Abstract
Meiosis is a critical phase in the life cycle of sexually reproducing organisms. Chromosome numbers are halved during meiosis, which requires meiosis-specific modification of chromosome behaviour. Furthermore, suppression of transposons is particularly important during meiosis to allow the transmission of undamaged genomic information between generations. Correspondingly, specialized genome defence mechanisms and nuclear structures characterize the germ line during meiosis. Survival of mammalian spermatocytes requires that the sex chromosomes form a distinct silenced chromatin domain, called the sex body. An enigmatic spherical DNA-negative structure, called the meiotic dense body, forms in association with the sex body. The dense body contains small non-coding RNAs including microRNAs and PIWI-associated RNAs. These observations gave rise to speculations that the dense body may be involved in sex body formation and or small non-coding RNA functions, e.g. the silencing of transposons. Nevertheless, the function of the dense body has remained mysterious because no protein essential for dense body formation has been reported yet. We discovered that the polycomb-related sex comb on midleg-like 1 (SCML1) is a meiosis-specific protein and is an essential component of the meiotic dense body. Despite abolished dense body formation, Scml1-deficient mice are fertile and proficient in sex body formation, transposon silencing and in timely progression through meiosis and gametogenesis. Thus, we conclude that dense body formation is not an essential component of the gametogenetic program in the mammalian germ line.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sexual reproduction in animals requires the generation of haploid gametes from diploid germ cells by the specialized cell division cycle of meiosis. The ploidy is halved because one round of pre-meiotic DNA replication is followed by two rounds of chromosome segregation during meiosis. Homologous chromosomes and sister chromatids segregate during the first and second rounds of meiotic nuclear divisions, respectively. Orderly segregation of homologous chromosomes requires that homologous chromosomes become physically linked through inter-homologue crossovers during the first meiotic prophase (Page and Hawley 2003; Petronczki et al. 2003). Crossovers are formed by a modified homologous recombination pathway specific to meiosis (Baudat and de Massy 2007; Hunter 2007). This entails the active generation of DNA double strand breaks (DSBs), the use of resulting DNA ends for homology search and the repair of DSBs with the use of homologous chromosomes as repair template. Importantly, homologous chromosomes must recognize each other and their proteinaceous cores/axes must closely juxtapose as part of the recombination process. This alignment of chromosome axes leads to the formation of a meiosis-specific chromatin structure, the synaptonemal complex, which assembles along each pair of homologous chromosomes as cells progress to the pachytene stage of prophase (Baudat and de Massy 2007; Hunter 2007; Page and Hawley 2003; Petronczki et al. 2003). Efficient crossover formation depends on synaptonemal complex formation during meiosis (Baudat and de Massy 2007; de Vries et al. 2005; Hunter 2007). Thus, each pair of homologous chromosomes must engage into synapsis to ensure correct chromosome segregation during the first meiotic division and to prevent generation of gametes with incorrect chromosome sets. Accordingly, asynapsis is monitored during meiosis, and persistent asynapsis seems to trigger elimination of meiocytes in mammals (Burgoyne et al. 2009; Daniel et al. 2011; Kogo et al. 2012a, b; Mahadevaiah et al. 2008; Wojtasz et al. 2012). Surveillance mechanisms that eliminate asynaptic meiocytes involve the recruitment of ATR activity specifically to unsynapsed chromosome regions. ATR activity leads to the accumulation of a phosphorylated form of histone H2AX, called γH2AX, and to meiotic silencing of unsynapsed chromatin (MSUC) (Burgoyne et al. 2009; Fernandez-Capetillo et al. 2003; Royo et al. 2013; Turner et al. 2005). It is thought that persistent asynapsis triggers oocyte elimination either due to direct activation of apoptotic signalling cascades by persistent ATR activity or due to inappropriate silencing of essential genes on unsynapsed chromosomes (Cloutier et al. 2015; Wojtasz et al. 2012). In contrast to oocytes, spermatocytes always contain unsynapsed chromosomes. The sex chromosomes are largely non-homologous; hence, they synapse only in their short homologous pseudoautosomal regions (PAR) during the pachytene stage of prophase, where autosomal chromosomes are fully synapsed. As a consequence, the unsynapsed regions of sex chromosomes are silenced (Baarends et al. 2005; Turner et al. 2005) and are incorporated into a distinct γH2AX-rich chromatin domain, called the sex body (Fernandez-Capetillo et al. 2003; Monesi 1965; Solari 1974). Crucially, the silencing of sex chromosomes is essential for progression of spermatocytes beyond mid-pachytene because there are genes on sex chromosomes whose expression is incompatible with spermatocyte survival at this stage (Royo et al. 2010, 2015). It is believed that the requirement for sex chromosome silencing provides an effective mechanism for the elimination of asynaptic spermatocytes (Burgoyne et al. 2009; Mahadevaiah et al. 2008). Abnormal autosomal asynapsis titrates ATR activity away from sex chromosomes resulting in imperfect silencing of sex chromosomes and the expression of genes that are toxic to spermatocytes in mid-pachytene. Thus, MSUC and sex body formation are important components of the surveillance mechanisms that safeguard the quality of gametogenesis in mammals. Interestingly, the sex body is accompanied by an enigmatic nuclear structure that was termed the dense body due to its high electron density in electron microscopy (Crackower et al. 2003; Dresser and Moses 1980; Marcon et al. 2008). The mouse dense body has low DNA content but it contains both microRNAs (miRNA) and PIWI-associated RNAs (piRNAs) (Marcon et al. 2008). The dense body was also reported to contain FKBP6 (Crackower et al. 2003), which is a protein involved in piRNA metabolism (Xiol et al. 2012). Although definitive localization to the dense body in spermatocytes was not established, nuclear foci were also detected for the piRNA-binding MILI, and MIWI proteins, which raised the possibility that these proteins are also components of the dense body (Beyret and Lin 2011). Thus, it is tempting to speculate that the dense body may contribute to the function of small RNA pathways that play diverse essential roles in spermatogenesis. Sex body formation is one of the important processes that involve microRNA pathways during spermatogenesis (Modzelewski et al. 2012). Thus, the dense body may be involved in the repression of sex chromosome-linked gene expression. Indeed, such function would be consistent with the close association of the dense body and the sex body in pachytene spermatocytes (Crackower et al. 2003; Dresser and Moses 1980; Marcon et al. 2008). Another tempting possibility is that the dense body is involved in meiotic down-regulation of transposons, given that the best documented function of piRNAs is the silencing of transposons during meiosis (Aravin et al. 2007; Carmell et al. 2007; De Fazio et al. 2011; Di Giacomo et al. 2013; Pillai and Chuma 2012; Reuter et al. 2011; Watanabe et al. 2006). This is a fundamentally important function because inappropriate activation of transposons leads to unscheduled DSB formation and a failure in the pairing of homologous chromosomes.
Despite the expectation that the dense body is important for successful meiosis and/or spermatogenesis in mammals, the function and the significance of dense body formation have remained mysterious, because up to now, no essential protein component of the dense body has been reported. We have screened for proteins that are preferentially expressed during meiosis in mice to identify new candidates that function in meiosis-specific processes that may be essential for the generation of haploid gametes in mice. We discovered that mouse SCML1 is a meiosis-specific protein that constitutes a key component of the meiotic dense body. Here, we report the functional analysis of Scml1-deficient mice.
