Abstract
Clustered DNA damage induced by 10, 20 and 30 MeV protons in pBR322 plasmid DNA was investigated. Besides determination of strand breaks, additional lesions were detected using base excision repair enzymes. The plasmid was irradiated in dry form, where indirect radiation effects were almost fully suppressed, and in water solution containing only minimal residual radical scavenger. Simultaneous irradiation of the plasmid DNA in the dry form and in the solution demonstrated the contribution of the indirect effect as prevalent. The damage composition slightly differed when comparing the results for liquid and dry samples. The obtained data were also subjected to analysis concerning different methodological approaches, particularly the influence of irradiation geometry, models used for calculation of strand break yields and interpretation of the strand breaks detected with the enzymes. It was shown that these parameters strongly affect the results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For a long time, it was generally accepted that biological effects of ionizing radiation are related to nuclear DNA damage in a single cell, directly traversed by the radiation. Unsuccessful repair of the induced DNA damage results in cell death, or in genomic instability of surviving cells and possibly in malignant cell transformation (Willers et al. 2004). First, this concept was challenged with evidence of bystander effects, showing that biological effects induced in cells that are not directly traversed by radiation, but are in proximity to hit cells (Hall and Hei 2003). Second, DNA as the only target from the point of view of carcinogenesis has been doubted, and it has been speculated that the main cellular target are proteins (Gebicki and Gebicki 1999; Watanabe 2007) or other cell compartments (Stefančíková et al. 2014). However, even if the relation of DNA damage to carcinogenesis is not as crystal clear as had been believed earlier, it still plays an important role.
The radiation action in a living cell takes place in four consecutive stages in time. In the physical stage, the energy of primary and secondary particles is transferred to molecules of the medium in a series of ionization and excitation events. Energy depositions on the DNA molecule and in the bound water layer result in directly induced primary DNA damage. Events in the surrounding environment lead to dissociation of water molecules and thus give rise to initial chemical products, including oxidative radical species. After this physicochemical stage, the chemical stage begins: The chemical species diffuse in the environment and undergo chemical reactions with each other and also with biological structures, leading to indirectly induced primary DNA damage. The majority of the indirect damage is attributed to hydroxyl radicals (von Sonntag 2006). The track gradually expands in space, and at about 1 μs, the spatial distribution of chemical species is close to homogenous. In living organisms, these primary DNA damages are then processed and possibly repaired during the biological stage.
DNA lesions consist of strand breaks and base damages, which can be isolated (single strand breaks—SSBs and single base damages) or clustered within a few DNA helical turns (double strand breaks—DSBs and clustered base damages). The clustered lesions constitute the most critical type of DNA damage with a risk of misrepair depending on the type and spatial distribution of lesions within a cluster (Asaithamby et al. 2011; Shikazono et al. 2009). The yield and the spectrum of the induced clustered damages in a given system depend on ionization density that can be linked to linear energy transfer (LET). Monte Carlo simulations predict that while the average number of induced individual DNA lesions per dose is independent on LET, their distribution within a DNA is LET dependent: With increasing LET, the lesions tend to cluster within a smaller number of clusters leading to a higher level of complexity (Semenenko and Stewart 2006; Georgakilas et al. 2013). An experimental confirmation of these predictions is not an easy task. Measurements of DNA clustered damage, comparing an increase in strand breaks detected with and without base excision repair enzymes (Sutherland et al. 2001), have several limitations (Pachnerová Brabcová et al. 2014), and therefore, the results regarding LET dependence are not conclusive (Leloup et al. 2005). However, it was proved that the clustered lesions are induced by both low-LET (Milligan et al. 2000; Gulston et al. 2002; Do et al. 2012) and high-LET radiation (Prise et al. 1999; Terato and Ide 2004; Hada and Georgakilas 2008; Pachnerová Brabcová et al. 2014) and the number of oxidized bases exceeds the number of strand breaks, particularly for sparsely ionizing radiation.
Proton radiation exhibits a specific combination of both sparsely and densely ionizing radiation properties. Lately, biological effects of protons came into the focus of interest as the dominant ionizing particles in both galactic cosmic rays and solar particle events. Hence, they are one of the major health risks for astronauts during prolonged space missions (Durante and Cucinotta 2008). Furthermore, proton radiation is increasingly used in tumor treatment for its selective dose delivery to the tumor while lowering an unwanted dose in surrounding healthy tissue (Girdhani et al. 2013).
Studies focused on the primary DNA damage by ionizing radiation often use a plasmid DNA as a simplified model system, in order to abstract from the eukaryotic cell complexity. Such system allows to omit or to restrict cell biological repair processes. The plasmid can be irradiated in dried form, where the indirect effects are fully suppressed, or in liquid water, where the indirect effects can be moderated by scavengers. Therefore, the contribution of indirect and direct effects can be distinguished. The size of plasmids, up to tens of kilo base pairs (kbp), allows an easy detection of DNA damage using conventional electrophoretic techniques.
