Abstract
Chromatin, once thought to serve only as a means to package DNA, is now recognized as a major regulator of gene activity. As a result of the wide range of methods used to describe the numerous levels of chromatin organization, the terminology that has emerged to describe these organizational states is often imprecise and sometimes misleading. In this review, we discuss our current understanding of chromatin architecture and propose terms to describe the various biochemical and structural states of chromatin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction—the language problem
The genetic information of eukaryotes is stored in the cell nucleus in the form of a nucleoprotein complex of DNA and histones, known as chromatin. Histones have been thought to have two functions; first, by virtue of being basic proteins, they serve to balance the negative charge of the DNA’s phosphate backbone. Second, through changing the trajectory of the DNA backbone, histones could contribute to the required effective shortening of the DNA in order to fit the genome into the small volume of the cell nucleus. Hence, from the earliest descriptions of chromatin by Flemming and Kossel (Flemming 1882; Kossel 1884) through to the modern period where chromatin is recognized to be composed of a repeating nucleosome subunit (Kornberg 1974; Olins and Olins 1974; Woodcock et al. 1976), chromatin was thought to play solely a structural role. This simple view of chromatin was dramatically changed in the late 1990s, particularly by the 1996 papers from the Allis lab, where it was shown that a well-studied transcription regulatory factor had a histone acetyltransferase activity (Brownell et al. 1996; Kuo et al. 1996). This finding showed that chromatin could no longer be considered only as a structural entity but as a fundamental participant in gene regulation. Though Dr. Vince Allfrey had proposed the potential of histone post-translational modifications (PTMs) for regulation many years before (Vidali et al. 1968), it was not until the late 1990s when it became widely accepted that chromatin offered a huge potential, particularly through fine structural changes at the nucleosome level, for modulating access to the DNA of large supramolecular complexes, such as transcription or replication machinery. In more recent years, with the recognition of the importance of sub-nuclear domains and non-random spatial relationships of specific gene loci in gene regulation, the scope of the importance of chromatin has expanded greatly. It is now recognized as both a structural entity as well as a fundamental regulatory complex.
Currently, numerous approaches are being employed to elucidate different aspects of chromatin structure and organization in the nucleus and to relate them to function. These studies contribute greatly to the understanding of chromatin organization, but as a result of the range of methodologies used to acquire and interpret the data and the variety of organizational levels of chromatin, there is frequent confusion in the terminology used to describe the observed chromatin characteristics. The terms used are often polysemes, causing them to be misinterpreted. Thus, we feel that the language used in the chromatin field is in need of clarification and perhaps requires establishing new definitions to describe the various structural and organizational properties of chromatin. Precise terminology will improve the communication between researchers who use vastly different experimental methodologies and will consequently lead to new insights into the structure and function of chromatin.
In this review, we will discuss some of the most widely used terms in the literature that describe experimental findings of chromatin’s structural and organizational properties. Since many of these terms can have a variety of meanings, we will provide our suggestions for revising this terminology in order to better appreciate the complexity of the organizational and functional properties of chromatin.
Our intention is solely to raise awareness of the importance of using clearly defined terminology to avoid confusion around the principles and concepts in the chromatin field. We therefore encourage that for each study precise definitions of the terminology used to describe the results will also be provided. We offer suggestions for definitions of commonly used terms that relate to chromatin structure and organization, though a transition to a unified lexicon will not occur overnight; but our goal is to bring awareness to the topic. It is important to note that by no means do our examples and suggestions detract from the significance of studies that have used certain terms to describe the findings in these studies. We believe that the need for clarification arose due to the immense expansion in chromatin research and the methodologies employed over the past several years.
Terms with multiple meanings—what do we mean?
Packing DNA into the nucleus
One of the first problems with our language about chromatin arises with the common answer to the question, “what is the role of chromatin?” The typical answer is, “to pack DNA into the small nuclear volume”. In some ways, this answer is true, but it is also very misleading. Indeed, the effective length of approximately 2 m of DNA must be massively shortened down to the scale of the nucleus, typically in the range of 10 μm in diameter. This represents a compaction ratio of 105. On the other hand, ‘to compact’ is not the most appropriate verb because it implies that ‘fitting’ the DNA genome into the nuclear volume is a difficult task in terms of geometry. In fact, fitting DNA into the nuclear volume is trivial. The volume of the DNA itself, if treated as a simple cylinder, is only about 6.4 μm3 (see Fig. 1). The volume of a typical cell nucleus, however, is much greater, approximately 500 μm3. Since the volume of the DNA is only 1.2 % that of the nuclear volume, compaction, i.e., getting the DNA ‘to fit’, is not the primary function of chromatin. Instead, chromatin’s histones must balance charge of the DNA’s phosphate backbone and reduce its effective length. Hence, the term ‘compaction’ is problematic.
In fact, wrapping DNA as chromatin actually increases the volume assigned to each base pair of DNA. The effective volume given over to 146 bp of DNA is hugely increased simply by wrapping it around a histone octamer. The volume of the DNA molecule of a single nucleosome is approximately 156 nm3, whereas the volume of the nucleosome core particle is approximately 570 nm3, nearly fourfold greater (see Fig. 1). Again, increasing the effective volume of an item through ‘packaging’ is typically the opposite of what is implied by the word ‘compaction’.
Higher order chromatin structure and organization
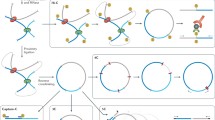
Chromatin does make a substantial contribution to the effective shortening of each DNA polymer that constitutes a chromosome. The nucleosome plays a modest role in this task through its ability to supercoil 1.8 turns of DNA. The length of 146 base pairs is shortened to either an 11-nm en face profile or a 6-nm side view profile dimension of the core particle. This achieves one order of magnitude compaction out of the five orders that are required. Further shortening has been proposed to be accomplished by the next higher order fibre structure, the so-called 30-nm fibre. Yet, to date, 30-nm fibres have been undisputedly observed only in very specialized cells, such as starfish sperm (Fussner et al. 2012; Woodcock 1994), or after nuclear swelling and disruption, and also under cell-free conditions (reviewed in Bian and Belmont 2012; Fussner et al. 2011a; Maeshima et al. 2010). Little supporting evidence for the existence of a 30 nm fibre in intact cell nuclei has materialized. Hence, the mechanism of DNA compaction by hierarchical levels of fibre structures must be re-examined. Indeed, what is meant by ‘higher order chromatin structure’ must be re-defined.
Recently, chromatin conformation capture techniques have identified topologically associated domains (TADs). A TAD is defined as a region of a chromosome that shares many interactions within it, but significantly fewer interactions with the adjacent and other more distal TADs (Dixon et al. 2012; Nora et al. 2012). These interactions are thought to be a result of folding and bending of the chromatin fibre (Dekker et al. 2013). Furthermore, TADs are fairly conserved throughout animal evolution and in different cells of an organism (Dixon et al. 2012). TADs, their boundaries and the interactions between them have been shown to correlate with replication domains and the replication timing programme (Pope et al. 2014; Dileep et al. 2015), thus providing a link between these structures and cellular function. Given that existing data do not support the existence of 30-nm fibres in interphase nuclei, together with the current notion that TADs form basic conformational modules of chromatin fibres, it is plausible that looping, bending and folding of chromatin fibres and formation of these ‘globule-like’ structures, separated by distinct insulators, are responsible for the higher order conformation needed for effective compaction of chromatin. While TADs are at the range of 1 Mb in size, higher resolution sequencing of Hi-C data recently provided data used to support the existence of smaller conserved domains in the genome (Rao Suhas et al. 2014). We therefore believe that these globular domains, their exact size yet to be determined, should be considered as higher order chromatin structures. These structures and their boundaries may be crucial for a higher order level of functional chromatin organization (as discussed and reviewed in Ciabrelli and Cavalli 2015; Nora et al. 2013; Sanyal et al. 2011).
