Abstract
The nucleus limited long-noncoding hsrω-n transcripts, hnRNPs, and some other RNA processing proteins organize nucleoplasmic omega speckles in Drosophila. Unlike other nuclear speckles, omega speckles rapidly disappear following cell stress, while hnRNPs and other associated proteins move away from chromosome sites, nucleoplasm, and the disappearing speckles to get uniquely sequestered at hsrω locus. Omega speckles reappear and hnRNPs get redistributed to normal locations during recovery from stress. With a view to understand the dynamics of omega speckles and their associated proteins, we used live imaging of GFP tagged hnRNPs (Hrb87F, Hrb98DE, or Squid) in unstressed and stressed Drosophila cells. Omega speckles display size-dependent mobility in nucleoplasmic domains with significant colocalization with nuclear matrix Tpr/Megator and SAFB proteins, which also accumulate at hsrω gene site after stress. Instead of moving towards the nuclear periphery located hsrω locus following heat shock or colchicine treatment, omega speckles rapidly disappear within nucleoplasm while chromosomal and nucleoplasmic hnRNPs move, stochastically or, more likely, by nuclear matrix-mediated transport to hsrω locus in non-particulate form. Continuing transcription of hsrω during cell stress is essential for sequestering incoming hnRNPs at the site. While recovering from stress, the sequestered hnRNPs are released as omega speckles in ISWI-dependent manner. Photobleaching studies reveal hnRNPs to freely move between nucleoplasm, omega speckles, chromosome regions, and hsrω gene site although their residence periods at chromosomes and hsrω locus are longer. A model for regulation of exchange of hnRNPs between nuclear compartments by hsrω-n transcripts is presented.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nucleus is a well-orchestrated dynamic information center in eukaryotic cells. Besides nuclear envelope, nuclear matrix, chromatin, and nucleolus, a variety of nuclear speckles/bodies, like splicing speckles, omega speckles, paraspeckles, OPT domains, Gems, PcG bodies, Cajal bodies, cleavage bodies, PML bodies, stress bodies/granules, perinucleolar compartments, etc., are also present in nucleoplasm (Matera et al. 2009; Mao et al. 2011; Lakhotia 2012; Sleeman and Trinkle-Mulcahy 2014). Many of these nuclear bodies contain, besides their specific sets of proteins, nucleus limited long noncoding RNAs (lncRNAs), which seem to help in maintaining their integrity and functions (Jolly and Lakhotia 2006; Spector and Lamond 2011; Ip and Nakagawa 2012; Lakhotia 2012). Live cell imaging has revealed remarkable spatiotemporal dynamicity of these nuclear bodies in relation to cellular conditions and cell type (Misteli et al. 1997; Phair and Misteli 2000; Platani et al. 2002; Gorisch et al. 2004; Spector and Lamond 2011; Mao et al. 2011). Abiotic and biotic stresses cause disturbances in the steady state influx and efflux of their constituents and thus affect their structure and organization (Lakhotia 2012; Mercer and Mattick 2013; Place and Noonan 2014).
The hsrω is a developmentally active, heat shock (HS) inducible, noncoding gene in Drosophila (reviewed in Lakhotia 2011) producing multiple lncRNAs (http://flybase.org). Its most extensively studied nucleus limited hsrω-n1 (hsrω-RB) and spliced hsrω-n2 (hsrω-RG) lncRNAs, together referred to as hsrω-n (Mallik and Lakhotia 2011), interact with variety of RNA processing proteins including Hrb87F/Hrp36, Hrb57A/Bancal, Hrb98DE/Hrp38, Squid/Hrp40, PEP, Rumpelstiltskin/Hrp59, NonA, and Sxl (Prasanth et al. 2000; Jolly and Lakhotia 2006; Ji and Tulin 2009; Onorati et al. 2011; Singh and Lakhotia 2012) to organize the omega speckles. In all cell types of Drosophila, these nucleoplasmic speckles disappear after HS or other stress while hnRNPs and several other proteins get clustered almost exclusively at hsrω/93D locus (Saumweber et al. 1980; Dangli et al. 1983; Lakhotia et al. 1999, 2012; Prasanth et al. 2000; Lakhotia 2011).
We have examined dynamics of GFP-tagged hnRNPs in normal and stressed cells. Our results show that omega speckles have restricted movement possibly because they interact with nuclear matrix as revealed by their association with Tpr/Megator and SAFB proteins. The stress-induced rapid clustering of hnRNPs requires continuing transcription at the hsrω locus. Unlike earlier presumption (Lakhotia et al. 1999; Prasanth et al. 2000; Lakhotia 2011) that stress causes individual nucleoplasmic omega speckles to fuse and the resulting larger clusters move to the hsrω locus, present time lapse imaging and photobleaching experiments revealed that omega speckles disappear in nucleoplasm rather than fusing with the hsrω gene site. In normal as well as stressed cells, the hnRNPs move rapidly and stochastically between different compartments; however, they appear to be also actively moved, possibly by nuclear matrix components, to the hsrω gene site in non-particulate form, where they are sequestered by hsrω transcripts. Interestingly, the hnRNPs accumulating at the hsrω locus still show a reduced stochastic efflux to nucleoplasmic pool. During recovery from HS, the hnRNPs aggregated at the hsrω locus are released as omega speckles, whose biogenesis requires the chromatin remodeling ISWI protein.
Materials and methods
Fly strains
Drosophila melanogaster wild-type (Oregon R +) and mutant stocks were reared on standard cornmeal-agar food medium at 23 ± 1 °C. Three GFP tagged hnRNP protein-trap homozygous viable stocks (Morin et al. 2001; Buszczak et al. 2007) viz. Hrb87F-GFP (CC00189), Squid-GFP (CB02655), and Hrb98DE-GFP (CC01563) were used to examine dynamics of hnRNPs in different nuclear domains of live cell nuclei. Since the GFP-coding region is inserted in an intron of the given endogenous gene in protein-trap lines (Morin et al. 2001), the GFP-tagged protein is generally expressed in the same pattern as the original gene. The hsrω 66/hsrω 66 was used as hsrω-null (Johnson et al. 2011; Lakhotia et al. 2012). Appropriate crosses were made to recombine the Hrb87F-GFP or Squid-GFP or Hrb98DE-GFP with hsrω 66 allele. ISWI 1 Bc/SM5; +/+ and ISWI 2; +/T(2;3)CyO, TM6B, Tb (Deuring et al. 2000) flies were crossed to obtain heterozygous ISWI 1/ISWI 2 progeny (ISWI null) which do not produce ISWI but survive till late larval stage because of the maternal contribution (Corona et al. 2007). The GFP expressing Act-GAL4/UAS-SAFB-GFP progeny larvae, obtained from cross between UAS-SAFB-GFP/UAS-SAFB-GFP (Alfonso-Parra and Maggert 2010) and Act-GAL4/CyO flies, were used to localize SAFB-GFP in live cells and for co-immunoprecipitation experiments.
Treatments and cytological methods
Late third instar larvae of the desired genotypes were collected in 1.5 ml moist filter paper lined microcentrifuge tubes and heat shocked in water bath at 37 ± 1 °C for desired duration. Parallel control larvae were similarly maintained at 24 ± 1 °C. For immunostaining, the larvae were dissected immediately after HS in Poels’ salt solution (PSS) (Tapadia and Lakhotia 1997) and the desired tissues were processed further (see below).
Actively wandering Act-GAL4/UAS-SAFB-GFP late third instar larval Malpighian tubules (MT) were dissected out in Graces’ insect medium and treated with DNase-free RNase A (1 mg/ml, Sigma-Aldrich, India) for 15 min at 23 °C prior to fixation and immunostaining.
Salivary glands (SG) of actively wandering late third instar Hrb87F-GFP, Squid-GFP, or Hrb98DE-GFP larvae were incubated in a hanging drop (see below) of Graces’ medium, freshly supplemented with colchicine (100 μg/ml) for live imaging of the distribution of Hrb87F-GFP by confocal microscopy at 1 min interval for 30 min.