Results
Identification of the full-length SCML1 protein
To identify proteins that are potentially involved in core functions of the first meiotic prophase, we analysed gene expression in the developing gonads of both sexes (our unpublished data) using microarrays of the NIA mouse 15 k complementary DNA (cDNA) set (Kargul et al. 2001; Tusher et al. 2001). One of the transcripts that were up-regulated in the gonads of both sexes at the time when meiocytes progressed to the first meiotic prophase mapped to the predicted sex comb on midleg-like 1 (Scml1) gene on the X chromosome (Online Resource 1.). RT-PCR analysis showed that Scml1 was not expressed in a set of 17 different somatic tissues (Fig. 1a) and that Scml1 was preferentially expressed in the female and male gonads at stages where gonads contained meiotic germ cells (Fig. 1b, c). We detected Scml1 expression in meiotic germ cells but not in somatic cells of female gonads at 16.5 days post coitum (dpc) (Fig. 1d). Therefore, we conclude that Scml1 expression is largely, if not completely, restricted to meiotic germ cells in mice. The NCBI-database predicted transcript of Scml1, XM_006544639.2, appears incomplete. The corresponding open reading frame (XP_003945662.2) starts from the very beginning of the predicted transcript, and the predicted mouse SCML1 protein is shorter in its N-terminus than the closely related predicted rat SCML1 protein (XP_006227380). Therefore, we used both 3′ and 5′ RACE on total RNAs of mouse testis to determine the sequence of the full-length Scml1 transcript (Online Resource 2, Supplementary Fig. S1). We reconfirmed the sequence of the full-length Scml1 transcript by RNA sequencing the messenger RNAs (mRNAs) of fetal ovaries (data not shown). The full-length transcript encodes a 501 amino acid-long protein that contains a “sterile-alpha” motif (SAM) at its C-terminus and 14 imperfect repeats of the T/P-V/I/M-D/N-L/N/C-S/N/T/A-Q/L/V-T/P/G-V/F/L/I-Q-Y/N-T-D/N/E 12-amino acid peptide. Although SCML1 proteins have been identified only in mammals, they are related to the polycomb group Drosophila sex-comb-on-midleg protein (Wu and Su 2008). Their most conserved region is the SAM domain (Wu and Su 2008), which is thought to mediate protein-protein interactions, oligomerization and/or RNA binding (Kim and Bowie 2003). In contrast, the 12-amino acid long repeats of mouse SCML1 have no predicted function and they appear to be absent from SCML1 proteins in mammals except the Muroidea superfamily. Thus, the repeats likely represent a recent modification of this protein in evolution (Online Resource 2, Supplementary Fig. S2).
SCML1 is expressed specifically in meiotic germ cells. a–d RT-PCR was used to detect expression of Scml1, a meiosis marker (Sycp3), a ‘house-keeping’ gene (S9), and in d, the expression of a germ cell marker Ddx4 and the soma specific Xist gene. a Total RNAs of testis and a somatic tissue mix were used as template in RT-PCRs. cDNAs were prepared from four RNA mixtures: (1) Somatic tissue mix: 1 μg of RNA mix of 59 ng total RNAs from 17 somatic tissues (see Materials and Methods for the tissue list). (2) Adult testis: 59-ng total testis RNAs from adult. (3) Somatic + adult testis: 1 μg of RNA mix of 59-ng total testis RNAs and 941 ng of somatic tissue mix. (4) Somatic + 5x adult testis: 1 μg of RNA mix of 295-ng total testis RNAs and 705 ng of somatic tissue mix. (5) no RT: no RT control with somatic + adult testis. Scml1 specific PCR products were amplified only from templates that contained testis cDNA. b, c Total RNAs of developing male (b) and female (c) gonads were used as templates in RT-PCRs. Germ cells begin to initiate entry into meiosis 7–11 days postpartum (dpp) in testes and 12.5–14.5 days post coitum (dpc) in ovaries. Most germ cells are in zygotene or pachytene stages of meiotic prophase at 16.5dpc in ovaries. d Total, somatic and germ cell populations of ovaries of 16.5 dpc fetuses were FACS sorted based on forward and side scattering (Wojtasz et al. 2009b). Total RNAs purified from these cell populations were used as templates in RT-PCRs
SCML1 localizes to the meiotic dense body
To gain insight into the possible functions of SCML1, we raised antibodies against the full-length SCML1, and affinity purified antibodies against a soluble C-terminal 208 amino acid-long fragment of the protein. We used our antibodies to detect SCML1 on cryosections of testes (Fig. 2). This revealed that robust anti-SCML1 staining appeared first in leptotene stage germ cells. Anti-SCML1 antibodies showed a diffuse nuclear staining and a single intense nuclear focus in each meiocyte at this stage. Following the progression of meiocytes to the pachytene stage diffuse nuclear staining disappeared, but the single focus detected by anti-SCML1 antibodies persisted in the nucleus of spermatocytes. These foci were present in germ cells throughout meiosis, and they disappeared from post-meiotic cells only after spermatids began to elongate.
SCML1 localizes to spermatogenic cells in testis. a, b DNA was detected by DAPI, and DDX4 (cytoplasmic germ cell marker) and SCML1 were detected by immunofluorescence on cryosections of adult wild-type testis. Epithelial cycle stage of testis tubules is shown. Each tubule contains several layers of germ cells at distinct spermatogenic stages. Enlarged inset is shown in the lower panel of both a and b. a In stage VII, preleptotene (pl) and late pachytene (pa) spermatocytes and post-meiotic round meiotic spermatids (sd) and spermatozoa (sp.) are shown. Yellow arrowheads mark SCML1 foci in a pachytene spermatocytes and a round spermatid in the enlarged inset. b In stage IX-X, leptotene (le), late pachytene (pa) spermatocytes and elongating spermatids (esd) are shown. Yellow arrowheads mark SCML1 foci in a leptotene and a pachytene spermatocyte and an elongated spermatid in the enlarged inset. Note that SCML1 is present throughout the nuclear volume in the leptotene/zygotene spermatocyte. SCML1 foci are still detectable in some of the b elongating spermatids but with lower intensity than in a round spermatids. a, b The cytoplasmic signal that is detectable at variable intensities in intertubular somatic cells (it) does not represent SCML1-specific staining because it does not depend on anti-SCML1 antibody (data not shown). DDX4 forms chromatoid body precursor granules (spermatocytes) and chromatoid bodies (spermatids), which are marked by white arrowheads in insets. Scale bars, 20 μm
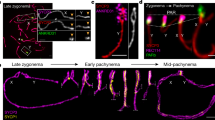
To better address the sub nuclear localization of SCML1 relative to other meiotic structures, we detected SCML1 also on nuclear surface spread spermatocytes (Fig. 3a–c). Anti-SCML1 antibodies stained weakly the unsynapsed axis in zygotene-early pachytene transition and also the chromatin loops of sex chromosomes in early mid-pachytene. In contrast, a round shape or ring-like structure was stained much stronger by anti-SCML1 antibodies in spermatogenic cells (Figs. 2 and 3). This structure associated with the sex body in pachytene cells (79.95 %, n = 419 cells) and grew in size as meiocytes progressed in prophase reaching a diameter of approximately 2 μm by the end of pachytene (n = 124 cells). This structure was also largely devoid of DAPI staining. These observations raised the possibility that our anti-SCML1 antibodies stained the meiotic dense body. To reconfirm this, we co-stained spermatocytes with anti-SCML1 and anti-FKBP6 antibodies (Fig. 3c). FKBP6 was reported to localize to the dense bodies and to chromosome axes in spermatocytes (Crackower et al. 2003). The aliquot of the published anti-FKBP6 antibody that was available to us did not stain robustly chromosome axes but apparently recognized the dense body in our nuclear surface spread preparations of spermatocytes. All the anti-SCML1 stained sphere-/ring-like structures were also stained by anti-FKBP6 antibodies (n > 500 cells), which supports the hypothesis that SCML1 localizes to dense bodies. To further test this hypothesis, we carried out immunotransmission electron microscopy (TEM) of spermatocytes in testis sections using the anti-SCML1 antibodies (Fig. 3d). We found that anti-SCML1 antibodies marked a homogenously granular electron-dense structure, which we identify as the dense body within the nucleus of spermatocytes. Importantly, anti-SCML1 labelling was distributed evenly in the dense bodies on ultrathin TEM sections. This indicated that the ring-like immunofluorescence staining pattern in surface spread spermatocytes was likely a staining artefact caused by an inability of the antibodies to access the centre of fixed dense bodies efficiently. Anti-SCML1-stained dense bodies exist beyond meiotic prophase as we also detected an anti-SCML1 marked structure in the vast majority of round spermatids (98.5 %, n = 197 spermatids) in single cell suspensions of testes (n = 200 spermatids).