In addition to its utilization for describing structured chromatin fibre folding beyond the 10-nm fibre, the term ‘higher order chromatin structure’ is frequently used to distinguish euchromatin from heterochromatin, where the latter would be characterized by ‘higher order’ levels of fibre folding. The distinction of these types of chromatin was first proposed by Emil Heitz in 1928, when a series of cytogenetical observations led him to conclude that there are two types of chromatin: ‘heterochromatin’, which remains condensed throughout the cell cycle, and ‘euchromatin’, which undergoes cycles of condensation and de-condensation throughout the cell cycle (Heitz 1928). Since then, our definition of these terms has expanded and heterochromatin has been further divided into constitutive and facultative heterochromatin. Constitutive heterochromatin is identical in all cells from a given species, while facultative heterochromatin is cell type and differentiation stage dependent. Recently, data from genome-wide association studies led to the idea that chromatin may be further categorized by a characteristic composition of non-histone chromosomal proteins, specific histone PTMs and specific sites of DNA methylation (Ernst and Kellis 2010; Filion et al. 2010; Kharchenko et al. 2011). The exact number of chromatin types described in the separate studies varies due to the algorithm used and other parameters. However, at least four main types can be described: one active and three repressive types (Ciabrelli and Cavalli 2015). Further, whereas heterochromatin was thought to be transcriptionally inert and euchromatin transcriptionally active, we now know that this distinction is not absolute—some transcription can occur in the heterochromatin (reviewed in Enukashvily and Ponomartsev 2013; Hall et al. 2012; Saksouk et al. 2015). Together, these new findings emphasize the fact that there is no simple distinction between two types of chromatin—heterochromatin and euchromatin—and therefore no clear correlation exists between a particular higher order chromatin structure and one of these chromatin types.
Another term commonly used interchangeably with higher order structure of chromatin is higher order organization of chromatin. An example of a higher order chromatin fibre organization is intrachromosomal loops that have been proposed by Trask et al. (Sachs et al. 1995; Yokota et al. 1995). These loops may be of various sizes and types, such as insulator-mediated interactions, polycomb-mediated long-range repressive interactions and the most well-known enhancer-promoter interactions (for a review, see Hou and Corces (2012)). A well-characterized example of this last type of interaction is the locus control region (LCR) of the β-globin locus, which makes physical contact with specific globin gene promoters located approximately 50 kb away (Carter et al. 2002; Tolhuis et al. 2002). These types of loops may be enclosed within a TAD, but may also occur between TADs. Unlike TADs, these loops may be cell type specific and have functional consequences for gene expression, and thus, we consider them to be a higher order organization of the chromatin fibre.
The next level of higher order organization relates to the sub-nuclear organization of chromatin into what are called active and inactive domains (Bickmore and van Steensel 2013; Dekker 2014; Sanyal et al. 2011). Both FISH (Shopland et al. 2006) and chromosome- conformation capture (3C)-based methods (Lieberman-Aiden et al. 2009; Sexton et al. 2012; Simonis et al. 2006; Yaffe and Tanay 2011) have confirmed the tendency of active gene-dense domains to colocalize and interact, both within and between chromosomes. Furthermore, 4C and Hi-C have also shown preferential interactions between inactive regions, mainly in cis (Libbrecht et al. 2015; Lieberman-Aiden et al. 2009; Sexton et al. 2012; Simonis et al. 2006). Yet to be confirmed, this compartmentalization into active and inactive chromatin domains may reflect a required spatial proximity of particular loci with specific nuclear bodies/factors/domains. Accordingly, higher order chromatin organization is used to describe the features of chromatin that are reflected by this sub-nuclear compartmentalization.
Two additional concepts in the chromatin field have also been referred to as higher order chromatin organization. The first of these is based on the observation that each chromosome occupies a discrete volume, forming a ‘chromosome territory’ (Bolzer et al. 2005). That the DNA fibre of a single chromosome occupies a discrete volume, much less than that of the nuclear volume, is not surprising because a property of a polymer is that its two ends tend to remain in relative close proximity as a result of twisting and tangling of the fibre (Gennes 1979). But what might be surprising, and reveals the importance of the three-dimensional (3D) organization of the genome, is that chromosome territories are non-randomly positioned. Radially distributed chromosomes, where gene-rich chromosomes are positioned internally, and gene-poor chromosomes toward the nuclear periphery have been observed (Boyle et al. 2001; Cremer et al. 2001; Croft et al. 1999). Also, particular chromosomes are positioned relative to other chromosomes, at least in particular cell types. This is reflected in the relatively common translocations between particular loci on separate chromosomes that give rise to specific cancers (Roukos and Misteli 2014). This level of higher order organization has been substantiated by 3C-based methods by confirming the existence of chromosome territories and the preferential association between certain sets of chromosomes (reviewed in Dekker 2014).
The second concept of higher order that does not pertain to the fibre structure itself is based on the discovery that the nucleus contains spatially distinct sub-compartments, each with distinct functions. As a result, specific gene loci and types of chromatin, heterochromatin vs. euchromatin, for example, will be specifically positioned in relation to these sub-compartments (Bickmore 2013; Cope et al. 2010). One of these sub-compartments is the nuclear lamina; the lamina-associated domains (LADs) are large genomic regions that are generally transcriptionally repressed and are located along the nuclear envelope (Guelen et al. 2008). Other nuclear structures that associate with specific loci include the nucleolus (Nemeth and Langst 2011; Olson and Dundr 2001), transcription factories (Osborne et al. 2004) and a variety of nuclear bodies, such as promyelocytic leukaemia (PML) nuclear bodies (Ching et al. 2013), Cajal bodies (Smith and Lawrence 2000) and histone locus bodies (Liu et al. 2006; Nizami et al. 2010). This spatial proximity between specific loci and nuclear factors or sub-compartments may be a consequence of either active movement directed by nuclear skeletal elements (Bridger 2011; Bridger et al. 2014; Chuang et al. 2006; Dundr et al. 2007) or may reflect a passive mass action and self-assembly of functional domains that stabilize once formed (reviewed in Chuang and Belmont 2007).
Thus, we propose that higher order chromatin structure should be used to describe the formation of conserved chromatin globules, folds or domains and their boundaries, while higher order chromatin organization will be used to describe specific looping of the chromatin fibre and the 3D organization of the genome in relation to interactions with sub-nuclear structures and trans interactions between chromatin fibres.
Condensation and compaction
The term ‘condensation’ has been primarily used in the context of transitions between the 10 nm, the 30 nm, and higher order chromatin fibres. Condensation from an ‘open’ 10-nm to a ‘closed’ 30-nm fibre has been seen as a critical step in gene silencing. Similarly, the term condensation of higher order fibres into the metaphase chromosome during prophase and early metaphase has been used extensively. However, since the in situ evidence for 30 nm and higher order fibres is lacking (Fussner et al. 2011a; Maeshima et al. 2010), the term condensation must take on a new meaning. Instead of a transition between lower and higher order fibre types, it can be understood as changes in other properties of the 10-nm fibre and/or a contraction in the distances between neighbouring fibres (Fussner et al. 2011a).
‘Compaction’ represents another term that has an ambiguous meaning and has frequently been synonymous with condensation. Chromatin compaction can be viewed at two separate levels. Linear compaction of a chromatin fibre can occur as a result of changes in nucleosome density, while spatial compaction of a chromatin domain is due to folding and looping of a fibre and changes in inter-fibre distances. As pointed out earlier, the primary role of chromatin is not ‘compaction’ of DNA in the sense of fitting it into the nucleus. Instead, chromatin as a dynamic entity can function to change the local concentration of DNA, by bringing particular gene loci into closer proximity, thus leading to a more compact state. Indeed, differences in the state of compaction of the genome can be readily seen when comparing a mouse embryonic stem cell (ESC) with a differentiated cell. ESCs, derived from the inner cell mass of the pre-implantation blastocyst of day 3.5, are pluripotent in that all cells of the embryo are derived from them. The chromatin distribution in an ESC is best described as a uniformly dispersed arrangement of 10-nm chromatin fibres, with little evidence of locally compact domains within the nucleoplasm and even along the nuclear envelope (Ahmed et al. 2010). In contrast, differentiated cells have varying degrees of compact chromatin along the nuclear envelope and domains of varying degrees of compaction throughout the nuclear volume. The degree of compaction into local domains does not necessarily reflect changes in nuclear volume. Instead, regions of the genome become more concentrated spatially in local volumes, perhaps reflecting a level of regulation brought about through changes in chromatin architecture.