To examine the effects of chemical inhibition of transcription on HS-induced changes in hnRNP distribution, SG of actively wandering late third instar Hrb87F-GFP homozygous larvae were dissected in Graces’ medium and incubated in actinomycin-D (10 μg/ml) or α-amanitin (5 μg/ml) containing hanging drop for 20 min at 23 °C followed by 30 min at 37 °C on confocal microscope heating stage (see below). Hrb87F-GFP distribution in live SG was monitored at 1 min interval throughout the treatment.
Confocal microscopy, live cell imaging, and image analysis
A Zeiss LSM510 Meta laser scanning confocal microscope with Plan-Apo 63X, 1.4-NA oil immersion objective was used for live cell imaging of hanging drop preparations (Reed et al. 2009; Szczepny et al. 2009; Millet and Gillette 2012). Desired tissue from late third instar larvae of various genotypes (see Results) was placed in a drop of Graces’ medium hanging from a coverslip mounted on a cavity-slide and sealed with nail polish. When required, HS was applied in situ using a heating stage (MC60, Linkam Scientific Instruments, UK) mounted on confocal microscope; its temperature can change from 23 to 37 °C or vice-versa in less than a minute. Time-lapse live cell images were collected using the ROI and time-series tools of the LSM 510 Meta software. Quantitative estimates of fluorescence intensity on defined regions were obtained with Profile and Histo tools.
Photobleaching experiments (FRAP and FLIP)
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) methods, based on irreversible loss of fluorescence after photobleaching (Axelrod et al. 1976; Klonis et al. 2002), was used to examine dynamics of GFP-tagged hnRNPs in live nuclei. The GFP expressing region of interest (ROI) in FRAP, a circle of fixed diameter (see Results), was rapidly photobleached by 200 iterations of exposure to Argon laser (488 nm) at 100 % power. Subsequent recovery of fluorescence in the photobleached region was examined at defined time intervals. As control, fixed cells were examined to confirm that fluorescence recovery in the ROI was indeed due to mobility of the GFP-tagged protein in live cells. FRAP experiments were carried out on desired tissues at 23 °C, during HS at 37 °C and during recovery from HS. The fluorescence signal from the ROI was normalized and data analyzed following published methods (Phair and Misteli 2000; Lippincott-Schwartz et al. 2001; Ishikawa-Ankerhold et al. 2012).
FLIP is a direct method to examine the connectivity and fluxes between different cellular compartments (Phair and Misteli 2000). Following acquisition of five control images, the Hrb87F-GFP fluorescence of ROI1 in desired tissue was continuously photobleached by Argon laser (488 nm) at 100 % power and images collected after each 200 iterations of photobleaching. The loss in fluorescence in another region of interest, the ROI2, over a period of time was concurrently measured. Fluorescence signal of ROI2 was normalized and data analyzed following Nissim-Rafinia and Meshorer (2011).
Immunostaining
Intact eye discs or SG or MT or SG polytene chromosome squash preparations from actively wandering late third instar larvae of desired genotypes and following the required treatment were processed for immunostaining as described (Singh and Lakhotia 2012; Lakhotia et al. 2012) using mouse anti-Hrb87F (P11, 1:20 dilution, Saumweber et al. 1980), mouse anti-Megator (BX34, 1:10 dilution, Zimowska and Paddy 2002), and rabbit anti-GFP (1:50 dilution, Sigma-Aldrich, India) primary antibodies. Secondary antibodies were conjugated with Cy3 (1:200, Sigma-Aldrich, India) or Alexa Fluor 488 (1:200; Molecular Probes, USA). Chromatin was counterstained with DAPI (1 μg/ml) for 10 min at room temperature.
Immunoprecipitation (IP), SDS-PAGE, western blotting, and RT-PCR
To detect physical interaction of Hrb87F with Megator and SAFB-GFP, respectively, total proteins from Act-GAL4/UAS-SAFB-GFP late third instar larvae were processed for immuno-precipitation (Prasanth et al. 2000) with anti-Megator (BX34) or anti-GFP antibody, respectively. The SAFB-GFP larval protein samples were incubated with 10 μl of the desired antibody for 2 h at 4 °C followed by overnight incubation with 50 μl protein-A agarose beads (Bangalore Genei, India) at 4 °C on 3600 rocker. As a control, beads were parallely incubated with antibody without protein sample. The beads were pelleted, washed five times with 1 ml of IP buffer, and the bound material was eluted either by addition of 50 μl SDS-PAGE sample buffer for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) or by addition of 200 μl DEPEC-water for reverse transcription polymerase chain reaction (RT-PCR). Samples for SDS-PAGE were boiled for 10 min and centrifuged at 6000 RPM for 2 min followed by separation of proteins in supernatant by 10 % SDS-PAGE and blotting on PVDF membrane. The western blots were probed (Prasanth et al. 2000) with anti-Hrb87F (P11, 1:200) or anti-GFP (1:500, Sigma-Aldrich, India) as primary and HRP-conjugated anti-mouse or anti-rabbit as secondary antibodies (1:1500), respectively.
For detecting the presence of hsrω-n transcripts in the above IP samples, DEPEC-water added samples were boiled for 10 min and centrifuged (6000 RPM for 2 min). The supernatant was mixed with 800 μl Trizol (Sigma-Aldrich, India) and 200 μl chloroform. The isolated RNA was used for cDNA synthesis (Singh and Lakhotia 2012), which was used for PCR with hsrω-n transcript-specific forward LP: 5′-GGCAGACATACGTACACGTGGCAGCAT-3′ and reverse R1 5′-TTGCGCTCACAGGAGATCAA-3′ primers (Lakhotia et al. 2012).
Results
As reported earlier (Prasanth et al. 2000), the hsrω-n transcripts show complete colocalization with Hrb87F and Squid in omega speckles. In addition, these hnRNPs are also present on many sites on chromosomes and in the nucleoplasm in non-speckled form. We used three GFP tagged hnRNP stocks, viz. w 1118; Hrb87F-GFP/Hrb87F-GFP, w 1118; Squid-GFP/Squid-GFP, and w 1118; Hrb98DE-GFP/Hrb98DE-GFP, to examine the dynamics of omega speckles in live cell nuclei. To confirm that GFP tagging did not affect localization of Hrb87F (Hrp36), Squid (Hrp40), or Hrb98DE (Hrp38), the endogenous Hrb87F was immunolocalized with these GFP tagged proteins (Fig. 1). Intensity profile analysis revealed that in each case, the distribution of Hrb87F completely overlapped with that of the Hrb87F-GFP (Fig. 1a), Squid-GFP (Fig. 1b), or Hrb98DE-GFP (Fig. 1c). This showed that GFP tagged proteins have similar distribution in the nucleus as their normal counterparts. Therefore, we used the GFP-tagged Hrb87F-GFP, Squid-GFP, and Hrb98DE-GFP to examine the behavior of these hnRNPs in live cell nuclei under normal conditions or after heat shock or colchicine treatment.
GFP tagging does not affect the distribution of hnRNPs in nucleus. a–c Colocalization of endogenous Hrb87F (red) with GFP-tagged hnRNPs (green) in MT principal cell nuclei of Hrb87F-GFP (a) or Squid-GFP (b) or Hrb98DE-GFP (c) larvae; intensity profile shown on right of each nucleus corresponds to fluorescence intensity along the white arrow across the nucleus. d–i Time-lapse (at 0.248 s interval) optical section images of a MT principal cell nucleus showing distribution of Hrb87F-GFP in diffused and speckled forms in nucleoplasm and on chromosome regions. d’–i’ are magnified views of the marked area in (d) at different time points (corresponding, respectively, to d–i) showing that two of the omega speckles (red and yellow arrowheads) continuously changed their position while another omega speckle (green arrowhead) did not move during this period
Omega speckles are dynamic in live cell nuclei
Different nuclear domains showed characteristic GFP fluorescence for each of the three hnRNPs in all examined cell types: (i) bright fluorescence at many chromatin sites (especially distinct in larval salivary gland (SG) polytene cells), (ii) low uniformly diffuse fluorescence all over the nucleoplasm, (iii) brightly fluorescing nucleoplasmic omega speckles, except in SG polytene nuclei, (iv) brighter fluorescence at the heterochromatic chromocenter, identifiable in nuclei by brighter DAPI fluorescence of irregular chromatin blocks, and (v) a more bright region on chromatin, which from previous RNA:RNA in situ hybridization results (Prasanth et al. 2000; Lakhotia 2011, 2012) is known to be the hsrω or 93D locus.