SCML1 localizes predominantly to the dense body in meiotic spermatocytes. a, b, c, e DNA was detected by DAPI, and SCML1, SYCP3 (chromosome axis marker), SYCP1 (a synaptonemal complex marker), γH2AX (b sex body marker) and FKBP6 (c dense body marker) were detected by immunofluorescence on nuclear surface spreads of a, b, c spermatocytes and e oocytes. Arrows mark reduced DAPI staining at the positions of the anti-SCML1 stained structures. Scale bars, 10 μm. (a upper panel) Zygotene-pachytene spermatocyte is characterized by full autosomal synapsis, but the synaptonemal complex transverse filament protein SYCP1 has not accumulated yet on the short homologous regions of sex chromosomes (arrowhead), despite apparent pairing of these so called PAR regions of X and Y chromosomes. At the more advanced mid-pachytene stage (a lower panel), SYCP1 is detected at the PAR region (arrowhead). Note the accumulation of SCML1 along sex chromosome axes (a upper panel) or on chromatin surrounding sex chromosome axes (a lower panel) at the zygotene-to-pachytene and mid-pachytene stages, respectively. (a lower panel, b) The round shape anti-SCML1 stained structure tends to associate with sex chromosomes as meiocytes progress to pachytene. b Cloud-like staining pattern in the SYCP3 channel is signal bled from the γH2AX channel, representing the unsynapsed chromatin of sex chromosomes. c The anti-SCML1 stained structure is also stained by anti-FKBP6 antibodies, which are known to mark the dense body. d Transmission electron microscopy (TEM) of a spermatocyte in a Tokuyasu cryo-section immunolabelled with anti-SCML1 antibody. Cytoplasm (CP) with organelles like mitochondria (m) is separated by the double membrane of nuclear envelop (NE) from nuclear content (N). Immunogold detection (arrow, dark spots of gold particles) reveals anti-SCML1 stain specifically at the dense body (db), a round homogenous nuclear structure of different density than the surrounding chromatin (CHR) in nucleus. Scale bar, 500 nm. e Anti-SCML1 recognizes a round structure in a DNA-poor region of pachytene oocyte at the 16.5 dpc developmental timepoint
Scml1 is also expressed in oocytes in the first meiotic prophase in fetal mice. This prompted us to examine the localization of SCML1 in oocytes of fetal mice. Although a dense body was not reported in oocytes previously, we detected an anti-SCML1 stained nuclear structure in pachytene oocytes (Fig. 3e). This female dense body is smaller than its counterpart in spermatocytes, but it is also characterized by poor DAPI staining. Thus, our results suggest that the dense body forms in meiosis in both sexes.
To test if the anti-SCML1 staining of the dense body is a genuine reflection of SCML1 localization to the dense body, we in vivo electroporated testes of mice with plasmids that expressed a Venus-tagged version of SCML1 (Online Resource 2, Supplementary Fig. S3a). The Venus-specific signal of Venus-SCML1 was always detected in a round shape structure that was identified as the dense body due to its low DAPI staining and its frequent association with the sex body. Lower level signal was also detected in the sex body. These observations reconfirmed the specificity of our anti-SCML1 antibodies and strongly supported the conclusion that SCML1 is a dense body component. Given these observations, we wondered if SCML1 might contribute to the formation of dense bodies. To test this, we ectopically expressed either Cherry (Online Resource 2, Supplementary Fig. S3b) or GFP-tagged versions (not shown) of SCML1 in mouse NIH 3T3 and human HeLa cell lines. We found that these tagged versions of SCML1 proteins formed spherical structures that resembled meiotic dense bodies. This indicated that SCML1 can form dense body-like nuclear structures even in the absence of other meiosis-specific proteins in the nuclei of non-meiotic/mitotic cells. Thus, SCML1 may be an important structural component of the meiotic dense bodies.
SCML1 is required for dense body formation
To address the functions of SCML1 during meiosis, we generated mice that lacked the seventh exon of the Scml1 gene (Online Resource 2, Supplementary Fig. S4). The seventh exon is a frame-shifting exon; therefore, its removal was expected to lead to the production of a truncated version of SCML1 that matched only the N-terminal 72 amino acids of the wild-type SCML1. RT-PCR with primers specific to exon 4 and 6 indicated that deletion of the seventh exon dramatically reduced the levels of the residual truncated transcripts (Online Resource 2, Supplementary Fig. S4c), possibly due to non-sense mediated mRNA decay. To test if full-length SCML1 protein was depleted from the testes of Scml1 Y/− mice, we detected SCML1 by Western blot analysis in wild-type and Scml1 Y/− testis extracts (Online Resource 2, Supplementary Fig. S4d-e). The predicted molecular weight of SCML1 is 57.5 kDa, yet our anti-SCML1 antibody detected a prominent protein-band approximately at 100 kDa size in wild-type but not in Scml1 Y/− testis extracts following SDS-gel electrophoresis. Given that a recombinant 6xHis-tagged version of SCML1 had similarly reduced electrophoretic mobility (Online Resource 2, Supplementary Fig. S4d), we concluded that the wild-type-specific ∼100 kDa protein-band represented full-length SCML1 and that Scml1 Y/− testes were depleted of full-length SCML1 protein. Accordingly, we could not detect spherical or ring-like anti-SCML1 staining pattern in Scml1 Y/− spermatocytes (Fig. 4) using either anti-full-length SCML1 sera or purified anti-C-terminal SCML1 fragment antibodies, which supported the idea that the exon 7-deletion allele of Scml1 is a null allele.
SCML1 is not detectable in the nucleus of spermatogenic cells of Scml1 Y/− mice. a, b DNA was detected by DAPI, SCML1 and either DDX4 (a cytoplasmic germ cell marker) or SYCP3 (b axis marker) was detected by immunofluorescence on cryosections of adult testes (a) or on nuclear surface spread pachytene spermatocytes (b) of indicated genotypes. a Enlarged insets of first and third row are shown in the second and fourth rows, respectively. Testis tubules at epithelial cycle stage V-VI are shown. Spermatogonia B (sg), pachytene stage spermatocytes (pa), spermatids (sd) and spermatozoa (sp.) are marked. Arrowheads mark chromatoid bodies. b Arrow marks reduced DAPI staining at the positions of the anti-SCML1 stained structure in the wild-type pachytene spermatocyte. XY marks the sex chromosomes. Scale bars, 10 μm
Scml1-deficient mice showed no obvious somatic or growth defects; average body weight of wild-type (36.05 g, n = 6) and Scml1-deficient (36.08 g, n = 6) adult (70–90 days of age) mice were not significantly different (p > 0.9999 Wilcoxon matched pair test). Scml1 −/− female and Scml1 Y/− male mice were also fertile and their litter sizes were similar to the litter sizes of their wild-type littermate controls. On average, we observed 12.2 (n = 25), 11.2 (n = 24) and 11.8 (n = 5) pups per litter in crosses of Scml1 +/+ × Scml1 Y/+, Scml1 +/+ × Scml1 Y/− and Scml1 −/− × Scml1 Y/−, respectively (Mann Whitney test calculated p values are 0.2084 and 0.6675 for the Scml1 +/+ × Scml1 Y/+ versus Scml1 +/+ × Scml1 Y/− and the Scml1 +/+ × Scml1 Y/+ versus Scml1 −/− × Scml1 Y/− comparison, respectively). This shows that SCML1 is not required for fertility in mice. Consistent with this observation, germ cells at all stages of spermatogenesis could be found in the adult Scml1 Y/− mice (Fig. 4a and not shown).
