 8, 10, 12, 14, or 16 mmol/L of indigo; 30, 40, 50, 60, or 70 μg/μl of humic acid; 0.6, 0.8, 1.0, 1.2, 1.4, or 1.6 mmol/L of calcium ion; 0.6, 0.8, 1.0, 1.2, or 1.4 μmol/L of EDTA; and 20, 30, 40, 50, or 60 μmol/L of hemoglobin.
Reproducibility
Fifty randomly selected samples from the population study (n = 247) were analyzed by three separate laboratories (Guangzhou Forensic Science Institute and two of its affiliated institutes) to validate the genotyping concordance. The allelic ladder of the 36-plex Y-STR system was detected by both an Applied Biosystems 3130xl Genetic Analyzer (upgraded to the 6-dye module) and a 3500xl Genetic Analyzer, to facilitate comparison of the genotyping results.
Parallel testing
Parallel testing was conducted using two blood FTA® Cards, two buccal swabs, two muscle samples, three nail samples, three salivary stains, and three hair root samples which had been obtained during daily practice. All the above samples were amplified separately using the 36-plex Y-STR system, Yfiler® Plus kit, and PowerPlex® Y23 system, and the genotype results were compared.
Casework samples
Thirty-five real casework samples were used to test the efficiency of the 36-plex Y-STR system. These samples included blood stain samples on different substrates, mixed seminal stains, cigarette butt, and epithelial fractions (swab of bottleneck, chopsticks, padlock, et al.).
Size precision and stutter calculation study
All samples from the population study (n = 247) were used to evaluate the sizing precision of the developed 36-plex Y-STR system. Sizing precision was assessed by running three full injection of allelic ladder. Allelic size was calculated for each allele with GeneMapper ID-X software and then compared with the allele size of corresponding ladder. All samples were analyzed in order to estimate the stutter ratios of the loci contained in the system. The stutter percentage was determined as the stutter peak height divided by the true peak height.
Population study and statistical analysis
A total of 247 unrelated Han males in Guangdong were detected using the developed 36-plex Y-STR system. Allele frequencies were determined using the direct counting method, where the frequency of i-th alleles in locus pi = the frequency of i-th allele /n, where n is the population sample number. The global discrimination capacity (DC) was ascertained by dividing the number of different haplotypes by the number of samples in the studied population [21]. The match probability (MP) was calculated as the sum of the squared haplotype frequencies. Genetic diversities (GD) and haplotype diversities (HD) were calculated according to Nei [22] as HD or GD = n(1 − ∑Pi2)/(n − 1), where n is the total number of samples and Pi is the relative frequency of the i-th allele or haplotype. Population Rst genetic distances and multidimensional scaling (MDS) analyses were obtained using the analysis of molecular variance (AMOVA) tool to assess the genetic structure and diversity among the following populations from the YHRD database: Henan Han [7, 23, 24], Shandong Han [25], Shanghai Han [26,27,28], Shenzhen Han [29], Liaoning Hui [30], Liaoning Mongolian [31], Fuzhou She [32], Qinghai Tibetan [33, 34], Xinjiang Uighur [35,36,37], Rennes French [38], Greek [39], Shizuoka Japanese [40], Daejeon Korean [41], Tripoli Libyan [42], and Singapore Malay [43]. To describe the relationships between populations, a neighbor-joining phylogenetic tree was constructed based on the Rst value using MEGA 6.0 software [44].
Quality control measures
All methods were carried out in accordance with the recommendations of International Society for Forensic Genetics (ISFG), as described by Parson on the analysis of genetic population data [45].
Results and discussion
PCR reaction: cycle number
The 36-plex Y-STR system was tested over a range of amplified cycle numbers. One nanogram of 9948 male DNA was amplified over 28, 29, 30, 31, and 32 cycles on an Applied Biosystems GeneAmp 9700. As expected, full profiles were observed, and an increase in cycle numbers generated higher peak heights in the 25-μL reaction system. The results indicated that a PCR cycle number of 30 was optimal for the developed 36-plex Y-STR system. This cycle number maximized assay sensitivity and minimized the occurrence of off-scale peaks, as demonstrated in Supplemental Fig. S1.