We propose that the terms compaction and condensation should be discriminated. Linear compaction reflects fine molecular changes in nucleosome and histone1 (H1) occupancy of a single chromatin fibre. Spatial compaction reflects differences in chromatin concentration throughout the interphase nucleus and could include one or more chromatin fibres. Cell type-specific configurations of chromatin compaction reflect chromatin’s role in regulating the gene expression profile of cellular activation and/or differentiation. Condensation, on the other hand, could refer to non-interphase physiological processes in which there is a global increase in the density of chromosomes, such as in the formation of mitotic chromosomes beginning in early prophase. These examples of global condensation are associated with levels and/or modifications of linker histones and the recruitment and/or activation of non-histone chromosomal proteins.
Open and closed chromatin
The terms ‘open’ and ‘closed’ chromatin have been used in a variety of ways and with many interpretations. Generally, ‘open’ chromatin refers to actively transcribed chromatin, while ‘closed’ refers to silent, transcriptionally repressed chromatin. As already noted, the 10-nm nucleosome fibre has been described as chromatin in an ‘open’ state, readily accessible to functional complexes such as transcription or replication machinery. In contrast, 30-nm or even higher order fibres have historically been described as being in a ‘closed’ state, inaccessible to large molecular complexes and machines.
Open and closed chromatins have also been used for referring to cytologically recognizable euchromatin and heterochromatin, respectively. Open chromatin is that which is de-compacted or dispersed and is transcriptionally active or poised for activity. It has thus gained access to the transcription apparatus. Closed chromatin, on the other hand, has been associated with heterochromatin, whether constitutive or facultative, and has been described as compact and transcriptionally repressed or non-permissive to transcription.
Furthermore, these ‘open’ and ‘closed’ states are often distinguished by sensitivity to nuclease digestion (Boyle et al. 2008) as well as by histone PTMs and CpG methylation that characterize euchromatin or heterochromatin, respectively (Kouzarides 2007). Interpretation of Hi-C data implies that the genome is partitioned into two spatial domains that correlate with ‘open’ and ‘closed’ chromatin, when characterizing them by these attributes (Lieberman-Aiden et al. 2009). Nevertheless, such models represent an over-simplification. Examples have been observed where a correlation between biochemical marks for ‘closed’ heterochromatin, H3K9me3 for example (Lachner and Jenuwein 2002), and a compact chromatin state break down. In mouse ESCs, for example, discrete domains of chromatin containing the H3K9me3 mark can be clearly observed by immuno-fluorescence microscopy (Meshorer et al. 2006) (Fig. 2). These clusters of constitutive heterochromatin in mouse cells, called chromocentres, contain major satellite repeats and other silenced genomic DNA (Probst and Almouzni 2011) and are considered closed chromatin domains. However, these domains in ESCs, when viewed by electron spectroscopic imaging (ESI), are not radially symmetric, compact chromatin structures, as found in differentiated cell types, such as mouse embryonic fibroblasts (MEFs) (Fussner et al. 2011b) (Fig. 2). Although all chromocentres are marked with the typical constitutive heterochromatin mark (H3K9me3), a tightly compacted ‘closed’ state is not directly correlated to this mark. In this context, these heterochromatin domains require other yet-unidentified factors to convert the domain into the typical compact, closed heterochromatin structure. Just the presence of this heterochromatin mark is not sufficient to confer a compact chromatin state that resembles typical heterochromatin from a cytological viewpoint. Some cultured human fibroblasts, such as Wi38 cells, also display H3K9Me3-rich foci, which have an intermediate level of DNA compaction, between the levels observed in mouse ESCs and MEFs (Fig. 2). This emphasizes that H3K9me3 chromatin domains, typically referred to as ‘closed’, have varied compaction states depending on the organism as well as the stage of differentiation. Closed chromatin is a biochemical state that reflects transcriptionally silenced chromatin but does not necessarily correlate with a compact organization.
Diverse chromatin ultrastructure underlying H3K9me3 domains. Top panels show overlays of low-magnification mass micrographs with correlating fluorescence images for H3K9me3 of the same physical section, for a mouse ESC, MEF and a human fibroblast (Wi38). White boxes in each of the top panels depict the approximate area of the ESI micrographs presented at a higher magnification in the bottom panels. In the ESI micrographs, chromatin is represented by intensities of yellow and protein-based structures are represented in cyan levels. Regions indicated by dashed lines represent approximate H3K9me3 domain boundaries determined by correlative fluorescence images. Scale bar, 0.5 μm. Bottom panels show line scans of phosphorus intensities of indicated H3K9me3 domains
While a correlation is not guaranteed between the presence of a silencing heterochromatin mark (H3K9Me3) and a structurally compact state, a correlation between transcription, the hallmark of open chromatin, and a dispersed chromatin state is also not guaranteed. In naïve lymphocyte nuclei, for example, ESI data reveal that almost all of the chromatin is in a highly compact state, reminiscent of heterochromatin in other cell types, and is compacted to at least the degree of metaphase chromosomes (Fig. 3, top left panel). Yet we know that many genes in these cells must be in a transcriptionally open state and have access to the transcriptional apparatus (Kouzine et al. 2013). Yet, blocks of compact heterochromatin separated by regions of euchromatin, as seen in most cell types, including MEFs, are not observed in naïve lymphocytes (see Fig. 3 top panels). Open, actively transcribing chromatin, however, is likely positioned on the immediate periphery of the bulk, densely packed chromatin (see arrows in the bottom left panel in Fig. 3) because this is where the RNA-protein complexes (RNPs) are observed. This is in agreement with previous findings from the Bernhard group in the 1970’s, which show using 3H-uridine labelling and transmission electron microscopy (TEM) that nuclear RNA is localized at the periphery of condensed chromatin, on so-called perichromatin fibrils (Fakan and Bernhard 1971; Fakan and Bernhard 1973). These RNP complexes are not observed in the interior of the compact chromatin domains throughout the nuclear volume. The RNPs on the periphery of the compact chromatin likely correspond to the so-called perichromatin granules described previously (Fakan 2004) (see arrowheads in the bottom left panel in Fig. 3). Hence, in lymphocytes, all chromatin, even the open euchromatin, has the appearance of closed heterochromatin. In contrast, MEFs have spatially discrete compact heterochromatin domains, including chromocentres and compact chromatin domains along the nuclear envelope, with the remaining chromatin being organized as dispersed 10-nm fibres (arrows in the right bottom panel of Fig. 3). All of this dispersed 10-nm chromatin, however, is not likely to be transcriptionally active because of the relatively large fraction of the genome that it would represent. In summary, we propose that the term ‘open chromatin’ should describe a biochemical state that is transcriptionally poised or active, but the term does not necessarily imply an organization of highly dispersed chromatin fibres.
Ultrastructure of ‘open’ chromatin. Top panels are ESI images of a mouse lymphocyte (left) and a MEF (right). Chromatin and RNPs are represented by intensities of yellow and protein-based structures are represented in cyan levels. White boxes depict the approximate area shown in a higher magnification and enlarged in the bottom panels. Arrows point to distinct chromatin fibres and arrowheads point to RNPs. The phosphorus density of RNPs is typically higher than that of chromatin and thereby appears as intense bright granules. Their concentration is significantly higher in lymphocyte compared to the MEF nuclei
Accessibility
The concept of genome accessibility represents another example of ambiguity in the language about chromatin. ‘Open’ active euchromatin is generally considered more accessible than ‘closed’ silent heterochromatin. However, there are many levels at which access must be considered. First, chromatin typically has a repressive effect on gene expression, in part through hindering access of macromolecular components. A nucleosome, for example, can block the interaction of a transcription factor with its cis-element binding site in the DNA (Hayes and Wolffe 1992). Also, nucleosomes can inhibit the initiation and progression of RNA polymerase along the DNA template, thereby slowing the rate of elongation of the nascent RNA (Izban and Luse 1991; Morse 1989). This ability to limit access can be relieved by nucleosome remodelling, which can slide the nucleosome upstream or downstream of the blocked transcription factor binding site, or can cause the DNA helix to rotate relative to the nucleosome, thereby exposing the factor binding site, or can displace the entire histone octamer from the DNA (for a comprehensive review on chromatin remodelling, see Clapier and Cairns (2009)). These structural changes that confer accessibility all occur at the level of the molecular dimensions of the nucleosome itself.