To test if dense bodies formed in the Scml1 Y/− spermatocytes, we compared dense body formation in wild-type and Scml1 Y/− pachytene spermatocytes using either DAPI, anti-FKBP6 or anti-γH2AX staining (Fig. 5). DAPI-negative spherical regions, most of which likely represented dense bodies, could be identified within the sex body or in the vicinity of the sex body in a majority of nuclear spreads of wild-type spermatocytes (79.95 % of pachytene cells have a DNA-negative region in the sex body or less than 5 μm away from the dense body, n = 419 cells) (Fig. 5b). In contrast, similar DAPI-negative regions were detected only in a small minority of Scml1 Y/− pachytene spermatocytes (average 22.16 %, n = 397 cells), and these DAPI-negative regions never or only rarely (average 0 and 7.05 %) localized within sex bodies or in the vicinity of sex bodies, respectively (Fig. 5b). Correspondingly, we observed γH2AX negative spherical regions in the sex bodies of a large fraction of wild-type spermatocytes (average 35 %, n = 249 cells). Similar γH2AX negative regions were never observed in Scml1 Y/− spermatocytes (n = 225, spermatocytes). Finally, whereas anti-FKBP6-stained dense bodies were detected in almost all nuclear spreads of wild-type pachytene spermatocytes (95.9 %, n = 244 cells, three mice), we found no anti-FKBP6-stained dense bodies in pachytene stage Scml1 Y/− spermatocytes (n = 233 cells, three mice) (Fig. 5d). In contrast, FKBP6 was not required for dense body formation (Marcon et al. 2008), and thus we detected SCML1 positive dense bodies in Fkbp6 −/− spermatocytes that were presumably equivalent to wild-type pachytene stage cells (data not shown, 74 out of 78 cells). The combination of these experiments showed that dense body formation was severely impaired if not completely prevented in the absence of SCML1.
Dense body formation is defective in the absence of SCML1. a, c DNA was detected by DAPI, and a γH2AX or c SYCP3 (chromosome axis marker) and FKBP6 (dense body marker) were detected by immunofluorescence on nuclear surface spreads of wild-type and Scml1 Y/− spermatocytes. Arrows mark round shape DAPI-negative regions that are identified as sites of dense bodies. Scale bars, 10 μm for full nuclei and 5 μm for cropped γH2AX marked sex body. a Four different categories of pachytene/diplotene cells are shown: (1) DAPI-negative dense body is inside the sex body (inside SB), (2) dense body is less than 5 μm away from the sex body (<5 μm), (3) clear DAPI-negative body is detected more than 5 μm away from the sex body (>5 μm), (4) no clear round shape dense body-like DAPI-negative region is detectable in the spermatocyte (NO DN). b Quantification of the fraction of spermatocytes that fall into the four categories listed in a wild-type and Scml1 Y/− spermatocytes. Summary of data from four experiments, averages and standard deviations are shown. Significance or lack of significance (n.s.) was calculated by Student’s t test. c The axes of the X and Y chromosomes are marked by X and Y, respectively. d Quantification of the fraction of Scml1 Y/+ and Scml1 Y/− pachytene spermatocytes that contain FKBP6 marked dense bodies
Spermatogenesis is apparently unaffected in the absence of SCML1 and dense bodies
Although Scml1-deficient mice were fertile, we wondered if progression through meiosis or spermatogenesis was affected in the absence of SCML1 and dense bodies. Therefore, we carefully compared testis weight and spermatogenesis in adult wild-type and Scml1 Y/− mice (Fig. 6). We found no significant difference between the absolute testis weight of wild-type and Scml1-deficient mice (Fig. 6a). To test if apoptosis was increased in the absence of SCML1, we quantified the fraction of testis tubules that contained spermatocytes with cleaved PARP1 in sections of testes. We used Spo11 −/− mice as positive controls, because Spo11 −/− spermatocytes are DSB formation-defective, which leads to their apoptosis in tubules at stage IV of the seminiferous epithelial cycle (Barchi et al. 2005; Baudat et al. 2000; Romanienko and Camerini-Otero 2000). Whereas significant fractions (7 out of 62) of tubules contained multiple apoptotic spermatocytes in Spo11 −/− mice, similar testis tubules with apoptotic spermatocytes were not found in Scml1 Y/− and wild-type mice (0 out of 204 and 0 out of 236 tubules, respectively, Fig. 6b). To assess spermatogenesis, we prepared nuclear surface spreads and quantified the fraction of cells at various stages of spermatogenesis. We found no significant difference between fractions of somatic cells and various stages of spermatogenic cells in wild-type and Scml1 Y/− mice (Fig. 6c, d n = 11,634, n = 12,365 in wild-type and Scml1 Y/− mice, respectively). The microRNA pathway has been implicated in the regulation of the initiation of spermatogenesis in mice (Modzelewski et al. 2012). Given the reported accumulation of some microRNAs in the dense body (Marcon et al. 2008), it was possible that SCML1 was involved in the regulation of the developmental timing of meiosis initiation. To test this hypothesis, we quantified non-meiotic and meiotic cell populations of testes of juvenile (8 days postpartum) wild-type and Scml1 Y/− mice at a developmental stage where the first spermatogenic cells started to enter meiosis (Fig. 6e). We found no significant difference in the fraction of meiotic cells at various early stages of meiosis in juvenile wild-type and Scml1 Y/− mice. Thus, the combination of these observations indicated that neither the correct developmental timing of meiosis in males nor correct progression through the spermatogenic process requires SCML1 and robust dense body formation in mice.
Spermatogenesis is not altered significantly by the disruption of Scml1. a Graph showing weight of testis pairs in adult (65-142dpp) Scml1 Y/+ and Scml1 Y/− mice. Each data point represents a testis pair/mouse. Median is shown. No significant (n.s.) difference was found between Scml1 Y/+ and Scml1 Y/− mice (p = 0.8457 Wilcoxon matched pairs test). b DNA was detected by DAPI, and cleaved PARP (apoptosis marker) was detected by immunofluorescence in cryosections of adult testes of indicated genotypes. While PARP positive apoptotic spermatocytes (ap) are readily detectable in DSB formation defective Spo11 −/− testes, apoptotic spermatocytes are not detected in the shown testis tubules of the Scml1 Y/+ and Scml1 Y/− mice. Scale bars, 50 μm. c, d Quantification of the c fractions of testicular cells that are non-meiotic, meiotic or post-meiotic or the d fractions of spermatocytes at the indicated stages in adult Scml1 Y/+ and Scml1 Y/− mice. e Quantification of the fractions of testicular cells that are non-meiotic or represent the indicated stages in spermatocytes in 8dpp Scml1 Y/+ and Scml1 Y/− mice. d, e Counts in preleptotene (pre-L), leptotene (Lep), zygotene (Zyg), pachytene (Pac), diplotene (Dip), first meiotic metaphase (Met I) stages and in non-meiotic cells (non-mei) are shown. (c–e) Total numbers of scored cells are indicated
Efficient silencing of transposons and sex body formation in the absence of SCML1 and dense bodies
Although our observations indicated that essential processes of meiosis do take place in the absence of SCML1, we wondered if partial defects can be observed in predicted microRNA or piRNA functions. MicroRNA pathways have been implicated in sex body formation (Modzelewski et al. 2012), and the dense body associates with the sex body during pachytene (Crackower et al. 2003; Marcon et al. 2008); therefore, dense body may play a role in sex body formation. Additionally, low levels of SCML1 associates with the sex body (Fig. 3a and Online Resource 2, Supplementary Fig. S3a) indicating a possible function in its formation. To examine sex body formation, we detected γH2AX in Scml1 Y/− spermatocytes. γH2AX-rich chromatin often expanded to synapsed autosomes and sex body formation was reported to be partially defective in the microRNA pathway mutant Ago4 −/− mice (Modzelewski et al. 2012). In contrast, we found no mislocalization of γH2AX in Scml1 Y/− spermatocytes and their sex bodies resembled sex bodies in wild-type spermatocytes (Fig. 7a). Likewise, another sex body marker, SUMO-1 (Vigodner and Morris 2005), decorated sex chromatin similarly in wild-type and Scml1 Y/− spermatocytes (Fig. 7a, n = 90 pachytene spermatocytes). Consistent with the idea that sex body formation is unaffected, we found that phosphorylated RNA polymerase II was depleted from sex chromatin in Scml1 Y/− spermatocytes (Fig. 7b, n = 80 pachytene spermatocytes). To further test if silencing of sex chromosomes was affected, we measured expression of three sex chromosome-linked genes in wild-type and Scml1 Y/− testes by real-time RT-PCR (Fig. 7c). Expression levels of these genes did not differ significantly in wild-type and Scml1 Y/− testis. Thus, we conclude that sex body formation and silencing of sex chromosomes does not require SCML1 and the meiotic dense body.