PCR reaction: annealing steps
Operating temperatures can vary slightly between different PCR thermocycler instruments and across different laboratories. The optimal annealing temperature of the 36-plex Y-STR system was determined by conducting the annealing step at 56, 58, 60, 62, 64, 66, and 68 °C, and the effect on the stability and accuracy of the genotyping results was studied. As seen in Fig. 2, accurate genotype profiles were observed at an annealing temperature between 56 and 62 °C. At 64°C, the peak heights of some loci were less than 1000RFU, and when the annealing temperature was increased to 66 or 68 °C, few loci were detected. Nonspecific peaks were not observed at an annealing temperature of 62 °C, which was therefore concluded to be the optimal, recommended, annealing temperature.
PCR reaction: master mix, primer mix and Taq DNA polymerase
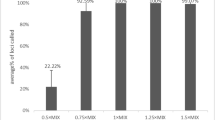
The concentration of the master mix, the primers mix, and of Taq polymerase used in PCR analysis are closely correlated to the quality and accuracy of the genotyping results. One nanogram of 9948 male DNA was amplified in different concentrations of reaction volume (0.5×, 0.75×, 1.0× (standard), 1.25×, 1.5×) in a 25-μL reaction system, and the genotyping results were compared. Results from the testing of the PCR master mix concentration revealed that over 50% of the loci were detected at a reaction mix concentration of 0.5×, and more than 20% were detected at a concentration of 1.5×. Full profiles were obtained from the other concentrations of master mix. Few loci were observed to have allelic drop-out at a primer concentration of 0.5×. All loci were detected from a primer concentration of 1.0× and showed balanced peak height. Variable concentrations of Taq DNA polymerase had little effect on the genotyping results. (Fig. S2, S3, S4).
Species specificity
Non-human genomic DNA samples from common animal species (dogs, pigs, cattle, sheep, cats, chickens, rats, fishes, and E. coli) were amplified using the 36-plex Y-STR system. The results did not reveal the presence of any allele peaks within the genotyping range (Fig. S5). On this basis, it was concluded that the developed STR system was robust and unlikely to be affected by the presence of genetic material from these animal species.
Sensitivity studies
The sensitivity of the 36-plex Y-STR system was tested with a range of DNA inputs from 0.03125 to 1 ng. When the DNA template concentration was between 1 and 0.125 ng, the peak height ratios of the majority of loci exhibiting the same fluorescent color were above 60%. When the amount of template DNA was reduced to 0.0625 ng, the peak height ratios of loci exhibiting the same fluorescent color were less than 60%, but no allelic dropout was observed. When the amount of template DNA was 0.03125 ng, allelic dropout occurred randomly at some loci, especially in those with longer DNA fragments such as DYS627, DYS460, DYS570, DYS449, and DYS518 (Fig. 3). On this basis, it was concluded that the 36-plex Y-STR system could satisfy the Chinese criteria for human fluorescent STR multiplex PCR reagents and provide reliable profiles at a threshold of 150RFU with DNA inputs higher or equal to 0.0625 ng. The sensitivity under these conditions were determined to be 0.0625 ng/25 μL.
Heat maps summarizing the sensitivity results of six serial dilutions (from 1 to 0.03125 ng) of control DNA 9948 (Stochastic threshold was 100RFU). Green represents peak height ratio > 60% within the same color. Yellow represents peak height ratio < 60% within the same color. Red represents allele drop-out
DNA mixtures
Forensic casework samples often comprise a mixture of DNA (male-male or male-female). To distinguish the major and minor male contributors, it is necessary to detect the performance of the 36-plex Y-STR system when applied to DNA mixtures. Male-male mixtures of 9948 DNA and 007 DNA in known ratios of 1:1, 1:2, 1:4, 1:8, and 1:15 were amplified to produce a total amount of 1-ng mixed DNA. Results showed that the peak heights of minor alleles decreased as the mixture ratio decreased. Full profiles were obtained for the minor contributor at non-overlapping and non-stutter positions with mixture DNA ratios ranging from 1:1 to 1:4 (Fig. 4). When the mixture ratio increased to 1:8, a general loss of alleles was observed. These findings showed that the developed 36-plex Y-STR system could satisfy the Chinese criteria for human fluorescent STR multiplex PCR reagents, which requires all alleles to be correctly detected for 1:4 DNA mixtures.