In addition to modulating access of a regulatory factor or of a macromolecular machine such as the transcription apparatus, accessibility can be considered at another level of spatial organization. Greater access to specific genetic loci could be achieved by modulating the degree of folding of the 10-nm fibre itself, changing the spacing between adjacent fibres and localizing to a specific nuclear environment. In many cell types, however, the spacing between the vast majority of chromatin fibres is sufficient so that access of macromolecules is not a limiting feature. Such cell types include many cancer cell lines as well as MEFs and ESCs (see Fig. 2). In these cells, spatial accessibility per se does not appear to regulate functional interactions, but rather other factors, such as chromatin shape (Abe et al. 2015), nucleosome occupancy or cofactor binding (Slattery et al. 2014). Nevertheless, there are cell types where nearly the entire fraction of the genome is organized into very compact domains, the lymphocyte being the best example, as described earlier (Fig. 3). In such nuclei, no evidence of on-going transcription in the interior of these compact domains is visible by ESI. Hence, loci that are transcribed are likely located in the periphery of these domains and in the channels or inter-chromatin space that is found between compact chromatin domains. From this location, a transcribed locus likely has significantly greater access to transcription factories or other nuclear bodies that regulate expression.
It is important, however, to distinguish between a gene gaining access by locating at the periphery of a chromatin domain or territory and looping out from a territory. In contrast to lymphocytes, MEFs, ESCs and many other cell types do not display large blocks of compact chromatin with inter-chromatin spaces between them. Hence, a locus that is to be transcribed has equal access to transcriptional machinery, whether the locus is in the interior or on the periphery of its chromosome territory. There is no role for modulating such ‘accessibility’ according to this meaning in such nuclei. Instead, an observed movement of a gene locus to the periphery of a chromosome territory or even extending far from the territory (e.g. Hox locus in induced ESCs (Chambeyron and Bickmore 2004)) serves to contact a specific nuclear structure or assembly of regulatory factors, such as a transcription factory, that are located there and is not to gain access from a compact domain.
Origins and evolution of the language about chromatin
Our understanding of chromatin organization, both under cell-free conditions and in intact nuclei, has been generated by a vast array of biochemical, physical and imaging techniques. As a result, descriptions and terms that arise and seem appropriate with one method may not have the same meaning when used in the context of a different technique. One current example of this problem concerns the spatial relationships of gene loci that are described by fluorescence in situ hybridization (FISH) vs. those that are described by chromatin conformation capture techniques. In a recent study, the authors present conditions in which a ‘dispersed’ chromatin domain visualized by FISH and measured by FISH probe distances does not correlate with the ‘compact’ view of this chromatin domain as measured by interaction frequencies in 5C (Williamson et al. 2014). In this section, we briefly describe how a variety of methods have provided insights into chromatin structure and have also led to confusion over the meaning of terms, including those discussed above.
Fluorescence microscopy—DNA stains
One of the most straightforward ways to visualize chromatin in the nucleus utilizes a fluorescent molecule that binds DNA, such as DAPI or the various Hoechst dyes (Kapuscinski 1995; Latt and Stetten 1976). Because DAPI does not bind all DNA equally, having a preference for A-T-rich regions, the staining intensity does not always reflect the concentration of DNA. Hence, the observation of DAPI-rich foci in mouse ESCs and reprogrammed mouse iPSCs may lead to the false conclusion that these structures are compact chromatin domains, as seen in MEFs, for example. However, the DAPI-rich foci in ESCs and iPSCs are typically not highly compact structures, often hardly noticeable above the chromatin density in the nucleoplasmic background (Fussner et al. 2011b).
Transgene arrays
The introduction of multi-copy transgene repeat arrays (e.g. lac, MMTV) into the genome and visualization with a GFP fusion that binds the array (e.g. lac repressor) has been used in numerous model systems to observe specific genomic loci and chromatin dynamics (Gunawardena and Rykowski 2000; Kato and Lam 2001; Matzke et al. 2005; Müller et al. 2001; Robinett et al. 1996). One application of such arrays has been to visualize changes in the degree of ‘compaction’ or ‘condensation’ associated with transcriptional activation (Dietzel et al. 2004; Tumbar et al. 1999) or binding of heterochromatin protein 1 (HP1) (Verschure et al. 2005). The changes in compaction/condensation are generally measured by changes in volume of the transgene array. The repetitive nature of the arrays, however, can lead to cis interactions within the array, trans interactions between multi-copy insertions of arrays, and associations with heterochromatin, thereby influencing the global chromatin organization (Pecinka et al. 2005). Hence, while these experiments can provide insightful data regarding changes in localization of chromatin domains in response to different stimuli, the interpretation of the structural organization of chromatin from such assays must be approached with care. Furthermore, configuration of the array as a particular fibre type, such as the 30-nm fibre, cannot be extrapolated from measured lengths of the arrays (Hu et al. 2009). Whereas distance measurements might be consistent with the contour length of a 30-nm configuration, they could also be consistent with a highly folded 10-nm chromatin fibre or where extensive looping of the 10 nm fibre occurs. The Belmont group has also provided structural information of lac arrays using electron microscopy (EM) and in vivo gold labelling (Kireev et al. 2008). The terms commonly used in the interpretation of data from these assays are ‘compaction’ or ‘condensation’/‘de-condensation’. These terms are used interchangeably, as well as other terms, such as ‘unfolding’ and ‘remodelling’. We suggest that the terms to describe the phenotype observed in these experiments are compaction and dispersal of the chromatin in the arrays. However, only imaging at the ultrastructure level of resolution can reveal the exact compaction or dispersal state that is attained due to the different perturbations.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) is a technique in which hybridization of a fluorescent probe to genomic sequences enables the sub-nuclear localization of a genomic locus as well as measurement of the spatial distance between loci. This assay has been commonly used to analyse ‘compaction’ or ‘condensation’ of chromatin by measuring the distance between specific FISH probes. Probes that are separated by 2 Mb or less show a linear relationship between the mean-square inter-probe distance in interphase (d2) and their genomic separation in kilobases (van den Engh et al. 1992). This method has been used to measure changes in chromatin during development and differentiation (Chambeyron and Bickmore 2004; Chambeyron et al. 2005; Morey et al. 2007) and to determine the role of specific epigenetic pathways in chromatin ‘compaction’ (Eskeland et al. 2010a; Eskeland et al. 2010b; Taylor et al. 2013). While this method can provide information on the spatial distance between loci, the changes in the underlying ultrastructure or folding of the chromatin fibre that results in the distance changes cannot be inferred. Moreover, the terms ‘compaction’ and ‘condensation’ have been used interchangeably when describing the same phenotype observed by FISH, such as the shortening of distances between probes in interphase nuclei. We propose that this observation should be termed compaction rather than condensation.
Another more global application of FISH to view chromatin organization has been the use of chromosome paints, which confirmed the concept of chromosome territories (for a review, see Cremer and Cremer 2010). FISH, in this context, has contributed to the use of the term higher order organization of chromatin. In this case, ‘higher order’ organization reflects the fact that the chromosomes in the interphase nucleus are organized into discrete domains, which are non-randomly distributed in the nuclear space. This level of higher order organization of chromatin in the interphase nucleus has been largely confirmed by Hi-C experiments (Lieberman-Aiden et al. 2009; Kalhor et al. 2012; Sexton et al. 2012; Zhang et al. 2012). It must be emphasized, however, that this method does not have the required resolution to define the higher order chromatin structure within the territories. It has, however, contributed greatly to the understanding of higher order organization of chromatin in the nuclear space.
Fluorescence lifetime imaging microscopy-Förster resonance energy transfer
The combination of fluorescence lifetime imaging microscopy (FLIM) and Förster resonance energy transfer (FRET) overcomes some of the problems encountered with regular FRET, providing better quantitative measurements independent of fluorophore concentrations. Lleres et al. present a FLIM-FRET-based method to measure chromatin ‘compaction’ in a cell line stably expressing H2B fused to mCherry and H2B fused to GFP. Increased FRET is observed when fluorescent histones from two different nucleosomes are closer to each other due to compaction of chromatin (Lleres et al. 2009; Visvanathan et al. 2013). Whereas the technique is certainly sensitive to nucleosome density, it cannot distinguish between nucleosomes that are on the same fibre (cis) or are on separate fibres (trans). Hence, the method cannot be used to imply changes in chromatin fibre structure, such as inter-conversions between fibre types. While FISH measures ‘compaction’ by measuring the distance between two specific loci, FLIM-FRET provides a more global view on ‘compaction’ of chromatin in the nucleus. However, just as in the case of FISH, this method cannot be used to infer the ultrastructural changes in the chromatin that lead to the difference in FRET measurements.