SCML1 is not needed for robust sex chromosome or transposon silencing. a, b DNA was detected by DAPI and γH2AX (sex body marker), and either a SUMO (sex body marker) or b phospho-S2-RNA polymerase II (marker of active transcription) was detected by immunofluorescence on nuclear surface spreads of pachytene spermatocytes of Scml1 Y/+ and Scml1 Y/− mice. Note that phospho-RNA polymerase II is absent from the sex body (outlined) of both Scml1 Y/+ and Scml1 Y/− spermatocytes. Scale bars, 10 μm. c, d Quantitative real-time RT-PCR was used to measure expression of c sex chromosome-linked genes and d indicated transposons. Averages of 6 independent measurements with standard deviation are shown. Expression values are normalized to the wild-type average. No significant difference was found in the expression of any of the examined genes/transposons in Scml1 Y/+ and Scml1 Y/− testes
The best documented function of piRNAs is the silencing of transposons during meiosis (Aravin et al. 2007; Carmell et al. 2007; De Fazio et al. 2011; Di Giacomo et al. 2013; Pillai and Chuma 2012; Reuter et al. 2011; Watanabe et al. 2006). piRNA targeted slicer activity of MIWI and MILI is important for the repression of transposons during spermatogenesis (De Fazio et al. 2011; Reuter et al. 2011). Given that some piRNAs accumulate in the dense body (Marcon et al. 2008), we used real-time RT-PCR to test if SCML1 and dense body formation is needed for silencing of transposons in adult testes. We did not find significant differences between the levels of LINE1-, IAP-, or SINEB1-specific transcripts in Scml1 Y/− and wild-type testes (Fig. 7d). Therefore, we conclude that SCML1 and the dense body are unlikely to contribute to the transposon silencing function of piRNAs.
Meiotic surveillance mechanisms of asynapsis do not require SCML1 and dense body in either sex
The association of dense body with the sex body raised the possibility that SCML1 and the dense body may play a role in synapsis surveillance mechanisms that involve meiotic silencing of unsynapsed chromatin. Our data indicated that SCML1 is not required for sex body formation in spermatocytes where synaptonemal complex formation and meiotic recombination proceeds normally (Fig. 7a–c). Nevertheless, we could not exclude the possibility that SCML1 had a function in the elimination of asynaptic meiocytes that are thought to undergo apoptosis due to either ineffective silencing of sex chromosomes or inappropriate silencing of essential genes in males and females, respectively. To address this possibility, we asked if SCML1 was required for the elimination of Spo11 −/− meiocytes that are defective in synaptonemal complex formation due to a lack of programmed DSBs. We examined spermatogenesis on sections of testes of adult mice, and we found that spermatocytes underwent apoptosis in epithelial cycle stage IV testis tubules in both Spo11 −/− and Spo11 −/− Scml1 Y/− mice (Fig. 8a, 730 and 586 tubules were examined in two mice of each genotype, and 17.39 and 19.28 % of tubules contained PARP positive cells, respectively). We also found that oocyte numbers were similarly reduced in ovaries of 6–7 weeks old Spo11 −/− and Spo11 −/− Scml1 −/− mice (on average, 1.33 and 1.04 oocytes per section, n = 51 and 68 non-adjacent ovary sections from two and three mice of respective genotypes) as compared to ovaries of wild-type and Scml1 −/− mice (on average, 30, 95 and 29.1 oocytes per section, n = 20 non-adjacent sections from two mice for each genotype, Fig. 8b). Thus, we conclude that elimination of asynaptic meiocytes does not require SCML1 in either sex.
SCML1 is not needed for the elimination of DSB formation defective meiocytes. DNA was detected by DAPI; a cleaved PARP (apoptosis marker) and b NOBOX (oocyte marker) were detected by immunofluorescence on cryosections of a testes (10 weeks) and b ovaries (6–7 weeks) in indicated genotypes. a Stage IV tubules are shown. Intermediate spermatogonia (InSg), metaphase stage spermatogonia (m), apoptotic spermatocytes (ap) and Sertoli cells (st) are indicated. Scale bars, 20 μm. b Primordial (pr), primary (pr) and antral (a) follicles are indicated in ovaries of Scml1 Y/+ and Scml1 Y/− mice. Scale bars, 50 μm
Discussion
Given that the sex body, microRNAs, piRNAs and FKBP6 all play crucial roles in spermatogenesis, it was expected that the dense body would have important role in spermatogenesis and/or meiosis. We identified mouse SCML1 as a meiosis-specific protein that forms part of the meiotic dense body. We discovered that an SCML1-rich structure also forms in oocytes, indicating that a dense body-like structure is present also in oocytes. Importantly, we found that in the absence of SCML1, anti-FKBP6-stained round shape structures ceased to exist and the fraction of spermatocytes with DNA-negative round shape structures was also strongly reduced. In fact, sex body-associated DNA-negative round shape structures were not observed at all in Scml1-deficient spermatocytes. Thus, our observations indicate that dense bodies do not form at all or they can form only with strongly altered characteristics, i.e. with altered protein composition and high DNA content. The loss of anti-FKBP6 staining was particularly intriguing because FKBP6 was known to play an important role in piRNA pathways that silence transposons in the male germline (Xiol et al. 2012). FKBP6 is thought to perform an essential function in transposon silencing by supporting the establishment of repressive methyl-marks on transposons in fetal male gonocytes (Xiol et al. 2012). In contrast, it was not addressed if FKBP6 also performed essential functions in spermatocytes in adult mice as a constituent of the dense body. The piRNA binding MILI protein plays an essential role in transposon silencing both in fetal gonocytes and in meiotic spermatocytes (Aravin et al. 2007; De Fazio et al. 2011; Di Giacomo et al. 2013). Thus, a late function for FKBP6 in the dense body was a real prospect despite the fact that FKBP6 was found to be dispensable for dense body formation. Surprisingly, we found that the loss of robust dense body formation in the absence of SCML1 did not disrupt key meiotic functions and/or known meiotic miRNA or piRNA functions. Thus, absence of SCML1 and the disruption of the dense body did not alter developmental timing of meiosis entry, progression through spermatogenesis, sex body formation and silencing of transposons. Furthermore, SCML1 and the dense body were not needed for fertility in either sexes. Thus, while we cannot exclude that the dense body and SCML1 play some roles in RNA or chromosome biology during meiosis and spermatogenesis, our observations indicate that these putative functions can be only minor and non-essential for gametogenesis. It is possible that other mammalian sex comb on midleg-like proteins, such as SCML2, SCML4 or SCMH1, may have overlapping functions with SCML1 and that these proteins may compensate for the loss of SCML1. However, we find this unlikely because SCML1 shows only very limited similarity to these sex comb on midleg-like proteins, and these proteins apparently cannot rescue dense body formation in Scml1-deficient mice.