Male:male mixture analysis. Male:male mixture samples (total of 1 ng input DNA) of DNA 9948 and 007 in ratios of 1:1, 1:2, 1:4, 1:8, and 1:15 were amplified using the 36-plex Y-STR system for 30 cycles. A minor contributor allele (alleles located in non-overlapping and non-stutter positions) is presented with the arrow
Inhibitors study
Crime scene DNA samples often contain inhibitors [46], which may interfere with the PCR amplification and can sometimes cause complete amplification failure. One nanogram of control DNA 9948 containing the most common inhibitors (hematin, indigo, humic acid, calcium ion, EDTA, and hemoglobin) was tested to evaluate the robustness of the 36-plex Y-STR system. As seen from the results presented in Table 2, complete DNA profiles were observed with concentrations of up to 50 μmol/L of hematin, 80 μmol/L of hemoglobin, 25 ng/μL of humic acid, 12 mmol/L of indigo, 0.9 mmol/L of calcium ion, and 0.9 mmol/L of EDTA. When the concentrations exceeded these levels, allelic drop-outs were observed in the loci with long fragment size alleles. The exception to this was calcium ion in which allelic drop-out was observed in the loci with short fragment size alleles (such as DYS19, DYS388, DYS447, and DYS533) when the concentration was increased to 1.2 or 1.5 mmol/L.
Reproducibility
A reproducibility study was performed to validate the reliability and accuracy of the developed 36-plex Y-STR system with correctly identifying DNA samples when used by different laboratories. Fifty samples were tested using the 36-plex Y-STR system in three participating laboratories. The results demonstrated that the genotypes of all samples were consistent with their known profiles (data not shown). To demonstrate the suitability of the system for use on different capillary electrophoresis platforms, both the Applied Biosystems 3130xl (upgraded to 6-dye module) and the 3500xl Genetic Analyzer were used to run the allelic ladder of the 36-plex Y-STR system. The results showed genotype consistency of the same locus across the two CE methods. For example, the electropherograms of the DYS448 allelic ladder were concordant between the two tested CE platforms (Fig. S6).
Parallel testing
To evaluate the compatibility of the different commercial kits available for Y-STR testing, parallel testing was performed using the 36-plex Y-STR system, Yfiler™ plus kit and PowerPlex® Y23 system to amplify control DNA 9948 and 15 samples. The results showed that the genotypes of control 9948 DNA and of the samples were detected consistently by the three systems, as shown in the Supplemental Table S2.
Casework samples
Thirty-five real forensic casework samples were processed using the 36-plex Y-STR system, and the results showed that full DNA profiles were successfully obtained from all samples with the exception of some epithelial cell fractions. Most of the 36 STR loci were successfully detected on swabs from toothbrushes and padlocks, while fewer loci were successfully detected on swab of wallet, and none of the loci were detected from swabs of brick (Table S3).
Stutter calculation and size precision study
Stutter peaks are common artifacts that may be caused by slippage of the Taq Polymerase during the elongation step [47, 48]. The stutter ratio of the 36-plex Y-STR system was evaluated with 247 DNA samples in population studies. The minimum and maximum stutter, the stutter mean and associated standard deviation (SD), and the recommended filter thresholds are shown in Table 3. The mean stutter ratios of all loci were lower than 15%, except those of DYS481 (17.68%) and DYS518 (16.24%).
Sizing precision is critical for accurate genotyping. Size precision of the 36-plex Y-STR system was evaluated by running three full injection of allelic ladder using a 3500xl Genetic Analyzer. Size variability was determined by calculating the standard deviation for each allele [49]. As expected, the fragment size increased with increasing standard deviation of the allele. Very little variation at each locus was seen in the size of the 36-plex Y-STR allelic ladder mix, and most allele deviations were nearly 0.06 base. The maximum SD was close to 0.1 base at locus DYS549 (Fig. 5). These results demonstrate that the 36-plex Y-STR system can ensure proper allele detection that is consistent and within the bin window.