Electron microscopy (cTEM, ESI, cryoEM)
Due to its high resolution, electron microscopy has played a pivotal role in the study of chromatin structure throughout the years. EM can be used to visualize both the global organization of chromatin in intact nuclei and the conformation of isolated chromatin fibres. Most studies of chromatin carried out by EM utilize either conventional transmission electron microscopy (cTEM) or electron spectroscopic imaging (ESI). Due to the low electron scattering property of biological samples, cTEM requires the use of heavy atom contrast agents such as uranium and lead salts, which is accompanied by two major disadvantages. First, since the heavy atom salts bind extensively, essentially coating the biological structures, spatial resolution is compromised. In addition, because the salts do not bind all biomolecular structures with equal affinity, the apparent density of certain structures relative to others may be exaggerated. This potential artefact pertains to imaging of chromatin in intact nuclei. The heterochromatin along the nuclear envelope or around the nucleolus often appears to be very dense, relative to other sub-nuclear components when imaged by cTEM. This does not reflect the true density of the heterochromatin or its biological density relative to that of sub-nucleolar compartments or nuclear bodies, for example. Furthermore, domains of compact chromatin fibres may appear indistinguishable from the nucleoplasm due to unpredictable stain affinities (reviewed in Rapkin et al. 2012).
Though cTEM correctly demonstrated compacted chromatin domains, such as those along the nuclear envelope, it has led to the concept of heterochromatin being extremely dense and therefore likely inaccessible to complexes such as the transcription apparatus. This is not necessarily true; hence, the chromatin that contrasts well with heavy atom salts should be described as compact, but not necessarily as inaccessible to macromolecular complexes.
To overcome some of the limitations of cTEM in chromatin structure studies, a specialized form of TEM called electron spectroscopic imaging (ESI) has been developed. This method exploits the element-specific energy loss of incident electrons that interact with the sample. With an imaging spectrometer, element-specific distributions or maps with high spatial resolution and sensitivity can be obtained (for an overview of ESI, see Bazett-Jones and Hendzel 1999; Dellaire et al. 2004). For studying chromatin structure in biological samples, phosphorus (P) and nitrogen (N) maps conveniently allow one to distinguish between chromatin-, RNA- and protein-based structures in situ.
Correlating fluorescence microscopy approaches with ESI permits the structural elucidation of specific sub-nuclear compartments or chromatin domains enriched in particular histone PTMs (Chandra et al. 2012; Fussner et al. 2011b). Most importantly in relation to chromatin, ESI provides a straightforward distinction between various degrees of chromatin compaction—from the compact chromatin domains around the nuclear periphery and nucleolus in somatic cells and down to dispersed 10-nm chromatin fibres (Bazett-Jones et al. 2008). Thus, ESI enables the following changes in different levels of chromatin compaction during diverse biological processes as well as studying the impact of various factors on chromatin compaction. ESI has revealed that chromatin domains enriched with heterochromatin marks, such as H3K9Me3, are not necessarily densely compacted (see Fig. 2) (Fussner et al. 2011b). Such chromatin is biochemically ‘closed’ to transcription, but not because it is inaccessible due to a compact configuration.
ESI, when combined with tomography, has also shown that even in the most compact regions of chromatin in mammalian cells, only 10-nm chromatin fibres are present, and not 30-nm or higher order fibres. This demonstrates that the transition from a transcriptionally closed to open chromatin does not involve a transition between fibre types. Instead, a change in the state of compaction can be understood as a combination of an increase in the bending and looping of the 10-nm fibre, an increase in the linear nucleosome density and a decrease in fibre-fibre distances (Fussner et al. 2012).
To overcome possible artefacts associated with chemical fixation and dehydration, typical in conventional processing for TEM, cryo-electron microscopy (cryoEM) methods have been developed. Besides avoiding aldehyde fixation, the major advantage of cryoEM is that the samples are imaged in a fully hydrated state. CryoEM methods can be applied to imaging chromatin in intact cells. Employing this method on both mitotic as well as interphase nuclei revealed only 10-nm chromatin fibres and could not detect any 30-nm fibres (for review see (Maeshima et al. 2014). Although cryoEM provided data on the structure of 30-nm fibres seen in extracted or reconstituted chromatin (Woodcock et al. 1993), the lack of evidence for such fibres in mitotic chromosomes infers that condensation of interphase chromatin into mitotic chromosomes does not mean transitions to higher order chromatin fibres. Instead, other mechanisms to condense chromatin may be involved, such as those dependent on histone H1 phosphorylation and recruitment or activation of non-histone chromosomal proteins, such as Condensin (Koshland and Strunnikov 1996). Electron microscopy methods have thus provided insight on the chromatin configuration that creates compact domains in the interphase nucleus and the configuration responsible for mitotic condensation. They have further provided data for studies on chromatin higher order organization and structure as well as accessibility. These methods have also demonstrated the importance of making the distinction between compaction state and a transcriptionally open or closed biochemical state.
Nuclease sensitivity
Sensitivity of chromatin to digestion by exposure of nuclei to exogenous nucleases has been used to infer features of chromatin structure. The two nucleases commonly used for revealing structural properties of chromatin are micrococcal nuclease (MNase) and deoxyribonuclease I (DNase I).
The MNase accessibility assay provides data on the localization of nucleosomes along the DNA strand. The principle assay using MNase measures nucleosome repeat length (NRL), which reflects nucleosome spacing. Using this method, Fan et al. demonstrated that a loss of linker histone H1 leads to a decrease in nucleosomal spacing (Fan et al. 2003). Digestion experiments with increasing concentrations of MNase (Hahn et al. 2013) or with increasing incubation times (Gilbert et al. 2007) provide diagnostic digestion patterns that have been used to imply changes in chromatin ‘compaction’.
DNase I hypersensitivity has been used to infer chromatin structure at the nucleosome level at promoter or enhancer elements. It is typically used to map cis regulatory elements of active genes. DNase I hypersensitive sites (DHSs) are believed to be depleted of nucleosomes or sites where nucleosome remodelling is prevalent. However, DNase I digestion has also been used to show that the chromatin of transcribed genes has an altered conformation when compared with non-transcribed genes (Weintraub and Groudine 1976). A study conducted by Bulger et al. on nuclease sensitivity and histone modifications of the mouse β-globin locus emphasizes the complexity of the relation between nuclease sensitivity and chromatin structure, as no direct relation was found between nuclease sensitivity and the histone modifications tested or changes in nuclease sensitivity and enhancer-blocking activity (Bulger et al. 2003). Recently, genome-wide mapping of DHSs has been carried out to map ‘open’ chromatin in several organisms (Boyle et al. 2008; Milon et al. 2014; Song et al. 2011; Zhang et al. 2012). Milon et al. performed high-throughput analysis for general chromatin sensitivity to DNase I in Drosophila and correlated their data to known genome annotations, such as histone PTMs, gene expression and LADs (Milon et al. 2014). Their goals were to find an association between chromatin ‘compactness’, as reflected by Dnase sensitivity, and certain chromatin modifications and to find the relation between chromatin ‘compactness’ and gene expression. Interestingly, they found that there are a number of loci in which the chromatin configuration is opposite to that predicted by the chromatin modifications or gene expression patterns.
The Dnase I hypersensitivity assay was further developed by Gerlitz and Bustin to measure ‘compaction’ or ‘condensation’ of chromatin fibres in situ (Gerlitz and Bustin 2010). In this in situ DNase I sensitivity assay, adherent cells were treated with DNase I and a decrease in nuclear size served as an indicator for the degree of digestion, thereby reflecting the degree of chromatin compaction. Using this assay, the authors showed that migrating cells are more resistant to DNase I digestion, thereby implying that they undergo global chromatin ‘compaction’.