One may wonder how it is possible that a prominent structure like the dense body has so little functional importance for gametogenesis. The answer may lie within the low conservation of the dense body’s key constituent, SCML1. SCML1 proteins have been identified only in mammals (Wu and Su 2008); hence, they are likely to be an evolutionarily recent duplication of the polycomb family member sex comb on midleg-like proteins. Interestingly, SCML1 appears to have undergone fast evolution and possibly positive selection in primates (Wu and Su 2008). This raises the possibility that SCML1 has a role in speciation and/or has non-essential functions. Our data is consistent with the hypothesis that SCML1 is a structural component that forms the scaffold of the dense body in mice. It is possible that the moderately conserved SAM domain of SCML1 is responsible for dense body formation. The SAM domain is described as a protein-protein interaction domain that may promote protein oligomerization and/or RNA binding (Kim and Bowie 2003). Thus, the SAM domain might confer properties that allow SCML1 to form sphere-like structures that accumulate RNAs in the nucleus of germ line cells. Given that the SAM domain is the most conserved domain of SCML1s in mammals, this hypothesis would predict that SCML1 should also form meiotic dense bodies in other mammalian species. Based on the similarity of their appearance, the DNA-negative RNA-positive dense body of mouse and the dense bodies of other species of Muroidea rodents (hamster and rat) were identified as equivalent structures (Marcon et al. 2008). Electron-dense structures have been also identified in the vicinity of sex chromosomes of spermatocytes in varied non-Muroidea mammalian species (Schmid et al. 1987), but it is not clear if these electron-dense nuclear bodies represent structures that are equivalent to the meiotic dense body of mice. Thus, it is uncertain if dense body formation is conserved beyond Muroidea.
Interestingly, we found that the Muroidea (hamster, mouse and rat) SCML1 diverged significantly from SCML1 orthologues of non-rodent mammals or even other rodent superfamilies and suborders. For example, there is only 22.9, 19.2 and 19.5 % identity between the amino acid sequence of full-length mouse SCML1 and the sequences of the corresponding orthologues in squirrel, guinea pig and human, respectively. This level of sequence divergence likely reflects functional divergence and/or low levels of constraints on the function of SCML1 proteins. The key distinguishing feature in the sequence of Muroidea SCML1 proteins is the presence of imperfect repeats of a 12 amino acid long peptide (4, 7, 14 and 16 repeats in Syrian hamster, Chinese hamster, mouse and rat, respectively, Online Resource 2, Supplementary Fig. S2 and data not shown) that is missing from SCML1 proteins of other mammals. Interestingly, these repeats are relatively rich in glutamine/asparagine residues and according to a recently developed prion-like domain finder software, prionW (Sabate et al. 2015; Zambrano et al. 2015), they may act as prion-like domains. We speculate that this feature may reflect an ability of these repeats to promote oligomerisation of SCML1, which may ultimately facilitate the formation of SCML1-based dense bodies in species of Muroidea. Thus, our observation raises the possibility that the fast evolving SCML1 protein may have acquired the ability to form dense bodies only in the Muroidea superfamily of rodents. The relatively recent emergence of these SCML1 properties in the Muroidea group may explain why the dense body has not acquired essential functions for spermatogenesis despite its apparent interactions with essential pathways of spermatogenesis.
Materials and methods
RNA isolation and RT-PCR
To test Scml1 expression in testis and ovaries, RNA was isolated and RT-PCR was performed as described earlier (Wojtasz et al. 2009a, b). Somatic and germ cell populations of fetal ovaries were FACS sorted based on forward and side scattering (Wojtasz et al. 2009b). The RNA of the somatic tissue mix in Fig. 1a originated from 17 distinct tissues: liver, brain, thymus, heart, lung, spleen, kidney, mammary gland, pancreas, placenta, salivary gland, skeletal muscle, skin, small intestine, spinal cord, tongue and uterus. To determine the sequence of the full-length Scml1 transcript, we used rapid amplification of cDNA ends PCR (RACE-PCR). We started off with a partial cDNA clone of Scml1 (NIA15k set: H3135E08.) and the GeneRacer Kit (L1502-0 1, Invitrogen) was used according to manufacturer’s instructions for RACE-PCR. Quantitative real-time RT-PCR was carried out using GoTaq® qPCR Master Mix kit (Promega) and qTOWER 2.0 Real-Time PCR machine (Analytik Jena AG) according to manufacturer’s recommendations. Expression was tested in duplicates in testes of three independent littermate pairs of Scml1 Y/+ and Scml1 Y/− mice. Gene-specific primers used for RT-PCRs: for Xist, Sycp3, Ddx4 (Mvh) and S9 as published before (Wojtasz et al. 2009b), for Scml1 in Fig. 1, and exon 7 in Supplementary Fig. S4, 5′-TGTACCTGGCTCTTCTACGATGC-3′ forward primer and 5′-TGAGCAGAGCCTCCAAGAAGG-3′ reverse primer, for 5′ RACE of Scml1, 5′-GGCAAAGGGATCATCATCAACTAC-3′, for Scml1 exon 4–6 in Supplementary Fig. S4, 5′-TGTCTGCCACTGGTGAAGAG-3′ forward primer and 5′-AGAAGGATCTGGAACGAGCA-3′ reverse primer, for Rps9, 5′-GGCCAAATCTATTCACCATGC-3′ forward primer and 5′-TAATCCTCTTCCTCATCATCAC-3′ reverse primer, for Hprt 5′-GTACAGCCCCAAAATGGTTA-3′ forward primer and 5′-GGCTTTGTATTTGGCTTTTCC-3′ reverse primer, for Pdha1, 5′-GGGACGTCTGTTGAGAGAGC-3′ forward primer and 5′-TGTGTCCATGGTAGCGGTAA-3′ reverse primer, for Rbmy, 5′-CAAGAAGAGACCACCATCCT-3′ forward primer and 5′-CTCCCAGAAGAACTCACATT-3′ reverse primer, for LINE1, 5′-GGACCAGAAAAGAAATTCCTCCCG-3′ forward primer and 5′-CTCTTCTGGCTTTCATAGTCTCTGG-3′ reverse primer, for IAP_3LTR, 5′-GCACATGCGCAGATTATTTGTT-3′ forward primer and 5′-CCACATTCGCCGTTACAAGAT-3′ reverse primer, SINE B1, 5′-CGCCTTTAATCCCAGCACTT-3′ forward primer and 5′-GGCTGTCCTGGAACTCACTC-3′ reverse primer
Generation of knockouts and genotyping
The Scml1 targeting construct (see Supplementary Fig. S4) was designed according to a multi-purpose allele strategy (Testa et al. 2004) and generated by recombineering methods. To modify the X chromosome-linked Scml1 locus, mouse R1 embryonic stem cells, which are male cells, were cultured (using mitomycin C-inactivated mouse embryonic fibroblasts as feeders) and electroporated with the linearized targeting construct using standard protocols. Southern blotting was used to identify correctly targeted embryonic stem cell clones: DNA was digested overnight with SacI (internal probe) or StuI (5′ or 3′-probes), and DNA fragments were separated on agarose gels for Southern blotting (data not shown). Correctly targeted ES clones containing Scml1 insertion alleles were transiently transfected with CAGGS-FLPe and CAGGs-Dre vectors (sequences available on request) to remove FRT- and Rox-flanked selection cassettes from the modified 6th and 7th introns, respectively. Correctly modified ES clones carrying Scml1 restored alleles, where exon7 is floxed but there are no large selection cassettes integrated, were identified by Southern blotting; genomic DNAs were digested by XbaI, separated by electrophoresis, blotted, and 3′-probes were used to detect clones that carry Scml1 restored alleles. Chimeras were generated by laser assisted C57BL/6 morula injections with Scml1 Y/restored ES cell clones. Progeny of the chimeric animals were crossed to the outbred wild-type CD-1® mouse line and to PGK-Cre (Lallemand et al. 1998) transgenic mice to generate Scml1 Y/deletion males and Scml1 deletion/deletion females. Given the loss of SCML1 protein in these mice, we consider them null mutants of Scml1 and refer to their genotypes as Scml1 Y/− and Scml1 −/− throughout the text. Mice were maintained on the outbred ICR (CD-1®) background. Mice were initially genotyped by Southern blotting (Supplementary Fig. S4) and by PCR in subsequent crosses using tail-tip genomic DNAs. Genotyping primers:
-
o382 Mes1-loxP1 5′-CACCAATACTGTCAACAACACC-3′
-
o383 Mes1-loxP2 5′-TTCCTATCACCTTATGATCACTCTG-3′
-
o516 Mes1_Rox2_Fv 5′-ACTCATCCCCATACGAAATCC-3′
-
o517 Mes1_Rox2_Rv 5′-AAGCAAAATGCCTGACTCC-3′
-
o597 LoxP-KO_Fw 5′-GTCGAGATAACTTCGTATAGCATA-3′
-
CreFw 5′-GCCTGCATTACCGGTCGATGCAACGA-3′
-
CreRv 5′-GTGGCAGATGGCGCGGCAACACCATT-3′
PCR product sizes: with Mes1-loxP1/Mes1-loxP2, wild-type allele template-650 bp, Scml1 restored template-850 bp; with Mes1_Rox2_Fw/Mes1_Rox2_Rv, wild-type allele template-508 bp, Scml1 deletion template-no specific product; with LoxP-KO_Fw/Mes1_Rox2_Rv, wild-type allele template-no specific product, Scml1 deletion template-308 bp; CreFw/CreRv were used to detect Cre recombinase (750 bp).