Population studies
The DNA of 247 unrelated Han males was genotyped by the 36-plex Y-STR system. The data was submitted to the Y-chromosome STR haplotype reference database (YHRD) under accession number YA004330. Relevant forensic parameters were investigated, among which, the gene diversity (GD) value is used to assess the polymorphism of Y-STR loci [50]. As seen in Fig. 6, the highest GD value among the 36 Y-STR loci was 0.9620 at locus DYS385, and the lowest was 0.3992 at locus DYS438. Compared with the Yfiler™ Plus kit, an additional 9 Y-STR loci were included in the 36-plex Y-STR system, and most of them were highly polymorphic, except locus DYS388. Haplotype diversity (HD) and discrimination capacity were calculated to evaluate system efficiency [51]. As presented in Supplemental Table S3, a total of 246 haplotypes were obtained from 247 individuals, from which the obtained HD value was 0.9998641 and the DC value was 0.995951. These results demonstrate that the 36-plex Y-STR system is highly genetically informative.
The five existing Y-STR marker sets (MHT, SWGDAM, PowerPlex® Y23, Yfiler® Plus, and AGCU Y24) were compared with the 36-plex Y-STR system in terms of their haplotype-based forensic parameters (haplotype diversity, discrimination capacity, and match probability) using the aforementioned 247 DNA samples obtained from unrelated individuals (Table 4). The 36-plex Y-STR system contained 99.19% of unique haplotypes and thus provided the highest power of forensic discrimination. Overall, the results showed that the 36-plex Y-STR system would be useful in the identification of related individuals.
In detecting Guangdong Han males, a null allele was observed at locus DYS448 (Fig. 7) in two samples; and triallelic genotypes were detected at locus DYS527a/b in five samples, locus DYS385a/b in three samples, and locus DYS387S1 in two samples. These findings were compared with the other commercial kits such as the Yfiler™ Plus kit and PowerPlex® Y23 kit, and the abnormal genotyping results were found to be consistent across kits (Fig. 8).
As shown in Fig. S7, a multidimensional scaling (MDS) plot based on 21 shared Y-STRs showed that Guangdong Han population and other Chinese Han populations tend to cluster together at the center of MDS plot. In the neighbor-joining tree (Fig. S8), Guangdong Han population was first clustered with Shenzhen Han population, and second with Henan Han and other Chinese Han populations. The degree of differentiation between the studied population and the reference population available in YHRD database was evaluated by calculation of the genetic distances (Rst) and p values (Table S4). The results indicated that Guangdong Han population was not significantly genetically different from Shandong Han, Shenzhen Han, or Fuzhou She populations (p > 0.0004, 120 pairs), but was significantly genetically different from the other compared populations (p < 0.0004).
Conclusion
This study has optimized and validated a novel 36-plex Y-STR system for forensic genetic testing, that incorporates all of the loci of the Yfiler® Plus kit and additionally includes a further nine Y-STR loci. Developmental validation studies that included PCR conditions as well as testing the cross-reactivity, sensitivity, anti-interference, and stability of the method and population data analysis have demonstrated it to be a sensitive, robust, and highly informative tool for use in forensic casework.
References
Jobling MA, Pandya A, Tyler-Smith C (1997) The Y chromosome in forensic analysis and paternity testing. Int J Legal Med 110(3):118–124
Roewer L (2009) Y chromosome STR typing in crime casework. Forensic Sci Med Pathol 5(2):77–84. https://doi.org/10.1007/s12024-009-9089-5
Ballantyne KN, Kayser M (2012) Additional Y-STRs in forensics: why, which, and when. Forensic Sci Rev 24(1):63–78