Examination of Hrb87F-GFP in more than 50 live cells each from larval tissues like MT, fat body, brain ganglia, imaginal discs, and gut revealed the number of omega speckles to range from 2 to 6 in larval diploid cells, 5 to 25 in fat body, and 50 to >1000 in the endoreplicated MT and gut cells. Diameter of mostly spherical omega speckles varied from 0.2 to 1.5 μm; some larger ones occasionally showed irregular shape. Larval SG nuclei generally did not show omega speckles: the hnRNPs as well as hsrω-n transcripts were distributed in a uniformly diffused non-particulate form throughout interchromatin space (not shown).
Late third instar larval MT, gut, brain, and eye-antennal disc live cells showed high dynamicity of the Hrb87F-GFP or Squid-GFP or Hrb98DE-GFP marked omega speckles as most of them changed their location in each consecutive 0.248 s intervals, although a small proportion of them did not move (Fig. 1d–i’). Single particle tracking (SPT) analysis, using Image J software (http://rsb.info.nih.gov/ij/), of the Hrb87F-GFP marked omega speckles in MT principal cells that remained visible in the same focal plane for at least eight consecutive 0.248 s intervals revealed their high mobility with an inverse correlation between speckle size and total distance traveled (not shown) as reported for other nuclear speckles (Misteli et al. 1997; Phair and Misteli 2000).
Omega speckles and nuclear matrix
The nuclear matrix-specific protein Tpr/Megator (Zimowska and Paddy 2002) and Hrb87F-GFP were co-immunolocalized in fixed MT cells. As reported (Zimowska and Paddy 2002), Megator was seen as nuclear lamina ring besides reticulated tracks in interchromatin space across the nuclear volume with superimposed brighter Megator puncta (Fig. 2a). Interestingly, while the peripheral omega speckles were within the Megator ring, those in inner nucleoplasm were always very close to or partially overlapped with the brighter Megator puncta (Fig. 2a) or followed the Megator reticulum. Distribution of omega speckles in relation to another nuclear matrix protein SAFB (Arao et al. 2000; Alfonso-Parra and Maggert 2010) was examined through Hrb87F immunostaining in SAFB-GFP expressing MT (Fig. 2b). Besides its presence on chromosome regions, SAFB-GFP was present in nucleoplasm in diffused as well as speckled form. The SAFB-GFP and Hrb87F speckles showed frequent partial or complete colocalization or close association (Fig. 2b).
Omega speckles interact with nuclear matrix. a–c Confocal optical sections showing distribution of Hrb87F-GFP and Megator (a), Hrb87F and SAFB-GFP in untreated (b), and in in vivo RNase treated (c) unstressed larval MT principal cell nuclei; insets in each show magnified images of overlapping speckles of the given two proteins; DAPI-stained chromatin (blue) is also shown in (c). d–f Profiles of Hrb87F-GFP (green) and Megator (red) fluorescence intensities (d) or of SAFB-GFP and Hrb87F (e, f) along the arrows drawn in a, b, and, c, respectively. g Co-immunoprecipitation of Hrb87F and Megator with BX34 (lane 2), and Hrb87F and SAFB-GFP with anti-GFP ab (lane 3); lane 1 corresponds to control sample without any larval protein while lane 4 is the input protein sample; Igh and Hrb87F on right mark the corresponding bands. h The blot in (g) reprobed with anti-GFP to detect the ∼132 kDa SAFB-GFP protein; the Igh band is also marked on right. i RT-PCR with the above immunoprecipitates using hsrω-n specific primers generated 598 bp amplicons in all samples, except the control IgG sample. j–m Polytene chromosome spreads from control (j, l) or heat shocked SG (k, m) immunostained for SAFB-GFP (j, k) or Megator (l, m); red boxes indicate the 93D region which is further enlarged in insets in each panel. Scale bar in (j) is applicable to (j–m)
In vivo RNase treatment is known to collapse the nuclear matrix with its constituents forming dense clusters/aggregates in nucleoplasm (Nickerson et al. 1989; Pederson 2000). Therefore, to further examine the relationship between omega speckles and nuclear matrix, live MT were treated with RNase followed by immunostaining to localize Hrb87F in SAFB-GFP expressing cells. Following in vivo RNase treatment, speckles of Hrb87F were no longer visible in nucleoplasm instead the Hrb87F as well as SAFB-GFP proteins formed partially or fully colocalized or closely associated large aggregates in interchromatin space in these nuclei (Fig. 2c).
The fluorescence intensity profiles in linescans of Megator and Hrb87F (Fig. 2d), SAFB-GFP, and Hrb87F in control (Fig. 2e) or in vivo RNase-treated cells (Fig. 2f) confirmed that omega speckles and Megator or SAFB-GFP puncta often overlapped or were close to each other.
Co-immuno-precipitation studies showed Hrb87F to be pulled down with Megator as well as with SAFB-GFP (Fig. 2g, h). In parallel with the immunofluorescence findings, our RT-PCR results with either of the IP samples revealed that the hsrω-n transcripts were also pulled down with Megator or SAFB-GFP (Fig. 2i). It is notable that Megator also co-immunoprecipitated with SAFB-GFP since the Megator-IP sample also showed the presence of SAFB-GFP (second lane in Fig. 2h).
Immunostaining for GFP in polytene chromosome spreads in control and heat shocked SAFB-GFP expressing SG revealed that in control cells this protein was present at many chromosome sites, including the 93D site (Fig. 2j). Its accumulation at the 93D puff site (red box in Fig. 2k) was significantly elevated after heat shock while other chromosome regions showed reduced but distinct presence.
Immunostaining of squashed polytene chromosomes with BX34 ab showed Megator to be mostly absent from chromosome regions except for faint but perceptible staining at 93D and a few other, but not all, puff sites in unstressed cells (Fig. 2l). Interestingly, HS distinctly enhanced the presence of Megator at 93D puff (Fig. 2m). No other chromosome regions showed detectable presence of Megator in heat shocked SG.
Dynamics of stress induced clustering of hnRNPs at 93D locus in live cells
HS induced time-dependent accumulation of Hrb87F-GFP, Hrb98DE-GFP, and Squid-GFP at 93D site in SG nuclei is shown in Fig. 3a–c. As noted above, distinct omega speckles were absent in unstressed SG nuclei with Hrb87F-GFP, Hrb98DE-GFP, and Squid-GFP being distributed uniformly through nucleoplasm; in addition, they were present on different chromosome bands including the 93D locus (Fig. 3a–c). All the three proteins started to accumulate at 93D site almost immediately after HS (Fig. 3a–c), and concomitantly, the GFP fluorescence at other chromosome regions started waning; in many nuclei, the chromocenter retained weak but detectable fluorescence. After 30 min, HS, Hrb87F-GFP, and Hrb98DE-GFP proteins were localized almost exclusively at 93D site but while bulk of Squid-GFP was present at 93D site, some chromosome bands continued to show Squid fluorescence even after 30 min HS. In all cases, nucleoplasm showed a very low diffuse presence of these proteins after 30 min HS (Fig. 3a–c).
HS or colchicine treatment induced clustering of hnRNPs at 93D in live cell nuclei of SG. Time lapse live cell images of Drosophila SG nuclei during HS (a–d) or colchicine treatment (e–h) at 5 min intervals (noted at upper corner in each column’s top row) for 30 min showing distribution of hnRNPs (Hrb87F (a, e), Hrb98DE (b, f), and Squid (c, d, g, h)) in hsrω + (a–c, e–g) or hsrω 66 (d, h) background. i, j Time-lapse live cell images of Hrb87F-GFP in SG nuclei at 10 min intervals (noted at upper corner in the i row) in presence of α-amanitin (i) or actinomycin-D (j) at 23 °C (left three images) followed by HS (right three images). The scale bar in a applies to a–h while that in i applies to i and j
Imaging of live SG cells revealed that in about 90 % nuclei (N = 206), the hsrω/93D locus, where the GFP tagged hnRNPs accumulated following HS, was located close to nuclear envelope. Its stable peripheral location was further confirmed by following the same nucleus for two rounds of HS and recovery. Two consecutive cycles of 20 min HS and 30 min recovery at 23 °C revealed identical kinetics of accumulation of hnRNPs at 93D locus and their dispersal without any change in position of hsrω/93D locus (see Supplementary Fig. S1). The absence of any net alteration in position of the 93D locus during these dynamic changes agrees with the classical view that transient changes in transcriptional activity do not entail chromosome repositioning (Hochstrasser and Sedat 1987; Yao et al. 2007; Zobeck et al. 2010).