Recently, Henikoff et al. have shown using successive salt extraction of MNase digested chromatin that actively transcribed chromatin is enriched in both the low-salt extracted and the insoluble chromatin. The authors suggest that these represent chromatin that is unbound or bound to protein complexes, respectively (Henikoff et al. 2009). These results imply the possibility that nuclease digestion assays may be enriching for specific chromatin; thus, the correlation between chromatin digestion and function should be done cautiously.
Results from nuclease digestion assays reveal structural changes in chromatin. Indeed, differences in nuclease sensitivity between cell types are due to diffusible factors that alter the chromatin (Chambers et al. 1984). However, to date, the exact nature of the changes that the chromatin undergoes is unclear. MNase sensitivity can measure nucleosome density, thereby providing information on linear compaction of chromatin. However, the changes may also be at one or more levels of chromatin structure, such as histone PTMs that lead to changes in nucleosome structure or remodelling. Alternatively, changes in nuclease sensitivity have been used to imply changes in fibre type (30 to 10-nm fibre transitions) as well as changes in global accessibility, such as in chromatin found in compact domains or in the interior of chromosome territories. Clearly, changes in nuclease sensitivity are based on real biochemical characteristics, but since the structural basis for changes in sensitivity is not clear and is not necessarily related to a compact state, the use of the term ‘compact’ when interpreting data from nuclease sensitivity experiments is not appropriate, unless using the term linear compaction to describe results from an appropriately designed MNase sensitivity assay.
Sucrose sedimentation
Sedimentation on a sucrose gradient is an additional method employed to determine structural properties of chromatin. Several studies have shown that chromatin of active genes sediments slower on a sucrose gradient than chromatin of silent genes, or than bulk chromatin (Fisher and Felsenfeld 1986; Kim and Clark 2002; Kimura et al. 1983). Moreover, mouse satellite repeats sediment faster than bulk chromatin (Gilbert and Allan 2001). However, a study by Gilbert et al. shows that in human cells there is no simple correlation between transcriptional activity and ‘open’ chromatin fibres, and rather that the correlation of compact vs. open chromatin, by virtue of their sedimentation properties, is with gene-poor vs. gene-rich regions, respectively (Gilbert et al. 2004). Hence, these results raise the need for refinement of the terms ‘open’ and ‘compact’ chromatin and reveals the gap between the structural use of these terms and the biochemical properties related to them.
Chromosome conformation capture techniques
In 2002, a major breakthrough came about in the field of chromatin organization with the development of the 3C technique by Job Dekker (Dekker et al. 2002), which maps interactions between chromatin loci. Since then, many techniques based on the original concepts have been developed to map chromatin interactions in the nucleus (for a review, see de Wit and de Laat 2012). It should be noted that the chromatin interactions mapped by these techniques are statistical associations and do not necessarily reflect contacts of every allele in the population at a given time point. The common term used to describe findings in all of these techniques is ‘higher order’ chromatin organization. In these approaches, ‘higher order’ means anything and everything from the loop formed between enhancer and promoter, through TADs, to cell type-specific compartments formed by particular TADs and chromosome territories (Bernardo et al. 2014; Dekker et al. 2013; Kurukuti et al. 2006; Sexton and Cavalli 2015).
Recently, ‘compaction’ has also been measured using Hi-C data. In a recent paper, Akhtar et al. have shown that chromatin compaction, as measured by Hi-C, is partially predictive for expression levels of integrated reporter transgenes (Akhtar et al. 2013). Compaction level, as interpreted from Hi-C data, is the rate of decay in contact probability between two loci with increasing genomic distance (Akhtar et al. 2013; Sexton et al. 2012). The steeper the decay function, the less compact the chromatin is. However, it is not clear to date whether interaction frequency measured using Hi-C is reflective of 3D chromatin organization (Dostie and Bickmore 2012; Williamson et al. 2014). There is yet much work to be done to understand the actual structure underlying such ‘compaction’ measurements.
Discussion
Chromatin exists in many structural states and at many levels of organization. The basic nucleosome subunit can itself occupy numerous structural states based on combinations of PTMs of the core histone N-terminal tails. The 10-nm fibre can also take on a variety of states, having regions of high or low nucleosome density, and by being more or less extensively folded and looped. And, at the sub-nuclear level, chromatin can be found in structurally distinct domains, with either tightly compacted fibres or with highly dispersed fibres. Hence, it is not surprising that the nomenclature used to describe chromatin contains many ambiguities, some of the common terms having multiple meanings.
In this review, we suggest a nomenclature that clarifies some of the concepts of chromatin structure (Table 1). These terms are briefly defined and explained here.
Packaging DNA—Chromatin packages DNA in the nuclear volume by two mechanisms: (1) balancing the negative charge on the DNA phosphate backbone with basic histones and (2) reducing the effective length of the DNA polymer. Chromatin does not ‘fit’ DNA into the nucleus.
Condensation—A global transition over an entire chromosome of chromatin to a concentrated state in the formation of mitotic chromosomes. Condensation can involve higher linear nucleosome density, higher levels of linker histones, modifications of histones, and recruitment of associating factors such as Structural Maintenance of Chromosomes (SMC) proteins.
Linear compaction—Higher DNA concentration per unit length of chromatin fibres through a higher linear nucleosome or H1 density.
Spatial compaction—Local sub-nuclear domains of higher chromatin density in interphase nuclei. The compaction can be achieved by higher degrees of folding, bending or looping of the 10-nm fibre by architectural proteins and closer spacing between fibres. A compact chromatin domain may contain loci from multiple chromosomes.
Open chromatin—Highly transcribed chromatin, in a biochemical state that favours transcription. This state should not be confused with highly dispersed chromatin fibres, which may or may not be highly transcribed.
Closed chromatin—Transcriptionally silent or having relatively low levels of transcriptional activity, in a biochemical state that favours silencing. It is not necessarily equivalent to a structurally compact domain.
Euchromatin—Transcriptionally active loci marked with specific histone PTMs, hypomethylated DNA sequences and unique histone variants.
Heterochromatin—Chromatin associated with low levels of transcriptional activity in all cell types (Constitutive) or in some cells only (Facultative). While marked with specific histone PTMs, DNA methylation states and containing specific histone variants, heterochromatin is not necessarily organized into a compact state. We suggest using the terms constitutive heterochromatin or facultative/polycomb-repressed heterochromatin distinctly, since these have been shown to have different biochemical, structural and 3D organizational properties.
Accessibility—Reflects the ability of RNA, histones or non-histone proteins to interact directly with a defined DNA sequence. It is typically regulated through histone remodelling, histone PTMs and DNA methylation.
Higher order chromatin structure—Folding, looping and bending of the 10-nm nucleosomal fibre into globular domains insulated by distinct borders (a.k.a domains, globules, TADs), which form discrete modules and appear to be conserved through evolution and development. Reference to transitions between 10-nm and higher order fibres only pertains to specialized cells, disrupted nuclei or cell-free conditions.
Higher order chromatin organization—The long-range distribution of chromatin in three-dimensional space includes the formation of long-range chromatin loops and chromosome territories as well as the localization of chromatin in relation to sub-nuclear structures and environments, such as nuclear bodies, nuclear envelope and transcription factories.
We hope that these suggestions will enhance the understanding and clarity surrounding concepts generated by continued advances in biochemical, physical and imaging studies of chromatin biology.