This study contributes to the discussion about clustered DNA damage induced by protons in one of the most extensively used plasmid model systems, pBR322 (Bolivar et al. 1977), middle-sized E. coli vector (4361 bp) with known nucleotide sequence. The presented clustered DNA damage data induced in dry and liquid samples were subjected to analysis concerning different methodological approaches and their influence on data comparability.
Materials and methods
DNA sample preparation
The plasmid DNA (pBR322, New England Biolabs Inc) supplied in 1× TE solution (10 mM Tris–HCl, pH 8.0, 1 mM EDTA) contained over 95 % of supercoiled form. It was verified with agarose gel electrophoresis, as described further. The original plasmid concentration of 1000 ng/μl was diluted with deionized water to a final concentration of 30 ng/μl.
The concentration of TE in the diluted samples was verified using a HPLC system consisting of binary pumps SDS20 and SDS30, vacuum degasser X-Act, automatic sampler AS 1000, column Macherey–Nagel Nucleosil 100-5 SA (250 × 4.4 mm, 10 μm particle size) and conductivity detector Shodex-CD5 connected via PCI Express card with SW Clarity 3.0. The mobile phase consisted of acetonitrile and water (90:10, v/v) with constant flow rate of 1 ml/min. Acetonitrile, 99 % HPLC grade, was purchased from Sigma-Aldrich, and deionized water was produced on a Millipore Direct Q3 water purification station (Merck, Czech Republic). The injection volume was 10 μl, and the retention time was 1.5 min. Calibration samples were prepared in concentration range from 1.95 × 10−3 to 2 × TE. The concentration in the diluted TE samples was calculated by averaging three measurements to (0.028 ± 0.006) × TE.
Three 5-μl aliquots, each containing 150 ng of plasmid DNA, were pipetted onto 20 × 20 mm2 glass coverslip and dried in nitrogen-filled desiccator cabinet for 60 min. The dry sample consisted of a layer of plasmid and buffer salts with a diameter of 3.40 ± 0.04 mm. The coverslips were attached to a Petri dish and mounted on a sample holder perpendicular to the beam axis, as shown in Fig. 1a.
For the preparation of liquid samples, the plasmid was diluted in 20 mM potassium phosphate buffer, pH 7.0, to concentration of 15 ng/µl. Residual TE buffer comprises hydroxyl radical scavenger Tris with scavenging capacity k × [S], where k = 1.5 × 105 M−1 s−1 (Prise et al. 1999) is the rate constant and [S] = 0.14 mM is the concentration of Tris in 0.014× TE buffer. The samples (30 µl) were placed in polypropylene microtubes and irradiated on a sample holder, as shown in Fig. 1b. After irradiation, three aliquots of 150 ng DNA were used for further analysis.
Proton irradiation
Plasmid DNA samples were irradiated at the U-120M isochronous cyclotron at the Nuclear Physics Institute CAS. Dry samples were irradiated by 10, 20 and 30 MeV protons. Doses up to 2000 Gy were delivered at a dose rate of 400 Gy/min. Liquid samples were irradiated by proton beams with 20 and 30 MeV energy with doses up to 20 Gy at 4 Gy/min.
Doses and dose rates were monitored using a PTW Unidos universal dosimeter (PTW, Germany) equipped with a NE 2581 ionization chamber (Nuclear Enterprises Ltd, UK) fixed at the sample holder. Uniformity of the beam was verified with Gafchromic films (XR-RV3, Ashland, USA) placed at the sample holder, irradiated with doses up to 10 Gy.
The cyclotron delivers the dose in pulses; for dose rate ~400 Gy/min, there are 150 macro-pulses per second, each beginning with a 0.333 ms group of micropulses with FWHM ~5.4 × 10−9 s width (Fig. 2). For liquid samples, the dose rate was decreased to 4 Gy/min by adjusting the parameters of the internal ion source. In comparison with high-peak-power laser-driven proton sources (Bin et al. 2012; Bolton et al. 2014; Kraft et al. 2010; Malka et al. 2008; Yogo et al. 2011; Zlobinskaya et al. 2014), the dose in this study is delivered in enormous number of bunches, thus serving as a low-dose-rate reference for investigations of dose rate effects in radiation biophysics.
Liquid samples were irradiated in the lower conical part and half-sphere tip of polypropylene microtubes. At the widest point, the diameter of the sample was 2.67 ± 0.06 mm and the tube walls was 0.87 ± 0.10 mm thick. With the sample described as water, the maximum entry and minimum exit energies for the sample volume were estimated using the PSTAR database (Berger et al. 2005). For 30 MeV beam, the energy inside a sample changed from 28.5 to 22.8 MeV, which corresponds to an increase in LET in water from 1.96 to 2.34 keV/μm. For 20 MeV, the energy and LET changes were more pronounced. The energy decreased from 17.7 to 5.9 MeV and LET increased from 3.10 to 6.95 keV/μm. As confirmed by a SRIM 2013 (Ziegler et al. 2010) calculation, for 20 and 30 MeV the whole volume was irradiated and the particles were stopped safely behind the sample (Fig. 3). This would not be the case for 10 MeV protons; therefore, only 20 and 30 MeV beams were used for liquid sample irradiation.