In vitro treatment of larval SG with colchicine or other amides singularly enhances transcription at the 93D puff while other chromosome sites are largely silenced (Lakhotia and Mukherjee 1984; Tapadia and Lakhotia 1997). Live imaging of SG exposed to colchicine (100 μg/ml) revealed that all the three GFP-tagged hnRNPs begin to increasingly localize at the 93D locus within 5–10 min of exposure to colchicine so that after 30 min, the cluster size became very large while the presence of these proteins at other chromosome regions and in nucleoplasm weakened (Fig. 3e–g). As after HS, Squid-GFP fluorescence persisted at several chromosome regions and in cytoplasm throughout colchicine treatment.
Clustering of hnRNPs at 93D site following stress is dependent upon hsrω gene’s transcription
In order to see if the aggregation of hnRNPs at 93D locus is dependent upon its DNA sequence or upon the increased presence of its transcripts, movement of Squid-GFP was examined in Squid-GFP hsrω 66 live SG nuclei. The hsrω 66 allele carries a deletion of 1598 bp spanning the hsrω promoter and 9 bp beyond transcription start site so that, in spite of the bulk of ∼21 kb transcribed region being intact, none of the hsrω transcripts are produced in unstressed or stressed cells (Johnson et al. 2011; Lakhotia et al. 2012; M. Mustafa and S. C. Lakhotia, unpublished). Interestingly, no chromosome site in Squid-GFP hsrω 66 nuclei showed an enhanced GFP fluorescence after HS (Fig. 3d) or colchicine (Fig. 3h) treatment, although other chromosome regions progressively lost Squid-GFP as in wild type.
To further confirm the transcription-dependent aggregation of hnRNPs at the 93D site during HS, Hrb87F-GFP hsrω + late third instar larval SGs were treated with α-amanitin or actinomycin-D to inhibit transcription and imaged during exposure to inhibitor for 20 min at 23 °C followed by 30 min at 37 °C (N = 9 nuclei in each case). The Hrb87F-GFP fluorescence either disappeared or weakened at many chromosome sites during the 20 min at 23 °C but no new GFP-positive chromosome bands appeared (Fig. 3i, j). Significantly, the stress induced characteristic accumulation of hnRNPs at 93D site was absent in transcriptionally inhibited nuclei since after 30 min heat shock, only a narrow band of fluorescence was detectable at 93D site, similar to that seen after the initial 20 min exposure to inhibitor at 23 °C (compare Fig. 3a with 3i and j).
Omega speckles do not directly move to the hsrω locus following HS but the sequestered hnRNPs are released during recovery as omega speckles
Distribution of Hrb87F-GFP was examined at shorter time intervals in live hsrω + MT principal cells and eye disc peripodial cells during HS (Fig. 4) and recovery (Fig. 5). Normally, Hrb87F-GFP is present in omega speckles, on chromatin regions and as a brighter spot on the 93D locus besides a diffuse distribution through nucleoplasm in both cell types (Fig. 4). Accumulation of Hrb87F-GFP at the 93D site started almost instantaneously after HS and with increasing duration, the cluster progressively enlarged (Fig. 4). Simultaneously, omega speckles either disappeared in nucleoplasm or a few of them aggregated to form larger speckles. No omega speckles or their larger clusters were seen moving to 93D locus in live cells during HS, suggesting that the nucleoplasmic hnRNP speckles mostly disappear within their local domains to release the associated proteins, which move in a non-particulate form to the 93D locus. This was further confirmed by higher resolution live images of the progressively increasing cluster of Hrb87F-GFP in peripodial cells during HS (Fig. 4c). The very bright cluster at the hsrω locus in stressed cells was surrounded by an irregular halo of fluorescence signal which was less intense than the cluster but brighter than rest of the nuclear area (Fig. 4c). Shape and size of the halo appeared very dynamic, although no omega speckles were seen to fuse with it. The halo surrounding the cluster could be the transition zone for continuing influx of hnRNPs which get associated at hsrω locus with its new transcripts.
HS induced clustering of hnRNPs at 93D locus does not involve fusion of nucleoplasmic omega speckles. Time lapse (1 min interval, noted at upper corner of each image) live cell projection images of MT principal cell (a) and eye disc peripodial cells (b) during HS showing dissociation of hnRNPs from chromatin, disappearance of omega speckles within the nucleoplasm with concomitant increased accumulation of Hrb87F-GFP at 93D locus; the chromatin-associated Hrb87F-GFP also gradually moves out. c Higher resolution images of the boxed area in (b) (1 min) to show the progressively enlarging aggregate of Hrb87F-GFP at different time intervals corresponding to those in (b). No omega speckles are seen at any time near the enlarging Hrb87F-GFP aggregate, which is surrounded by a dynamically streaming halo of less intense fluorescence
Hrb87F-GFP is released from the hsrω locus during recovery from HS as omega speckles in MT principal cell nucleus. Time lapse (1 min interval, noted at upper right corner of each panel) live cell projection images of MT principal cell nucleus showing emergence of omega speckles from 93D locus into nucleoplasm during recovery following HS
There was almost no change in the Hrb87F-GFP fluorescence intensity at the 93D site during the first 5–10 min recovery from HS (Fig. 5a–k). After 10 min recovery, size of 93D cluster started to decrease. Interestingly, unlike the manner of accumulation during HS, the Hrb87F-GFP appeared to dissociate in speckled form (Fig. 5l–j’). During early stages of recovery, a few strings of omega speckles dissociated from 93D cluster to move away in nucleoplasm. With increasing recovery, greater numbers of omega speckles were released from the 93D cluster which spread farther into nucleoplasm. Interestingly, the halo of weaker fluorescence surrounding the bright cluster, as seen during HS (Fig. 4c), was not seen during recovery (Fig. 5). Along with the 93D cluster, the nucleoplasmic omega speckle aggregates, occasionally present in heat shocked cells, also could be seen to release smaller omega speckles which moved independently in nucleoplasm. Several of the released omega speckles progressively disappeared with time, perhaps as their constituents were released into the nucleoplasm. Very little Hrb87F-GFP appeared to associate with chromosomal sites till 30 min recovery from HS.
ISWI chromatin remodeler is required for optimal aggregation of hnRNPs at hsrω locus upon HS and release of omega speckles during recovery
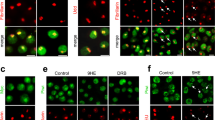
It is known that ISWI, the ATPase subunit of several chromatin remodelers (Corona and Tamkun 2004; Corona et al. 2007), interacts with hsrω-n transcripts and that omega speckles are not properly formed in ISWI-null cells (Onorati et al. 2011). Therefore, we examined the movements of Hrb87F-GFP in live MT cells of ISWI 1/ISWI 2 (ISWI-null) larvae. As can be seen in examples of Hrb87F-GFP fluorescence patterns in fixed (Fig. 6a–c’) and live cells (Fig. 6d–aa), the smaller omega speckles were nearly absent in unstressed ISWI-null cells. Instead, larger speckles, often arranged in strings or streaks (arrows in Fig. 6), were visible, especially in older larvae in which the maternal ISWI contribution is progressively dwindling (Corona et al. 2007). The accumulation of hnRNPs at the hsrω gene locus in heat shocked ISWI-null cells (arrow head in Fig. 6a–c’) generally appeared to be less than in wild-type cells since the area of the hnRNP cluster at 93D site in fixed MT principal cell nuclei after 30 min HS in WT and ISWI 1/ISWI 2 was 7.8 ± 0.5 μm2 (N = 15) and 3.2 ± 0.3 μm2 (N = 15), respectively. Individual or strings of omega speckles persisted in nucleoplasm even after 30 min at 37 °C in ISWI 1/ISWI 2 cells (Fig. 6b, b’, d–i). The release of hnRNPs from the hsrω site during recovery was severely thwarted in ISWI-null MT cells since most of the protein remained at the hsrω gene locus even after >1 h recovery, and only a few persisting large omega speckles, connected with a faint thread of fluorescence (arrows in Fig. 6), were seen in the vicinity of the hsrω locus (Fig. 6c, c’, j–aa). These results suggest that the absence of zygotically synthesized ISWI in ISWI 1/ISWI 2 cells partially affects HS induced clustering of hnRNPs at 93D site and normal biogenesis of omega speckles requires optimal ISWI levels.