References
Abe N et al (2015) Deconvolving the recognition of DNA shape from sequence. Cell 161:307–318. doi:10.1016/j.cell.2015.02.008
Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP (2010) Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 5:e10531. doi:10.1371/journal.pone.0010531
Akhtar W et al (2013) Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154:914–927. doi:10.1016/j.cell.2013.07.018
Bazett-Jones DP, Hendzel MJ (1999) Electron spectroscopic imaging of chromatin. Methods 17:188–200. doi:10.1006/meth.1998.0729
Bazett-Jones DP, Li R, Fussner E, Nisman R, Dehghani H (2008) Elucidating chromatin and nuclear domain architecture with electron spectroscopic imaging. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 16:397–412. doi:10.1007/s10577-008-1237-3
Bernardo TJ, Dubrovskaya VA, Xie X, Dubrovsky EB (2014) A view through a chromatin loop: insights into the ecdysone activation of early genes in Drosophila nucleic acids research doi:10.1093/nar/gku754
Bian Q, Belmont AS (2012) Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol 24:359–366. doi:10.1016/j.ceb.2012.03.003
Bickmore WA (2013) The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14:67–84. doi:10.1146/annurev-genom-091212-153515
Bickmore WA, van Steensel B (2013) Genome architecture: domain organization of interphase chromosomes. Cell 152:1270–1284. doi:10.1016/j.cell.2013.02.001
Bolzer A et al (2005) Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3:e157. doi:10.1371/journal.pbio.0030157
Boyle AP et al (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132:311–322. doi:10.1016/j.cell.2007.12.014
Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA (2001) The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10:211–219
Bridger JM (2011) Chromobility: the rapid movement of chromosomes in interphase nuclei. Biochem Soc Trans 39:1747–1751. doi:10.1042/bst20110696
Bridger JM et al (2014) The non-random repositioning of whole chromosomes and individual gene loci in interphase nuclei and its relevance in disease, infection, aging, and cancer. Adv Exp Med Biol 773:263–279. doi:10.1007/978-1-4899-8032-8_12
Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843–851
Bulger M, Schübeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M (2003) A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol Cell Biol 23:5234–5244. doi:10.1128/MCB.23.15.5234-5244.2003
Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P (2002) Long-range chromatin regulatory interactions in vivo. Nat Genet 32:623–626. doi:10.1038/ng1051
Chambers SA, Cognetti G, Shaw BR (1984) Diffusible factors are responsible for differences in nuclease sensitivity among chromatins originating from different cell types. Exp Cell Res 154:213–223
Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18:1119–1130. doi:10.1101/gad.292104
Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA (2005) Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132:2215–2223. doi:10.1242/dev.01813
Chandra T et al (2012) Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47:203–214. doi:10.1016/j.molcel.2012.06.010
Ching RW, Ahmed K, Boutros PC, Penn LZ, Bazett-Jones DP (2013) Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J Cell Biol 201:325–335. doi:10.1083/jcb.201211097
Chuang C-H, Belmont AS (2007) Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol 18:698–706. doi:10.1016/j.semcdb.2007.08.012
Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS (2006) Long-range directional movement of an interphase chromosome site. Current biology : CB 16:825–831. doi:10.1016/j.cub.2006.03.059
Ciabrelli F, Cavalli G (2015) Chromatin-driven behavior of topologically associating domains. J Mol Biol 427:608–625. doi:10.1016/j.jmb.2014.09.013
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. doi:10.1146/annurev.biochem.77.062706.153223
Cope NF, Fraser P, Eskiw CH (2010) The yin and yang of chromatin spatial organization. Genome Biol 11:204. doi:10.1186/gb-2010-11-3-204
Cremer M et al (2001) Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 9:541–567
Cremer T, Cremer M (2010) Chromosome territories. Cold Spring Harb Perspect Biol 2:a003889. doi:10.1101/cshperspect.a003889
Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
de Wit E, de Laat W (2012) A decade of 3C technologies: insights into nuclear organization. Genes Dev 26:11–24. doi:10.1101/gad.179804.111
Dekker J (2014) Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin 7:25. doi:10.1186/1756-8935-7-25
Dekker J, Marti-Renom MA, Mirny LA (2013) Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 14:390–403. doi:10.1038/nrg3454
Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295:1306–1311. doi:10.1126/science.1067799
Dellaire G, Nisman R, Bazett-Jones DP (2004) Correlative light and electron spectroscopic imaging of chromatin in situ. Methods Enzymol 375:456–478
Dietzel S, Zolghadr K, Hepperger C, Belmont AS (2004) Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J Cell Sci 117:4603–4614. doi:10.1242/jcs.01330
Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM (2015) Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program Genome Research doi:10.1101/gr.183699.114
Dixon JR et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. doi:10.1038/nature11082
Dostie J, Bickmore WA (2012) Chromosome organization in the nucleus—charting new territory across the Hi-Cs. Curr Opin Genet Dev 22:125–131. doi:10.1016/j.gde.2011.12.006
Dundr M et al (2007) Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179:1095–1103. doi:10.1083/jcb.200710058
Enukashvily NI, Ponomartsev NV (2013) Mammalian satellite DNA: a speaking dumb. Adv Protein Chem Struct Biol 90:31–65. doi:10.1016/b978-0-12-410523-2.00002-x
Ernst J, Kellis M (2010) Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28:817–825. doi:10.1038/nbt.1662
Eskeland R, Freyer E, Leeb M, Wutz A, Bickmore WA (2010a) Histone acetylation and the maintenance of chromatin compaction by polycomb repressive complexes. Cold Spring Harb Symp Quant Biol 75:71–78. doi:10.1101/sqb.2010.75.053
Eskeland R et al (2010b) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38:452–464. doi:10.1016/j.molcel.2010.02.032
Fakan S (2004) The functional architecture of the nucleus as analysed by ultrastructural cytochemistry. Histochem Cell Biol 122:83–93. doi:10.1007/s00418-004-0681-1
Fakan S, Bernhard W (1971) Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res 67:129–141. doi:10.1016/0014-4827(71)90628-8
Fakan S, Bernhard W (1973) Nuclear labelling after prolonged 3H-uridine incorporation as visualized by high resolution autoradiography. Exp Cell Res 79:431–444. doi:10.1016/0014-4827(73)90463-1
Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI (2003) H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 23:4559–4572
Filion GJ et al (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224
Fisher EA, Felsenfeld G (1986) Comparison of the folding of beta-globin and ovalbumin gene containing chromatin isolated from chicken oviduct and erythrocytes. Biochemistry 25:8010–8016
Flemming W (1882) Zellsubstanz, Kern und Zelltheilung. F. C.W. Vogel, Leipzig
Fussner E, Ching RW, Bazett-Jones DP (2011a) Living without 30nm chromatin fibers. Trends Biochem Sci 36:1–6. doi:10.1016/j.tibs.2010.09.002
Fussner E et al (2011b) Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J 30:1778–1789. doi:10.1038/emboj.2011.96
Fussner E et al (2012) Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep 13:992–996. doi:10.1038/embor.2012.139
Gennes PG (1979) Scaling concepts in polymer physics, 1st edn. Cornell University Press, Ithaca, NY, USA
Gerlitz G, Bustin M (2010) Efficient cell migration requires global chromatin condensation. J Cell Sci 123:2207–2217. doi:10.1242/jcs.058271
Gilbert N, Allan J (2001) Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci U S A 98:11949–11954. doi:10.1073/pnas.211322798
Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566. doi:10.1016/j.cell.2004.08.011
Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA (2007) DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol 177:401–411. doi:10.1083/jcb.200607133
Guelen L et al (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951. doi:10.1038/nature06947
Gunawardena S, Rykowski MC (2000) Direct evidence for interphase chromosome movement during the mid-blastula transition in Drosophila. Current biology : CB 10:285–288
Hahn M et al (2013) Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev 27:859–872. doi:10.1101/gad.210377.112
Hall LE, Mitchell SE, O’Neill RJ (2012) Pericentric and centromeric transcription: a perfect balance required. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 20:535–546. doi:10.1007/s10577-012-9297-9
Hayes JJ, Wolffe AP (1992) Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci U S A 89:1229–1233
Heitz E (1928) Das Heterochromatin der Moose. Jahrb Wiss Bot 69:762–818
Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K (2009) Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19:460–469. doi:10.1101/gr.087619.108
Hou C, Corces VG (2012) Throwing transcription for a loop: expression of the genome in the 3D nucleus. Chromosoma 121:107–116. doi:10.1007/s00412-011-0352-7
Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS (2009) Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol 185:87–100. doi:10.1083/jcb.200809196
Izban MG, Luse DS (1991) Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 5:683–696. doi:10.1101/gad.5.4.683
Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L (2012) Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30(1):90–8. doi:10.1038/nbt.2057
Kapuscinski J (1995) DAPI: a DNA-specific fluorescent probe. Biotechnic & histochemistry : official publication of the Biological Stain Commission 70:220–233