Kayser M (2017) Forensic use of Y-chromosome DNA: a general overview. Hum Genet 136(5):621–635. https://doi.org/10.1007/s00439-017-1776-9
Gopinath S, Zhong C, Nguyen V, Ge J, Lagace RE, Short ML, Mulero JJ (2016) Developmental validation of the Yfiler((R)) plus PCR amplification kit: an enhanced Y-STR multiplex for casework and database applications. Forensic Sci Int Genet 24:164–175. https://doi.org/10.1016/j.fsigen.2016.07.006
Gao HM, Wang C, Han SY, Sun SH, Xiao DJ, Wang YS, Li CT, Zhang MX (2017) Analysis of the 19 Y-STR and 16 X-STR loci system in the Han population of Shandong province, China. Genet Mol Res 16(1). https://doi.org/10.4238/gmr16019573
Shi M, Liu Y, Zhang J, Bai R, Lv X, Ma S (2015) Analysis of 24 Y chromosomal STR haplotypes in a Chinese Han population sample from Henan Province, Central China. Forensic Sci Int Genet 17:83–86. https://doi.org/10.1016/j.fsigen.2015.04.001
Thompson JM, Ewing MM, Frank WE, Pogemiller JJ, Nolde CA, Koehler DJ, Shaffer AM, Rabbach DR, Fulmer PM, Sprecher CJ, Storts DR (2013) Developmental validation of the PowerPlex(R) Y23 system: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci Int Genet 7(2):240–250. https://doi.org/10.1016/j.fsigen.2012.10.013
Hedman M, Pimenoff V, Lukka M, Sistonen P, Sajantila A (2004) Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages. Forensic Sci Int 142(1):37–43. https://doi.org/10.1016/j.forsciint.2003.07.003
Hedman M, Neuvonen AM, Sajantila A, Palo JU (2011) Dissecting the Finnish male uniformity: the value of additional Y-STR loci. Forensic Sci Int Genet 5(3):199–201. https://doi.org/10.1016/j.fsigen.2010.03.007
Iacovacci G, D'Atanasio E, Marini O, Coppa A, Sellitto D, Trombetta B, Berti A, Cruciani F (2017) Forensic data and microvariant sequence characterization of 27 Y-STR loci analyzed in four Eastern African countries. Forensic Sci Int Genet 27:123–131. https://doi.org/10.1016/j.fsigen.2016.12.015
Li S, Chen L, Wang Y, Xu Q, Liu H, Li Y, Liu C (2017) Developmental validation of a 6-dye STR kit with 27 loci. Int J Legal Med 132:335–342. https://doi.org/10.1007/s00414-017-1586-6
Zhang W, Xiao C, Yu J, Wei T, Liao F, Wei W, Huang D (2017) Multiplex assay development and mutation rate analysis for 13 RM Y-STRs in Chinese Han population. Int J Legal Med 131(2):345–350. https://doi.org/10.1007/s00414-016-1489-y
Ballantyne KN, Keerl V, Wollstein A, Choi Y, Zuniga SB, Ralf A, Vermeulen M, de Knijff P, Kayser M (2012) A new future of forensic Y-chromosome analysis: rapidly mutating Y-STRs for differentiating male relatives and paternal lineages. Forensic Sci Int Genet 6(2):208–218. https://doi.org/10.1016/j.fsigen.2011.04.017
Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, Herrmann S, Herzog B, Hidding M, Honda K, Jobling M, Krawczak M, Leim K, Meuser S, Meyer E, Oesterreich W, Pandya A, Parson W, Penacino G, Perez-Lezaun A, Piccinini A, Prinz M, Schmitt C, Roewer L et al (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110(3):125–133 141–129
SWGDAM Report on the Current Activities of the Scientific Working Group on DNA Analysis Methods Y-STR. http://www.fbi.gov/about-us/lab/forensicsciencecommunications/fsc/july2004/index.htm/standards/2004_03_standards03.htm. http://www.fbi.gov/about-us/lab/forensicsciencecommunications/fsc/july2004/index.htm/standards/2004_03_standards03.htm
Wang Y, Liu C, Zhang CC, Li R, Liu H, Ou XL, Li HX, Sun HY (2016) Analysis of 24 Y-STR haplotype data in a Chinese Han population from Guangdong Province. Int J Legal Med 130(3):689–691. https://doi.org/10.1007/s00414-015-1301-4
He G, Chen P, Zou X, Chen X, Song F, Yan J, Hou Y (2017) Genetic polymorphism investigation of the Chinese Yi minority using PowerPlex(R) Y23 STR amplification system. Int J Legal Med 131(3):663–666. https://doi.org/10.1007/s00414-017-1537-2