Optimal aggregation of Hrb87F-GFP at the 93D site during HS and its removal during recovery requires ISWI. a–c’ Confocal projection images of fixed MT cells of Hrb87F-GFP expressing ISWI-null late third instar larvae: two examples each of unstressed (a, a’), 30 min heat shocked (b, b’), and 30 min HS followed by 2 h recovered (c, c’) cells show the variable appearance of omega speckles and the cluster at hsrω locus. d–aa Time-lapse (5 min intervals) confocal projection images of Hrb87F-GFP expressing ISWI-null MT principal cell during 30 min HS (d–i) and recovery at 23 °C (j–aa). Arrow heads in (a–c’) indicate the hsrω locus; asterisk indicates the chromocenter region with brighter Hrb87F-GFP fluorescence; arrows point to strings of omega speckles
We noticed that the chromocenter region in unstressed ISWI − cells (* in Fig. 6a–c’) showed a little higher presence of Hrb87F-GFP than in wild type. Furthermore, unlike in ISWI + cells, Hrb87F-GFP was also present in the nucleolus. Significance of this change in distribution of Hrb87F in ISWI-null cells is not clear but may be related to the altered chromatin organization and nuclear functions that follow the absence of this important chromatin remodeler.
Mobility of Hrb87F-GFP across different nuclear compartments in unstressed and heat shocked live cells
FRAP and FLIP analyses were carried out in unstressed and heat shocked cells expressing Hrb87F-GFP in hsrω wild type (hsrω +) or hsrω-null (hsrω 66) background to examine if hnRNPs can move between (i) different chromosomal sites, (ii) hsrω/93D site, (iii) omega speckles, and (iv) diffuse nucleoplasmic component. We confirmed that chemically fixed GFP expressing cells neither showed any recovery in FRAP experiments nor lost any fluorescence signal at other sites following continuous photobleaching at a site.
Movements of Hrb87F-GFP between different nuclear locations were examined by FRAP in control and heat shocked peripodial cells. In controls, the ROI was nucleoplasm (Supplementary Fig. 2A), while after HS, ROI was either nucleoplasm or the 93D locus (Supplementary Figs. 2B–C). The “nucleoplasm” ROI in peripodial cells may have included some chromatin regions since they could not be resolved in unstained cells. Figure 7a and Table 1 show that 95 to 100 % fluorescence was recovered within 25 s of photobleaching of “nucleoplasm” ROI indicating rapid mobility of Hrb87F-GFP molecules. The time required for 50 % fluorescence recovery (t1/2) for the nucleoplasmic ROI was comparable in unstressed (0.83 ± 0.12 s) and heat shocked (0.87 ± 0.09 s) cells, suggesting a fast and temperature-independent movement of Hrb87F-GFP. Interestingly, FRAP kinetics were different when ROI for photobleaching in heat shocked cells was the 93D hnRNP cluster. The t1/2 at the 93D locus after 30 min HS was 1.74 ± 0.14 s (Table 1), suggesting a slower efflux of Hrb87F-GFP from the 93D site under HS conditions. This is also supported by the much greater photobleaching achieved at the 93D ROI than at the nucleoplasmic ROI (Table 1, Fig. 7, Supplementary Fig. 2). Comparable values were obtained in SG nuclei (not shown).
The Hrb87F is continuously exchanged between different nuclear compartments. FRAP curves for ROI in nucleoplasm (Nucl) or hsrω gene (93D) in peripodial hsrω + control (Con) or heat shocked (HS) nuclei (a) and for ROI at 74EF/75B puffs in hsrω + or hsrω 66 SG nuclei in control or after 30 min HS or after 30 min recovery from HS (Rec) (b). c FLIP curves for control or heat shocked hsrω + peripodial cells with different ROI1 and ROI2 regions (Nucl = nucleoplasm; 93D = hsrω gene region). d FLIP curves for nucleoplasmic ROI1 and ROI2 regions in control and heat shocked hsrω + or hsrω 66 peripodial cells. The Y-axis shows the normalized relative fluorescence intensity while the X-axis shows time (in s) from the beginning of FRAP (a, b) or FLIP (c, d) experiments. e, f Confocal projection images showing DAPI (red) and Hrb87F (green) fluorescence in heat shocked SG polytene chromosomes (telomere to 93D region of 3R only is shown) from hsrω +/hsrω + (e) and hsrω 66/hsrω 66 (f) larvae. White lines with arrowhead near 93D in (e, f) indicate the paths used for profile analysis, which is shown on right side of each image as fluorescence intensity values (Y-axis, red tracings for DAPI and green for Hrb87F fluorescence) along the chromosome length (X-axis). The mean (±S.E., N = 10) Hrb87F/DAPI fluorescence intensity ratios for 100 F-93 F chromosome region are also noted at bottom in (e, f)
The effect of the absence of hsrω transcripts on exchange of Hrb87F-GFP at developmental 74EF/75B puffs (Supplementary Fig. 2D) was examined by FRAP in Hrb87F-GFP hsrω + and Hrb87F-GFP hsrω 66 homozygous (hsrω-null) larval control, heat shocked, and recovering SG nuclei. Data in Table 1 and Fig. 7b show that the t1/2 in cells with two copies of the wild-type hsrω alleles was comparable with that in hsrω-null cells, indicating independence of general rate of free movement of Hrb87F-GFP from temperature and the presence or absence of hsrω transcripts. Remarkably, however, while more than 90 % fluorescence was recovered in heat shocked and recovering cells with wild-type hsrω alleles, the fluorescence recovery in Hrb87F-GFP hsrω 66 cells in both cases was significantly less (rows 8 and 9 in Table 1, Fig. 7b), indicating that in stressed cells, a fraction of protein does not move out of the chromosome regions in the absence of hsrω transcripts. A greater photobleaching at the ROI in heat shocked hsrω-null cells, reflected by the smaller Bf value (Table 1, rows 5 and 8), also indicates reduced efflux and influx of Hrb87F-GFP from the developmental puff sites in hsrω-null cells. Slow removal of hnRNPs from chromosomal regions in heat shocked hsrω-null cells was also confirmed by comparison of the Hrb87F-GFP fluorescence persisting on chromosome regions in squash preparations of chemically fixed hsrω wild type and hsrω-null heat shocked SG (Fig. 7e, f). Measurement of Hrb87F/DAPI fluorescence intensity ratios in hsrω + and hsrω-null polytene chromosome spreads after 30 min HS through profile analysis (Fig. 7e, f) clearly revealed greater amounts of Hrb87F along the hsrω-null polytene chromosomes than seen in wild type after similar HS; the mean Hrb87F/DAPI fluorescence ratio (relative amount of Hrb87F protein per unit DNA) in hsrω-null cells was also found to be nearly thrice that in hsrω + cells (Fig. 7e, f). A comparable elevated presence of Hrb87F on chromosome regions in heat shocked hsrω-null polytene chromosome spreads was also reported in an earlier publication from our laboratory (Fig. 6 in Lakhotia et al. 2012). These results suggest that hsrω transcripts are necessary for efficient removal of hnRNPs from chromosome regions.