Kato N, Lam E (2001) Detection of chromosomes tagged with green fluorescent protein in live Arabidopsis thaliana plants. Genome Biol 2:RESEARCH0045
Kharchenko PV et al (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471:480–485. doi:10.1038/nature09725
Kim Y, Clark DJ (2002) SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc Natl Acad Sci U S A 99:15381–15386. doi:10.1073/pnas.242536699
Kimura T, Mills FC, Allan J, Gould H (1983) Selective unfolding of erythroid chromatin in the region of the active beta-globin gene. Nature 306:709–712
Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS (2008) In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat Meth 5:311–313, http://www.nature.com/nmeth/journal/v5/n4/suppinfo/nmeth.1196_S1.html
Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184:868–871
Koshland D, Strunnikov A (1996) Mitotic chromosome condensation. Annu Rev Cell Dev Biol 12:305–333. doi:10.1146/annurev.cellbio.12.1.305
Kossel A (1884) Uber einen peptonartigen bestandteil des zellkerns. Z Phys Chem 8:511–515
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705. doi:10.1016/j.cell.2007.02.005
Kouzine F et al (2013) Global regulation of promoter melting in naive lymphocytes. Cell 153:988–999. doi:10.1016/j.cell.2013.04.033
Kuo MH et al (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269–272. doi:10.1038/383269a0
Kurukuti S et al (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A 103:10684–10689. doi:10.1073/pnas.0600326103
Lachner M, Jenuwein T (2002) The many faces of histone lysine methylation. Curr Opin Cell Biol 14:286–298. doi:10.1016/S0955-0674(02)00335-6
Latt SA, Stetten G (1976) Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. Journal of Histochemistry & Cytochemistry 24:24–33. doi:10.1177/24.1.943439
Libbrecht MW, Ay F, Hoffman MM, Gilbert DM, Bilmes JA, Noble WS (2015) Joint annotation of chromatin state and chromatin conformation reveals relationships among domain types and identifies domains of cell type-specific expression genome research doi:10.1101/gr.184341.114
Lieberman-Aiden E et al (2009) Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science (New York, NY) 326:289–293. doi:10.1126/science.1181369
Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG (2006) The Drosophila melanogaster Cajal body. J Cell Biol 172:875–884. doi:10.1083/jcb.200511038
Lleres D, James J, Swift S, Norman DG, Lamond AI (2009) Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol 187:481–496. doi:10.1083/jcb.200907029
Maeshima K, Hihara S, Eltsov M (2010) Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol 22:291–297. doi:10.1016/j.ceb.2010.03.001
Maeshima K, Imai R, Tamura S, Nozaki T (2014) Chromatin as dynamic 10-nm fibers. Chromosoma 123:225–237. doi:10.1007/s00412-014-0460-2
Matzke AJM, Huettel B, van der Winden J, Matzke M (2005) Use of two-color fluorescence-tagged transgenes to study interphase chromosomes in living plants. Plant Physiol 139:1586–1596. doi:10.1104/pp.105.071068
Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10:105–116. doi:10.1016/j.devcel.2005.10.017
Milon B et al (2014) Map of open and closed chromatin domains in Drosophila genome. BMC Genomics 15:988. doi:10.1186/1471-2164-15-988
Morey C, Da Silva NR, Perry P, Bickmore WA (2007) Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134:909–919. doi:10.1242/dev.02779
Morse RH (1989) Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J 8:2343–2351
Müller WG, Walker D, Hager GL, McNally JG (2001) Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol 154:33–48. doi:10.1083/jcb.200011069
Nemeth A, Langst G (2011) Genome organization in and around the nucleolus. Trends in genetics : TIG 27:149–156. doi:10.1016/j.tig.2011.01.002
Nizami Z, Deryusheva S, Gall JG (2010) The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2:a000653. doi:10.1101/cshperspect.a000653
Nora EP, Dekker J, Heard E (2013) Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 35:818–828. doi:10.1002/bies.201300040
Nora EP et al (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385. doi:10.1038/nature11049
Olins AL, Olins DE (1974) Spheroid chromatin units (v bodies). Science 183:330–332
Olson MOJ, Dundr M (2001) Nucleolus: structure and function. In: eLS. John Wiley & Sons, Ltd. doi:10.1002/9780470015902.a0005975.pub3
Osborne CS et al (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36:1065–1071. doi:10.1038/ng1423
Pecinka A, Kato N, Meister A, Probst AV, Schubert I, Lam E (2005) Tandem repetitive transgenes and fluorescent chromatin tags alter local interphase chromosome arrangement in Arabidopsis thaliana. J Cell Sci 118:3751–3758. doi:10.1242/jcs.02498
Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gülsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515:402–405. doi:10.1038/nature13986
Probst AV, Almouzni G (2011) Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet 27:177–185. doi:10.1016/j.tig.2011.02.002
Rao Suhas SP et al (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. doi:10.1016/j.cell.2014.11.021
Rapkin LM, Anchel DR, Li R, Bazett-Jones DP (2012) A view of the chromatin landscape. Micron 43:150–158. doi:10.1016/j.micron.2011.11.007
Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135:1685–1700
Roukos V, Misteli T (2014) The biogenesis of chromosome translocations. Nat Cell Biol 16:293–300. doi:10.1038/ncb2941
Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE (1995) A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci U S A 92:2710–2714
Saksouk N, Simboeck E, Déjardin J (2015) Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8:3. doi:10.1186/1756-8935-8-3
Sanyal A, Baù D, Martí-Renom MA, Dekker J (2011) Chromatin globules: a common motif of higher order chromosome structure? Curr Opin Cell Biol 23:325–331. doi:10.1016/j.ceb.2011.03.009
Sexton T, Cavalli G (2015) The role of chromosome domains in shaping the functional genome. Cell 160:1049–1059. doi:10.1016/j.cell.2015.02.040
Sexton T et al (2012) Three-dimensional folding and functional organization principles of the Drosophila. Genome Cell 148:458–472. doi:10.1016/j.cell.2012.01.010
Shopland LS et al (2006) Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174:27–38. doi:10.1083/jcb.200603083
Simonis M et al (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38:1348–1354. doi:10.1038/ng1896
Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R (2014) Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci 39:381–399. doi:10.1016/j.tibs.2014.07.002
Smith KP, Lawrence JB (2000) Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell 11:2987–2998
Song L et al (2011) Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21:1757–1767. doi:10.1101/gr.121541.111
Taylor GC, Eskeland R, Hekimoglu-Balkan B, Pradeepa MM, Bickmore WA (2013) H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res 23:2053–2065. doi:10.1101/gr.155028.113
Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10:1453–1465
Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145:1341–1354
van den Engh G, Sachs R, Trask BJ (1992) Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science 257:1410–1412
Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R (2005) In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol 25:4552–4564. doi:10.1128/MCB.25.11.4552-4564.2005
Vidali G, Gershey EL, Allfrey VG (1968) Chemical studies of histone acetylation. The distribution of epsilon-N-acetyllysine in calf thymus histones. J Biol Chem 243:6361–6366
Visvanathan A, Ahmed K, Even-Faitelson L, Lleres D, Bazett-Jones DP, Lamond AI (2013) Modulation of higher order chromatin conformation in mammalian cell nuclei can be mediated by polyamines and divalent cations. PLoS One 8:e67689. doi:10.1371/journal.pone.0067689
Weintraub H, Groudine M (1976) Chromosomal subunits in active genes have an altered conformation. Science 193:848–856
Williamson I et al (2014) Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev 28:2778–2791. doi:10.1101/gad.251694.114
Woodcock CL (1994) Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J Cell Biol 125:11–19
Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N (1993) A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A 90:9021–9025
Woodcock CL, Safer JP, Stanchfield JE (1976) Structural repeating units in chromatin. I. Evidence for their general occurrence. Exp Cell Res 97:101–110
Yaffe E, Tanay A (2011) Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 43:1059–1065. doi:10.1038/ng.947
Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ (1995) Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J Cell Biol 130:1239–1249
Zhang W, Zhang T, Wu Y, Jiang J (2012) Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24:2719–2731. doi:10.1105/tpc.112.098061
Acknowledgments
We would like to thank Lindsy Rapkin for the critical reading of the manuscript and constructive comments. The work is supported by operating funds from the Natural Sciences and Engineering Research Council and the Canadian Institutes of Health Research. D.P.B.-J. holds the Canada Research Chair in Molecular and Cellular Imaging.
Conflict of interest
The authors declare that they have no competing interests.
Compliance with Ethical Standards
ᅟ
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
Operating grants from the Natural Sciences and Engineering Research Council (Canada) and the Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Even-Faitelson, L., Hassan-Zadeh, V., Baghestani, Z. et al. Coming to terms with chromatin structure. Chromosoma 125, 95–110 (2016). https://doi.org/10.1007/s00412-015-0534-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-015-0534-9