Animal experiments
Mice carrying Spo11-null allele were described earlier (Baudat et al. 2000). Whenever possible, experimental animals were compared with littermate controls or with age-matched non-littermate controls from the same colony. Each conclusion in the manuscript was based on at least two experiments and at least two mice of each genotype. To overexpress Venus-tagged version of SCML1 in spermatocytes, we injected 6–8 μl of an expression vector (5 μg/μl) that carried Venus-Scml1 under the control of a CMV promoter into the rete testis of live juvenile mice (16dpp) according to published protocol (Shibuya et al. 2014; Shoji et al. 2005). One hour after injection, testes were held between tweezer type of electrodes (CUY650P5, Nepagene) and in vivo electroporation was carried with 4 times 35 V pulse for 50 ms with 950 ms intervals in-between and 4 times in the other direction with polarity switch function (Nepa21, Nepagene). Spermatocytes were collected 24 h after electroporation for the detection of Venus-SCML1 in spermatocytes. All animals were used and maintained in accordance with the German Animal Welfare legislation (‘Tierschutzgesetz’). All procedures pertaining to animal experiments were approved by the Governmental IACUC (‘Landesdirektion Sachsen’) and overseen by the animal ethics committee of the Technische Universität Dresden. The license numbers concerned by the present experiments with mice are DD24-5131/207/18 (in vivo testis electroporation) and DD24-5131/287/1 and 24-9168.24-1/2006-13 (tissue collection without prior in vivo experimentation).
Antibodies
To produce a recombinant 6xHis-tagged version of SCML1 for antibody generation, full-length mouse Scml1 open reading frame was cloned into pDEST17 bacterial expression vector. 6xHis-SCML1 was expressed in E. coli BL21 tRNA strain and was purified on Ni NTA Sepharose beads (Cat#17-5318-01, GE Healthcare) under denaturing conditions (8 M urea) followed by poly acrylamide gel purification. Homogenized gel slices containing 6xHis-SCML1 were used for immunization of two guinea pigs. Because the full-length SCML1 protein was highly insoluble in non-denaturing buffers, we cloned fragments of Scml1 open reading frame into pDEST17 vector and tested the solubility of SCML1 fragments. A 6xHis-tagged 208 amino acid-long peptide encompassing the SCML1 sequence between amino acids 294I and 501Q was soluble in non-denaturing buffers. SCML1 fragment coupled to NHS-activated Sepharose 4 fast flow beads (Cat#17-0906-01, GE Healthcare) were used to affinity purify SCML1 antibodies. Non-purified serum (anti-full-length SCML1) and affinity purified antibodies (anti-C-terminal SCML1 fragment) from both guinea pigs gave similar immunofluorescence staining patterns, and affinity purified antibodies showed similar immunoblot patterns (data not shown). Both non-purified and affinity purified antibodies were used for the detection of SCML1 by immunofluorescence (1:500), and affinity purified antibodies were used for immunoblot (1:500–1000). In addition to antibodies that were previously described (Daniel et al. 2011; Fukuda et al. 2010; Wojtasz et al. 2009a, 2012), we used rabbit anti-FKBP6 antibody (IF: 1:50), rabbit anti-phospho S2-RNA polymerase II (Abcam: ab5095, IF 1:500), mouse anti-SUMO-1 (developed by M. Matunis, was obtained from the Developmental Studies Hybridoma Bank, 1:250) and goat anti-GFP (30 μg/ml, purchased from MPI-CBG Dresden protein facility).
Cell culture
NIH 3T3 and HeLa cell lines were seeded on polylysine treated coverslips in 12-well culture dishes. Lipofectamine (Invitrogen) was used according to manufacturer’s recommendations to transfect these cells with vectors that expressed either nuclear Cherry or Cherry-tagged SCML1 under CMV promoter (sequences available upon request). Cells were cultured for 48 h after transfection, and then they were fixed in PBS pH 7.4, 3.6 % formaldehyde for the visualization of Cherry and SCML1.
Immunofluorescence microscopy
Preparation and immunostaining of testis-ovary cryosections and nuclear surface spreads of meiocytes were carried out as described before (Peters et al. 1997; Wojtasz et al. 2009a).
Immunogold labelling of ultrathin cryosections for electron microscopy
Mouse testes were fixed with 4 % paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.4) and processed for Tokuyasu cryosectioning (Slot and Geuze 2007; Tokuyasu 1980). Tissue pieces were washed in PB, infiltrated in graded series of gelatine (1, 3, 7 and 10 % gelatin in PB), cooled on ice, dissected into small cubes (0.5 × 0.5 × 0.5 mm), incubated in 2.3 M sucrose/water for 24 h at 4 °C, mounted on pins (Leica # 16701950) and plunge frozen in liquid nitrogen. Seventy-nanometre sections were cut on a Leica UC6 + FC6 cryo-ultramicrotome and picked up in methyl cellulose/sucrose (1 part 2 % methyl cellulose (MC), Sigma M-6385, 25 cP + 1 part 2.3 M sucrose). For immunogold labelling, grids were placed upside down on drops of PBS in a 37 °C incubator for 20 min, washed with 0.1 % glycine/PBS (5 × 1 min), blocked with 1%BSA/PBS (2 × 5 min) and incubated with anti-SCML1 (guinea pig antibody, 1:100) for 1 h. After washes in PBS (4 × 2 min), sections were incubated with Protein A conjugated to 10-nm gold for 1 h, washed in PBS (3 × 5 s, 4 × 2 min) and post-fixed in 1 % glutaraldehyde (5 min). Sections were washed in distilled water (6 × 1 min), stained with neutral uranyloxalate (2 % uranylacetate (UA) in 0.15 M oxalic acid, pH 7.0) for 5 min, washed in water and incubated in MC containing 0.4 % UA for 5 min. Grids were looped out, the MC/UA film was reduced to an even thin film and air dried. Finally, the sections were analysed on a Morgagni 268 (FEI) at 80 kV and images were taken with a MegaView III digital camera (Olympus).
Preparation of protein extracts
Preparation of total, TritonX-100-soluble and TritonX-100-insoluble testis extract fractions was performed as published previously (Wojtasz et al. 2012).