A 2 μm circle, the ROI1, was continuously photobleached for FLIP in control and heat shocked peripodial cells (Table 2 and supplementary Fig. 3) while loss of fluorescence at another ROI2 region was concurrently measured. Fluorescence in ROI2 gradually disappears if the GFP-tagged protein is exchanged between ROI1 and ROI2 (Phair and Misteli 2000). Fluorescence loss was uniform throughout the nucleoplasm following FLIP in unstressed nuclei with more than 90 % signal disappearing within 68 s (Table 2, Fig. 7c, and Supplementary Fig. 3). Movement of Hrb87F-GFP protein in heat shocked cells was assessed by FLIP using two different ROIs (2 μm diameter). In first case, a nucleoplasmic region (ROI1) was photobleached and fluorescence intensity was measured at another nucleoplasmic region and at 93D site (ROI2, rows 2 and 3 in Table 2). In another case, 93D site (ROI1) was photobleached and fluorescence intensity was measured at separate nucleoplasmic region and at the same 93D region (ROI2). Kinetics of fluorescence loss was significantly different between them (Fig. 7c, rows 2–5 in Table 2). Photobleaching at 93D ROI1 in heat shocked nuclei showed faster loss of fluorescence from nucleoplasmic as well as 93D areas, but photobleaching in nucleoplasm of heat shocked nuclei resulted in much slower fluorescence loss at 93D ROI2 site (Table 2), supporting FRAP data that the efflux of Hrb87F-GFP protein from the HS induced 93D aggregate is slower than from nucleoplasmic sites.
FLIP analysis was also carried out in control and heat shocked (Table 2; Fig. 7d and Supplementary Fig. 4) Hrb87F-GFP expressing hsrω + and hsrω-null peripodial cells. Fluorescence signal was lost relatively faster in control as well as heat shocked Hrb87F-GFP hsrω 66 nuclei, suggesting that association of hnRNPs with the nuclear hsrω-n RNA slows down their net movements so that in hsrω-null nuclei, the Hrb87F-GFP can move faster. The greater loss of fluorescence at ROI2 after 68 s (T68) in heat shocked hsrω 66 than in hsrω + nuclei seems to be related to the complete absence of stress induced sequestering of hnRNPs at 93D site in hsrω-null cells.
Discussion
Expression patterns and nuclear distribution of Hrb87F-GFP, Hrb98DE-GFP, or Squid-GFP in the protein-trap lines (Morin et al. 2001; Buszczak et al. 2007) were identical to those of the respective untagged protein and since no mutant phenotype is seen in any of these lines, we believe that the GFP tagging has not affected functions and interactions of these proteins. Consequently, their live imaging is expected to faithfully report the dynamics of the corresponding untagged hnRNP. Although each hnRNP has some unique functions in different cell types (Han et al. 2010; Piccolo et al. 2014), the three hnRNPs share some properties including nuclear presence, association with the active chromatin sites, presence in the hsrω-nuclear transcript-dependent omega speckles, and their almost complete aggregation at the hsrω locus following stress (Prasanth et al. 2000; Jolly and Lakhotia 2006; Lakhotia 2011, 2012). Thus, information obtained with one GFP tagged hnRNP can in general be applicable to others, especially in relation to hsrω-n transcripts and omega speckles.
Our FRAP and FLIP studies showed continuous and rapid exchange of hnRNPs between nucleoplasm, omega speckles, hsrω/93D locus, and other developmentally active chromosome sites. Such stochastic mobility of regulatory proteins between different nuclear compartments is significant for cell’s ability to dynamically and rapidly respond to continuously changing internal and external environments. Differences in the FRAP and FLIP curves for hnRNPs located in nucleoplasm and those at hsrω gene site in unstressed and stressed cells indicate that while the movement in nucleoplasm may be stochastic, the directional movements towards or away from the hsrω/93D site involve additional factors. Although FRAP/FLIP studies with rapidly moving omega speckles were not possible, we believe that efflux of hnRNPs from omega speckles is also regulated rather than completely stochastic.
A significant interaction of omega speckles with nuclear matrix is indicated by (i) close association or colocalization of omega speckles with Megator and SAFB nuclear matrix-associated proteins, (ii) co-aggregation of hnRNPs and SAFB in RNase-treated nuclei, (iii) co-IP of Hrb87F with Megator and SAF-B and presence of hsrω-n transcripts in the Megator as well as SAF-B IP samples, and (iv) accumulation of Megator and SAFB at hsrω locus following HS. While the hnRNP and nuclear matrix association is known for long (He et al. 1991; Zimowska et al. 1997), we now show that omega speckles too are associated with nuclear matrix components. This association reflects an important role of nuclear matrix in omega speckle and hnRNP dynamics.
Cell stress and transcription inhibition affect a variety of nuclear speckles (Spector and Lamond 2011; Lakhotia 2012). However, the rapid relocation of omega speckle associated proteins in stressed cells to the 93D site is unique. This more than three decades old observation (Saumweber et al. 1980; Dangli and Bautz 1983; Prasanth et al. 2000) has remained intriguing. Present observations shed some light on processes underlying the rapid and synchronous relocation of many different proteins to one specific chromosomal site. Contrary to earlier suggestion (Prasanth et al. 2000; Lakhotia 2011, 2012) that cell stress may cause smaller omega speckles to form larger clusters in nucleoplasm which finally move and get sequestered at the hsrω locus, we did not find any omega speckles moving towards the hsrω site in control, HS, or colchicine-treated live cells. Instead, they always disappeared within nucleoplasm. High resolution imaging of heat shock cells strongly indicated that hnRNPs and other colocalizing proteins, stream into hsrω gene site from all directions as sub-microscopic entities possibly added by as yet unidentified nuclear matrix tracks for rapid and directional movement. Earlier studies from our laboratory (Lakhotia et al. 2012; Singh and Lakhotia 2012) have shown that when Hrb87F or hsrω-n transcripts are absent, neither hsrω-n transcripts nor Hrb87F, respectively, exist as nucleoplasmic speckles. Therefore, disappearance of hnRNP speckles following heat shock or otherwise implies that the associated hsrω-n transcripts are also simultaneously released from the disappearing omega speckles. Since the level of hsrω-n transcripts in nucleoplasm does not concomitantly increase (Prasanth et al. 2000), we believe that these transcripts are turned over with the release of hnRNPs from omega speckles, which thus disappear. Such continuous turnover of hsrω-n transcripts is also supported by the maintenance of a steady state level of these transcripts (Bendena et al. 1989) in spite of continued new transcription at the gene locus in unstressed cells (Mukherjee and Lakhotia 1979).
The effect of down- or upregulation of the hsrω transcripts on accumulation of hnRNPs at hsrω site during HS (Mallik and Lakhotia 2011; Lakhotia et al. 2012), and the removal of hnRNPs from the 93D site by in vivo RNase treatment (Singh and Lakhotia 2012), suggested the association of these proteins with the gene’s transcripts. Present findings of complete absence of hnRNP aggregation at 93D site in hsrω 66 chromosome (Johnson et al. 2011) and in α-amanitin or actinomycin-D-treated wild-type cells establish that the hnRNPs released from chromosomes and omega speckles and being moved to the hsrω gene site are retained there by the actively synthesized new transcripts. The slow removal of chromosomal hnRNPs during HS in hsrω-null cells (Mallik and Lakhotia 2011; Lakhotia et al. 2012, and present results) indicates that hsrω nuclear transcripts also act as “sponge” for the released hnRNPs.
Based on our present live cell studies and earlier observations, a model for movements of different hnRNPs in relation to the hsrω transcripts in normal, stressed, and recovering cells is presented (Fig. 8). In normal and stressed cells, the hnRNPs move between different nuclear compartments by stochastic as well as regulated active processes. Movements in and out of omega speckles, chromosome regions, and hsrω site are more likely to be regulated processes, possibly involving nuclear matrix components. Continuing developmental transcription at hsrω site entails greater presence of hnRNPs at this site at all times. Some hnRNPs may leave the hsrω site by stochastic events, but majority leave only as organized omega speckles, which seem to require ISWI for their biogenesis (Onorati et al. 2011 and present observations). As discussed above, we suggest that the release of hnRNPs (and other proteins) from the omega speckles is related to breakdown of the omega speckles-associated hsrω-n transcripts so that as the omega speckle disappears, the hnRNPs move into the non-particulate nucleoplasmic pool.