SWGDAM. Validation Guidelines for DNA Analysis Methods. http://swgdam.org/SWGDAM_Validation_Guidelines_APPROVED_Dec_2012.pdf. http://swgdam.org/SWGDAM_Validation_Guidelines_APPROVED_Dec_2012.pdf
Walsh PS, Metzger DA, Higushi R (2013) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10(4): 506-13 (April 1991). BioTechniques 54(3):134–139
Nunez C, Baeta M, Ibarbia N, Ortueta U, Jimenez-Moreno S, Blazquez-Caeiro JL, Builes JJ, Herrera RJ, Martinez-Jarreta B, de Pancorbo MM (2017) 17 to 23: a novel complementary mini Y-STR panel to extend the Y-STR databases from 17 to 23 markers for forensic purposes. Electrophoresis 38(7):1016–1021. https://doi.org/10.1002/elps.201600313
Nei M, Tajima F (1981) DNA polymorphism detectable by restriction endonucleases. Genetics 97(1):145–163
Wang L, Chen F, Kang B, Zheng H, Zhao Y, Li L, Zeng Z (2016) Genetic population data of Yfiler Plus kit from 1434 unrelated Hans in Henan Province (Central China). Forensic Sci Int Genet 22:e25–e27. https://doi.org/10.1016/j.fsigen.2016.02.009
Bai R, Liu Y, Zhang J, Shi M, Dong H, Ma S, Bai RF, Shi M (2016) Analysis of 27 Y-chromosomal STR haplotypes in a Han population of Henan province, Central China. Int J Legal Med 130(5):1191–1194. https://doi.org/10.1007/s00414-016-1326-3
Xu J, Li L, Wei L, Nie Z, Yang S, Xia M, Ma T, Sun H, Zhao X, Ping Y, Zhou H, Xue F, Zhao Z, Jin L, Li S (2016) Genetic analysis of 17 Y-STR loci in Han population from Shandong Province in East China. Forensic Sci Int Genet 22:e15–e17. https://doi.org/10.1016/j.fsigen.2016.01.016
Zhou Y, Shao C, Li L, Zhang Y, Liu B, Yang Q, Tang Q, Li S, Xie J (2018) Genetic analysis of 29 Y-STR loci in the Chinese Han population from Shanghai. Forensic Sci Int Genet 32:e1–e4. https://doi.org/10.1016/j.fsigen.2017.11.003
Li L, Yu G, Li S, Jin L, Yan S (2016) Genetic analysis of 17 Y-STR loci from 1019 individuals of six Han populations in East China. Forensic Sci Int Genet 20:101–102. https://doi.org/10.1016/j.fsigen.2015.10.007
Zhang S, Tian H, Wang Z, Zhao S, Hu Z, Li C, Ji C (2014) Development of a new 26plex Y-STRs typing system for forensic application. Forensic Sci Int Genet 13:112–120. https://doi.org/10.1016/j.fsigen.2014.06.015
Wang M, Wang Z, Zhang Y, He G, Liu J, Hou Y (2017) Forensic characteristics and phylogenetic analysis of two Han populations from the southern coastal regions of China using 27 Y-STR loci. Forensic Sci Int Genet 31:e17–e23. https://doi.org/10.1016/j.fsigen.2017.10.009
Guo F (2017) Population genetics for 17 Y-STR loci in Hui ethnic minority from Liaoning Province, Northeast China. Forensic Sci Int Genet 28:e36–e37. https://doi.org/10.1016/j.fsigen.2017.02.011
Guo F (2015) Population genetics for 17 Y-STR loci in Mongolian ethnic minority from Liaoning Province, Northeast China. Forensic Sci Int Genet 17:153–154. https://doi.org/10.1016/j.fsigen.2015.05.008
Bai R, Liu Y, Lv X, Shi M, Ma S (2016) Genetic polymorphisms of 17 Y chromosomal STRs in She and Manchu ethnic populations from China. Forensic Sci Int Genet 22:e12–e14. https://doi.org/10.1016/j.fsigen.2016.01.011
Cao SBP, Zhu W, Chen D, Wang H, Jin B, Zhang L, Liang W (2018) Genetic portrait of 27 Y-STR loci in the Tibetan ethnic population of the Qinghai province of China. Forensic Sci Int Genet 34:e18–e19. https://doi.org/10.1016/j.fsigen.2018.02.005
Zhu B, Wu Y, Shen C, Yang T, Deng Y, Xun X, Tian Y, Yan J, Li T (2008) Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic group residing in Qinghai province of China. Forensic Sci Int 175(2–3):238–243. https://doi.org/10.1016/j.forsciint.2007.06.012
Ou X, Wang Y, Liu C, Yang D, Zhang C, Deng S, Sun H (2015) Haplotype analysis of the polymorphic 40 Y-STR markers in Chinese populations. Forensic Sci Int Genet 19:255–262. https://doi.org/10.1016/j.fsigen.2015.08.007