References
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747
Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA (2005) Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25:1041–1053
Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S (2005) Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 25:7203–7215
Baudat F, de Massy B (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosom Res 15:565–577
Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998
Beyret E, Lin H (2011) Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev Biol 355:215–226
Burgoyne PS, Mahadevaiah SK, Turner JM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10:207–216
Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12:503–514
Cloutier JM, Mahadevaiah SK, ElInati E, Nussenzweig A, Toth A, Turner JM (2015) Histone H2AFX links meiotic chromosome asynapsis to prophase I oocyte loss in mammals. PLoS Genet 11, e1005462
Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, Mo R, Hui CC, Nieves E, Cohen PE, Osborne LR, Wada T, Kunieda T, Moens PB, Penninger JM (2003) Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300:1291–1295
Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, Keeney S, Toth A (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13:599–610
De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D (2011) The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480:259–263
de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19:1376–1389
Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O’Carroll D (2013) Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50:601–608
Dresser ME, Moses MJ (1980) Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). IV Light and electron microscopy of synapsis and nucleolar development by silver staining. Chromosoma 76:1–22
Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A (2003) H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4:497–508
Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C (2010) A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res 316:158–171
Hunter N (2007) Meiotic recombination. In: Molecular genetics of recombination. Springer-Verlag, Berlin Heidelberg
Kargul GJ, Dudekula DB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MS (2001) Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet 28:17–18
Kim CA, Bowie JU (2003) SAM domains: uniform structure, diversity of function. Trends Biochem Sci 28:625–628
Kogo H, Tsutsumi M, Inagaki H, Ohye T, Kiyonari H, Kurahashi H (2012a) HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells 17:897–912
Kogo H, Tsutsumi M, Ohye T, Inagaki H, Abe T, Kurahashi H (2012b) HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes Cells 17:439–454
Lallemand Y, Luria V, Haffner-Krausz R, Lonai P (1998) Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res 7:105–112
Mahadevaiah SK, Bourc’his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS (2008) Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 182:263–276
Marcon E, Babak T, Chua G, Hughes T, Moens PB (2008) miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosom Res 16:243–260
Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE (2012) AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell 23:251–264
Monesi V (1965) Differential rate of ribonucleic acid synthesis in the autosomes and sex chromosomes during male meiosis in the mouse. Chromosoma 17:11–21
Page SL, Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785–789
Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom Res 5:66–68
Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423–440
Pillai RS, Chuma S (2012) piRNAs and their involvement in male germline development in mice. Development, growth & differentiation
Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS (2011) Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–267
Romanienko PJ, Camerini-Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6:975–987
Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20:2117–2123
Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, Bradley A, Brown EJ, Turner JM (2013) ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev 27:1484–1494
Royo H, Seitz H, ElInati E, Peters AH, Stadler MB, Turner JM (2015) Silencing of X-linked MicroRNAs by meiotic sex chromosome inactivation. PLoS Genet 11, e1005461
Sabate R, Rousseau F, Schymkowitz J, Ventura S (2015) What makes a protein sequence a prion? PLoS Comput Biol 11, e1004013
Schmid M, Johannisson R, Haaf T, Neitzel H (1987) The chromosomes of Micromys minutus (Rodentia, Murinae). II Pairing pattern of X and Y chromosomes in meiotic prophase. Cytogenet Cell Genet 45:121–131
Shibuya H, Morimoto A, Watanabe Y (2014) The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet 10, e1004821
Shoji M, Chuma S, Yoshida K, Morita T, Nakatsuji N (2005) RNA interference during spermatogenesis in mice. Dev Biol 282:524–534
Slot JW, Geuze HJ (2007) Cryosectioning and immunolabeling. Nat Protoc 2:2480–2491
Solari AJ (1974) The behavior of the XY pair in mammals. Int Rev Cytol 38:273–317
Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF (2004) A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38:151–158
Tokuyasu KT (1980) Immunochemistry on ultrathin frozen sections. Histochem J 12:381–403
Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37:41–47
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121
Vigodner M, Morris PL (2005) Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev Biol 282:480–492
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–1743
Wojtasz L, Daniel K, Toth A (2009a) Fluorescence activated cell sorting of live female germ cells and somatic cells of the mouse fetal gonad based on forward and side scattering. Cytometry A 75A:547–553
Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu HL, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5
Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Remenyi A, Turner JMA, Toth A (2012) Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26:958–973
Wu HH, Su B (2008) Adaptive evolution of SCML1 in primates, a gene involved in male reproduction. BMC Evol Biol 8:192
Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS (2012) A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47:970–979
Zambrano R, Conchillo-Sole O, Iglesias V, Illa R, Rousseau F, Schymkowitz J, Sabate R, Daura X, Ventura S (2015) PrionW: a server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res 43:W331–W337
Acknowledgments
We thank Rolf Jessberger for sharing ideas, anti-SYCP3 antibody and departmental support, Mary Ann Handel for histone H1T antibody, Nadine Maiwald for the purification of anti-SCML1 antibodies and Daniel Tränkner for lab support and help with Southern blotting. We thank the DIGS-BB program for supporting I.D. CRTD seed grant supported A.G-R. The Deutsche Forschungsgemeinschaft (DFG; grants: SPP1384:TO421/4-1 and 4–2, TO421/3-1, 3–2, 5–1, 6–1, 7–1, 8–1 and 8–2), and MeDDrive 2010 from the TU Dresden Faculty of Medicine supported F.P., K.D., A.G-R., L.W., I.D. and A.T. IMBA, the Austrian Ministry of Sciences, and the Austrian Academy of Sciences supported J.M.P. Wellcome Trust/Cancer Research UK supported A.S. The Max Planck Society supported B.H. An EFRE grant (EuropäischerFondsfürRegionaleEntwicklung) supported J.F. and A.F.S.
Author contributions
Most experiments were carried out by F.P. (Figs. 2, 3b, c, 4, 5, 6, 7, 8, S2 and S4c, e) and K.D. (Figs. 1, 3a, e, S1 and S4d). K.D. identified and cloned the full-length Scml1 open reading frame, generated anti-SCML1 antibodies, devised experiments together with A.T. and contributed to day-to-day supervision of F.P. and A.R. during the initial stages of characterization of Scml1-deficient mice. The Scml1-deficient ES cells and mice were generated by A.G-R. (Fig. S4a, b) with the assistance and supervision of J-F.F. Further experiments were performed by T.K. (Fig. 3d), L.W. (Supplementary Fig. S3b) and I.D. (Supplementary Fig. S3a). Knockout construct was generated by J.F. and A.F.S.; J.P. supplied FKBP6 antibodies and experimental material; A.S. and B.H. provided support for the discovery of Scml1; A.T. wrote the manuscript. All authors were involved in discussions and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All experiments performed in this study involving animals were in accordance with the ethical standards of the Technische Universität Dresden. This article does not contain any studies with human participants performed by any of the authors.
Funding
This work was supported by DFG grants: SPP1384:TO421/4-1 and 4-2, TO421/3-1, 3-2, 5-1, 6-1, 7-1, 8–1 and 8-2, MeDDrive 2010 from the TU Dresden Faculty of Medicine, DIGS-BB program, IMBA, the Austrian Ministry of Sciences, and the Austrian Academy of Sciences, Wellcome Trust/Cancer Research UK, The Max Planck Society and an EFRE grant (EuropäischerFondsfürRegionaleEntwicklung).
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Frantzeskos Papanikos, Katrin Daniel and Angelique Goercharn-Ramlal contributed equally to this work.
Rights and permissions
About this article
Cite this article
Papanikos, F., Daniel, K., Goercharn-Ramlal, A. et al. The enigmatic meiotic dense body and its newly discovered component, SCML1, are dispensable for fertility and gametogenesis in mice. Chromosoma 126, 399–415 (2017). https://doi.org/10.1007/s00412-016-0598-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-016-0598-1