Dynamics of hnRNPs and omega speckles in Drosophila cell nuclei. A model for the movements of hnRNPs (exemplified with Hrb87F) in unstressed (a), heat shocked (b, c, after 10 and 30 min HS, respectively) cells and in those recovered from HS for 30 min (d). Chromatin is shown as a double helix; 93D, L1, and L2 indicate the hsrω and two developmentally active gene loci, respectively, with which hnRNPs are associated. The diffuse nucleoplasmic hnRNPs and hsrω-n transcripts are shown as independent entities but it is possible that they may remain together without forming larger visible entities. Paired and oppositely oriented arrows close to the 93D, L1, and L2 gene sites reflect efflux (E) and influx (I) of hnRNPs from a given locus with thickness of the arrow indicating relative quantity of efflux or influx through stochastic and/or regulated movements. Thick arrows in nucleoplasm indicate the stochastic and/or regulated movement of hnRNPs in stressed cells towards the 93D site while dotted thick arrows indicate the emergence of omega speckles from this site. Thin arrows in nucleoplasm point to break down of omega speckles to release hnRNPs and hsrω transcripts in the nucleoplasm; the transcripts are often concomitantly degraded. For simplicity, the nuclear matrix, ISWI, and other factors that regulate directed active transport of hnRNPs are not shown
Cell stress modifies the above steady-state dynamics of these proteins resulting in disappearance of nucleoplasmic omega speckles, release of chromosomal hnRNPs, and an increased influx of hnRNPs to the hsrω site. Stress induced release from chromosome sites and the disappearing omega speckles enrich nucleoplasmic hnRNPs, which diffuse stochastically or, more likely, are actively and directionally transported by components of nuclear matrix to hsrω site, where they are trapped by the concurrently increasing new hsrω transcripts. With continuing stress, balanced stochastic outward and inward movements establish a steady state. Elevated levels of Hsp83 at the 93D site in stressed cells (Morcillo et al. 1993) may facilitate formation of very large ribonucleoprotein (RNP) particles at 93D site (Derksen and Willart 1976; Dangli et al. 1983). Activity of ISWI, which accumulates at this site in stressed cells (Singh and Lakhotia 2012), may also be regulated so that biogenesis of omega speckle during stress remains inhibited. These aspects need further studies.
As cells recover from stress, the hnRNPs and other proteins are rapidly released from the 93D site for restoration of developmental transcriptional activity. Since both the hsrω-nuclear transcripts and Hrb87F are essential for the formation of omega speckles (Mallik and Lakhotia 2011; Singh and Lakhotia 2012), we believe that during recovery, as also in normal cells, the new hsrω transcripts and the accumulated Hrb87F protein together prime biogenesis of omega speckles by facilitating other protein partners to associate so that, with the help of ISWI, the individualized omega speckles are released into nucleoplasm. Other studies in our laboratory (Deoprakash Chaturvedi and S. C. Lakhotia, unpublished) suggest that some other components of the chromatin remodeling complexes like Nurf301, Nurf 38, and GCN5 also affect omega speckle biogenesis.
The hsrω transcripts are among the earliest known essential and functionally conserved noncoding RNAs (Lakhotia and Mukherjee 1982; Lakhotia and Singh 1982; Garbe et al. 1986) that were identified to be essential for organization of a class of nuclear speckles (Lakhotia et al. 1999; Prasanth et al. 2000). Present studies provide new insights into dynamicity of hnRNPs in different compartments of nuclei and their intriguing behavior during cell stress. However, several questions remain to be understood. While it is shown that omega speckles are not formed if either hsrω-n transcripts or Hrb87F is absent (Mallik and Lakhotia 2011; Singh and Lakhotia 2012), it is not known if the other hnRNPs and other omega speckle proteins associate directly with the hsrω-n transcripts or Hrb87F or they bind cooperatively to the hsrω-n and Hrb87F complex. We need to understand the specific roles of Tpr/Megator, SAFB, and other nuclear matrix components in the dynamics of hnRNPs and omega speckles. The site of PARylation and dePARylation of hnRNPs (Ji and Tulin 2009) and the role of omega speckles or hsrω-n transcripts in these events also need to be examined. The recently annotated (http://flybase.org) ∼21 kb hsrω-F transcripts, which also presumably remain nuclear, add further complexity in nuclear dynamics of hnRNPs etc. Novel experimental approaches are needed to elucidate the mechanism and the factor/s regulating the release of hnRNPs and other associated proteins from omega speckles with concomitant breakdown of hsrω-n transcript and disappearance of omega speckles. Future studies on live cells in which the hsrω-n transcripts and the other associated proteins are fluorescently tagged will be useful.
Present observations on dynamics of hnRNPs in normal and stressed Drosophila cells have bearing upon dynamics of other lncRNA-dependent nuclear speckles. Of particular interest in this context is the formation of nuclear stress bodies at the transcriptionally activated Sat III genomic loci in stressed human cells since the Sat III lncRNAs are believed to be functional analogues of the hsrω-n transcripts (Jolly and Lakhotia 2006; Lakhotia 2012).
References
Alfonso-Parra C, Maggert KA (2010) Drosophila SAF-B links the nuclear matrix, chromosomes, and transcriptional activity. PLoS One 5:e10248
Arao Y, Kuriyama R, Kayama F, Kato S (2000) A nuclear matrix-associated factor, SAF-B, interacts with specific isoforms of AUF1/hnRNP D. Arch Biochem Biophys 380:228–236
Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW (1976) Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J 16:1055–1069
Bendena WG, Garbe JC, Traverse KL, Lakhotia SC, Pardue ML (1989) Multiple inducers of the Drosophila heat shock locus 93D (hsr omega): inducer-specific patterns of the three transcripts. J Cell Biol 108:2017–2028
Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A et al (2007) The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175:1505–1531
Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677:113–119
Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW (2007) ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5:e232
Dangli A, Bautz EK (1983) Differential distribution of nonhistone proteins from polytene chromosomes of Drosophila melanogaster after heat shock. Chromosoma 88:201–207
Dangli A, Grond C, Kloetzel P, Bautz EK (1983) Heat-shock puff 93D from Drosophila melanogaster: accumulation of a RNP-specific antigen associated with giant particles of possible storage function. EMBO J 2:1747–1751
Derksen J, Willart E (1976) Cytochemical studies on RNP complexes produced by puff 2-48BC in Drosophila hydei: uranyl acetate and phosphotungstic acid staining. Chromosoma 55:57–68
Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M et al (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5:355–365
Garbe JC, Bendena WG, Alfano M, Pardue ML (1986) A Drosophila heat shock locus with a rapidly diverging sequence but a conserved structure. J Biol Chem 261:16889–16894
Gorisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P (2004) Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci U S A 101:13221–13226
Han SP, Tang YH, Smith R (2010) Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430:379–392
He DC, Martin T, Penman S (1991) Localization of heterogeneous nuclear ribonucleoprotein in the interphase nuclear matrix core filaments and on perichromosomal filaments at mitosis. Proc Natl Acad Sci U S A 88:7469–7473
Hochstrasser M, Sedat JW (1987) Three-dimensional organization of Drosophila melanogaster interphase nuclei II Chromosome spatial organization and gene regulation. J Cell Biol 104:1471–1483
Ip JY, Nakagawa S (2012) Long non-coding RNAs in nuclear bodies. Develop Growth Differ 54:44–54
Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP (2012) Advanced fluorescence microscopy techniques–FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17:4047–4132
Ji Y, Tulin AV (2009) Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res 37:3501–3513
Johnson TK, Cockerell FE, McKechnie SW (2011) Transcripts from the Drosophila heat-shock gene hsr-omega influence rates of protein synthesis but hardly affect resistance to heat knockdown. Mol Genet Genomics 285:313–323
Jolly C, Lakhotia SC (2006) Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res 34:5508–5514
Klonis N, Rug M, Harper I, Wickham M, Cowman A, Tilley L (2002) Fluorescence photobleaching analysis for the study of cellular dynamics. Eur Biophys J 31:36–51
Lakhotia SC (2011) Forty years of the 93D puff of Drosophila melanogaster. J Biosci 36:399–423