Bian Y, Zhang S, Zhou W, Zhao Q, Siqintuya ZR, Wang Z, Gao Y, Hong J, Lu D, Li C (2016) Analysis of genetic admixture in Uyghur using the 26 Y-STR loci system. Sci Rep 6:19998. https://doi.org/10.1038/srep19998
Shan W, Ablimit A, Zhou W, Zhang F, Ma Z, Zheng X (2014) Genetic polymorphism of 17 Y chromosomal STRs in Kazakh and Uighur populations from Xinjiang, China. Int J Legal Med 128(5):743–744. https://doi.org/10.1007/s00414-013-0948-y
Ramos-Luis E, Blanco-Verea A, Brion M, Van Huffel V, Sanchez-Diz P, Carracedo A (2014) Y-chromosomal DNA analysis in French male lineages. Forensic Sci Int Genet 9:162–168. https://doi.org/10.1016/j.fsigen.2013.12.008
Katsaloulis P, Tsekoura K, Vouropoulou M, Miniati P (2013) Genetic population study of 11 Y chromosome STR loci in Greece. Forensic Sci Int Genet 7(3):e56–e58. https://doi.org/10.1016/j.fsigen.2013.02.001
Mizuno N, Nakahara H, Sekiguchi K, Yoshida K, Nakano M, Kasai K (2008) 16 Y chromosomal STR haplotypes in Japanese. Forensic Sci Int 174(1):71–76. https://doi.org/10.1016/j.forsciint.2007.01.032
Lee HY, Park MJ, Chung U, Lee HY, Yang WI, Cho SH, Shin KJ (2007) Haplotypes and mutation analysis of 22 Y-chromosomal STRs in Korean father-son pairs. Int J Legal Med 121(2):128–135. https://doi.org/10.1007/s00414-006-0130-x
Triki-Fendri S, Sanchez-Diz P, Rey-Gonzalez D, Ayadi I, Alfadhli S, Rebai A, Carracedo A (2013) Population genetics of 17 Y-STR markers in West Libya (Tripoli region). Forensic Sci Int Genet 7(3):e59–e61. https://doi.org/10.1016/j.fsigen.2013.02.002
Yong RY, Lee LK, Yap EP (2006) Y-chromosome STR haplotype diversity in three ethnic populations in Singapore. Forensic Sci Int 159(2–3):244–257. https://doi.org/10.1016/j.forsciint.2005.05.010
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Gusmao L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM (2006) DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int J Legal Med 120(4):191–200
Hedman J, Nordgaard A, Rasmusson B, Ansell R, Radstrom P (2009) Improved forensic DNA analysis through the use of alternative DNA polymerases and statistical modeling of DNA profiles. BioTechniques 47(5):951–958. https://doi.org/10.2144/000113246
Walsh PS, Fildes NJ, Reynolds R (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res 24(14):2807–2812
Leclair B, Fregeau CJ, Bowen KL, Fourney RM (2004) Systematic analysis of stutter percentages and allele peak height and peak area ratios at heterozygous STR loci for forensic casework and database samples. J Forensic Sci 49(5):968–980
Du W, Chen L, Liu H, Qiu P, Li F, Gao J, Zhou Y, Wang B, Liu C (2017) Developmental validation of the HomyGene19+14Y system. Int J Legal Med 131(3):605–620. https://doi.org/10.1007/s00414-016-1505-2
Li S, Chen L, Wang Y, Xu Q, Liu H, Li Y, Liu C (2018) Developmental validation of a 6-dye STR kit with 27 loci. Int J Legal Med 132(2):335–342. https://doi.org/10.1007/s00414-017-1586-6
Pickrahn I, Muller E, Zahrer W, Dunkelmann B, Cemper-Kiesslich J, Kreindl G, Neuhuber F (2016) Yfiler((R)) Plus amplification kit validation and calculation of forensic parameters for two Austrian populations. Forensic Sci Int Genet 21:90–94. https://doi.org/10.1016/j.fsigen.2015.12.014
Funding
This work was supported by a grant from the Natural Science Foundation of China (grant no. 81501627), Innovative Training Program for College Students (grant no. 201612121083), and Open project of Key Laboratory of Forensic Genetics in Ministry of Public Security (2015FGKFKT03).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Du, W., Feng, P., Huang, H. et al. Technical note: developmental validation of a novel 6-dye typing system with 36 Y-STR loci. Int J Legal Med 133, 1015–1027 (2019). https://doi.org/10.1007/s00414-018-1864-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-018-1864-y