Lakhotia SC (2012) Long non-coding RNAs coordinate cellular responses to stress. WIREs RNA 3:779–796
Lakhotia SC, Mukherjee T (1982) Absence of novel translation products in relation to induced activity of the 93D puff in Drosophila melanogaster. Chromosoma 85:369–374
Lakhotia SC, Mukherjee T (1984) Specific induction of the 93D puff in polytene nuclei of Drosophila melanogaster by colchicine. Indian J Exp Biol 22:67–70
Lakhotia SC, Singh AK (1982) Conservation of the 93D puff of Drosophila melanogaster in different species of Drosophila. Chromosoma 86:265–278
Lakhotia SC, Ray P, Rajendra TK, Prasanth KV (1999) The non-coding transcripts of hsr-omega gene in Drosophila: do they regulate trafficking and availability of nuclear RNA-processing factors? Curr Sci 77:553–563
Lakhotia SC, Mallik M, Singh AK, Ray M (2012) The large noncoding hsromega-n transcripts are essential for thermotolerance and remobilization of hnRNPs, HP1 and RNA polymerase II during recovery from heat shock in Drosophila. Chromosoma 121:49–70
Lippincott-Schwartz J, Snapp E, Kenworthy A (2001) Studying protein dynamics in living cells. Nat Rev Mol Cell Biol 2:444–456
Mallik M, Lakhotia SC (2011) Pleiotropic consequences of misexpression of the developmentally active and stress-inducible non-coding hsromega gene in Drosophila. J Biosci 36:265–280
Mao YS, Sunwoo H, Zhang B, Spector DL (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13:95–101
Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17:639–647
Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20:300–307
Millet LJ, Gillette MU (2012) Over a century of neuron culture: from the hanging drop to microfluidic devices. Yale J Biol Med 85:501–521
Misteli T, Caceres JF, Spector DL (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523–527
Morcillo G, Diez JL, Carbajal ME, Tanguay RM (1993) HSP90 associates with specific HS puffs (hsr omega) in polytene chromosomes of Drosophila and Chironomus. Chromosoma 102:648–659
Morin X, Daneman R, Zavortink M, Chia W (2001) A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A 98:15050–15055
Mukherjee T, Lakhotia SC (1979) 3H-uridine incorporation in the puff 93D and in chromocentric heterochromatin of heat shocked salivary glands of Drosophila melanogaster. Chromosoma 74:75–82
Nickerson JA, Krochmalnic G, Wan KM, Penman S (1989) Chromatin architecture and nuclear RNA. Proc Natl Acad Sci U S A 86:177–181
Nissim-Rafinia M, Meshorer E (2011) Photobleaching assays (FRAP & FLIP) to measure chromatin protein dynamics in living embryonic stem cells. J Vis Exp 52: doi: 10.3791/2696
Onorati MC, Lazzaro S, Mallik M, Ingrassia AM, Carreca AP, Singh AK, Chaturvedi DP, Lakhotia SC, Corona DF (2011) The ISWI chromatin remodeler organizes the hsromega ncRNA-containing omega speckle nuclear compartments. PLoS Genet 7:e1002096
Pederson T (2000) Half a century of "the nuclear matrix". Mol Biol Cell 11:799–805
Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404:604–609
Piccolo LL, Corona D, Onorati MC (2014) Emerging Roles for hnRNPs in post-transcriptional regulation: what can we learn from flies? Chromosoma. doi:10.1007/s00412-014-0470-0
Place RF, Noonan EJ (2014) Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian HS response. Cell Stress Chaperones 19:159–172
Platani M, Goldberg I, Lamond AI, Swedlow JR (2002) Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol 4:502–508
Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC (2000) Omega speckles—a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci 113:3485–3497
Reed BH, McMillan SC, Chaudhary R (2009) The preparation of Drosophila embryos for live-imaging using the hanging drop protocol. J Vis Exp. doi:10.3791/1206
Saumweber H, Symmons P, Kabisch R, Will H, Bonhoeffer F (1980) Monoclonal antibodies against chromosomal proteins of Drosophila melanogaster: establishment of antibody producing cell lines and partial characterization of corresponding antigens. Chromosoma 80:253–275
Singh AK, Lakhotia SC (2012) The hnRNP A1 homolog Hrp36 is essential for normal development, female fecundity, omega speckle formation and stress tolerance in Drosophila melanogaster. J Biosci 37:659–678
Sleeman JE, Trinkle-Mulcahy L (2014) Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 28:76–83
Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3:a000646
Szczepny A, Hogarth CA, Young J, Loveland KL (2009) Identification of Hedgehog signaling outcomes in mouse testis development using a hanging drop-culture system. Biol Reprod 80:258–263
Tapadia MG, Lakhotia SC (1997) Specific induction of the hsr omega locus of Drosophila melanogaster by amides. Chromosome Res 5:359–362
Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT (2007) Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28:978–990
Zimowska G, Paddy MR (2002) Structures and dynamics of Drosophila Tpr inconsistent with a static, filamentous structure. Exp Cell Res 276:223–232
Zimowska G, Aris JP, Paddy MR (1997) A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110:927–944
Zobeck KL, Buckley MS, Zipfel WR, Lis JT (2010) Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40:965–975
Acknowledgments
We thank Dr. H. Saumweber (Germany) for P11 and BX34 antibodies, Dr. Keith A. Maggert (USA) for UAS-SAFB-GFP stock, and Drs. Stephen W. Mckechnie (Australia) for hsrω 66, D. Corona (Italy) for ISWI 1 Bc/SM5 and ISWI 2; +/T(2;3)CyO, TM6B, Tb, Allan C. Spradling, (USA) for Hrb87F-GFP, and Alain Debec (France) for Squid-GFP and Hrb98DE-GFP stocks. This work was supported by the CEIB-II grant from Department of Biotechnology, Govt. of India and by the Board of Research in Nuclear Sciences (Department of Atomic Energy, Govt. of India) through Raja Ramanna Fellowship to SCL. We thank the Department of Science & Technology, Govt. of India (New Delhi) and the Banaras Hindu University for Confocal Microscopy facility in our laboratory. AKS has been supported as Senior Research Fellow by the Council of Scientific & Industrial Research (New Delhi) and as Research Associate by the Department of Biotechnology, Govt. of India.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Stable peripheral location of the hsrω gene site and reproducible accumulation and redistribution of hnRNPs during repeated HS and recovery periods. Time lapse live cell confocal images of Hrb87F-GFP expressing hsrω + SG nucleus at 5 min intervals (noted at upper left corner of each panel) during repeated HS and recovery periods. The on-stage incubation temperature for each row is indicated on left. (GIF 120 kb)
Fig. S2
FRAP of Hrb87F-GFP in nucleoplasm and at the 93D site in wild type cells. (A) FRAP in unstressed control peripodial cell at nucleoplasmic ROI (red circles). (B, C) FRAP after 30 min HS at nucleoplasmic ROI (B) or at 93D cluster ROI (C). (D) FRAP at ROI in the 74EF/75B developmental puffs in unstressed SG polytene nucleus. In all cases, confocal images of the same optical section are shown at prebleach stage, just after bleaching (t = 0.0 s) and at different time points (in s) thereafter (noted in top row of each column). Scale bar applies to all images. (GIF 221 kb)
Fig. S3
Hrb87F-GFP is exchanged between different nuclear compartments. (A) FLIP in unstressed control peripodial cell with ROI1 (red circle) and ROI2 (blue circle) being different sites in the nucleoplasm. (B) FLIP in 30 min heat shocked peripodial cell with ROI1 being nucleoplasmic (red circle), ROI2 being aggregate at 93D site (green circle) and ROI3 being a nucleoplasmic area (pink circle). (C) FLIP in 30 min heat shocked peripodial cell with the 93D cluster as ROI1 (red circle), ROI2 being the same region at 93D cluster (sky blue circle) and ROI3 being an area in nucleoplasm (purple circle). Each row shows confocal images of same optical section at different time points (in s) noted in top row of each column. The scale bar in 1st column of top row applies to all images. (GIF 171 kb)
Fig. S4
Association of hnRNPs with hsrω transcripts slows down their movement. FLIP in unstressed Hrb87F-GFP hsrω + (A) unstressed Hrb87F-GFP hsrω 66 peripodial cell (B) and heat shocked Hrb87F-GFP hsrω + peripodial cell (C). Green circles indicate photobleached nucleoplasmic ROI1 while red circles indicate nucleoplasmic ROI 2 region used to measure loss of fluorescence at different time points (in s) indicated in top row of each column. Genotype of cells is indicated on left of each row. The scale bar in 1st column of top row applies to all images. (GIF 203 kb)
Rights and permissions
About this article
Cite this article
Singh, A.K., Lakhotia, S.C. Dynamics of hnRNPs and omega speckles in normal and heat shocked live cell nuclei of Drosophila melanogaster . Chromosoma 124, 367–383 (2015). https://doi.org/10.1007/s00412-015-0506-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-015-0506-0