In order to limit the indirect effect of ionizing radiation on the sample, a set of dry samples were irradiated as well. The relative humidity in the beam room was 38–43 %, which corresponds to eight water molecules per nucleotide in a dry DNA sample (Swarts et al. 1992).
Real doses delivered to the dry samples were calculated from the doses in water as measured by an ionization chamber by correcting for density and material composition. The LET for 10, 20 and 30 MeV protons in water and in the dry sample was determined using SRIM 2013. The dry samples consist of 150 ng pBR322, 70 ng water and 222 ng TE. The stoichiometric composition, when omitting small P content, was estimated to be C25H64N6O20Na, and the density 1.407 g/cm3 was taken into account (Śmiałek et al. 2013). The calculated LET for dry samples was 1.373× higher than the LET for liquid samples. This correction was taken into account in data evaluation.
Gamma irradiation
For comparison, the dry and liquid samples were also irradiated with 60Co source under the same conditions as described for proton irradiation. The dose rate of the source was ~6 Gy/min. The average LET of the secondary electrons produced by 60Co in water was about 0.2 keV/μm (ICRU Report 16 1970). For the dry samples, similarly as in the case of protons, a 1.373× increase in LET was taken into account.
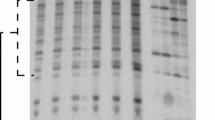
Agarose gel electrophoresis
Irradiated dry samples were recovered from the coverslips with 10 µl of deionized water. All samples were analyzed by agarose gel electrophoresis (AGE). The samples were run on a neutral 1 % agarose under 100 V for 80 min in 0.5× TAE buffer (20 mM Tris, 10 mM acetic acid, 0.5 mM EDTA, pH 8.0). Gels were stained with fluorescent dye SYBR Green I (Sigma-Aldrich) and imaged on a UV transilluminator table (UVT-20 ME, Herolab) with a Panasonic DMC-GF2 digital camera. Excitation wavelength was filtered out using an appropriate band-pass filter (Wratten 15, Sigma-Aldrich). The acquired images were transformed to gray scale (8 bits), and peaks corresponding to supercoiled (S), open-circular (C) and linear (L) plasmid forms were integrated using an in-house-developed software Luthien.
Enzymatic treatment
Selected samples were treated with the E. coli base excision repair enzymes Fpg (formamimopyrimidine DNA glycosylase) and Nth (endonuclease III), both New England Biolabs Inc, in 37 °C dry bath to reveal oxidized purine and pyrimidine bases, which manifest as additional strand breaks detected with AGE. Fpg samples were incubated with 0.8 U Fpg and specific reaction buffer containing 10 mM bis–Tris–propane–HCl, 10 mM MgCl2 and 1 mM dithiothreitol (DTT) at pH 7.0 supplied with 100 μg/ml bovine serum albumin for 60 min. Nth samples were incubated with 1 U Nth and specific reaction buffer containing 20 mM Tris–HCl, 1 mM EDTA and 1 mM DTT at pH 8.0 for 30 min. The enzymatic activity was quenched with 2 µl of DNA loading dye (Thermo Scientific) containing 60 % glycerol, 0.03 % xylene cyanol, 0.03 % bromophenol blue, 60 mM EDTA and 10 mM Tris–HCl at pH 7.6, and the samples were allowed to cool at 4 °C for 30 min before run on AGE.
The optimal enzyme concentrations were found by the following procedure. Two sets of samples with 100 ng of pBR322 dissolved in deionized water were prepared. The first set was irradiated by 60Co with dose of 5 Gy, and the second remained non-irradiated. Both sets were treated with increasing concentrations of Fpg (up to 5 U) and Nth (up to 10 U), and open-circular fractions were analyzed with AGE. The titration curves for Fpg are shown in Fig. 4. Plasmid open-circular fractions increased with enzyme concentration because of the specific cleavage of damaged bases until plateau regions were reached. The overlapping range for both sets was within 0.7–2.4 U. For concentrations higher than 2.4 U, non-specific cleavage in non-irradiated samples can be induced due to excess amount of enzyme. As a safe value, 0.8 U was chosen for the experiment.
Titration curves for the Fpg enzyme: damage detected in the set irradiated by γ-rays (full symbols) and non-irradiated (open symbols) as a function of the Fpg enzyme concentration. Gray zone represents the safe zone with maximal specific and minimal un-specific cleavage. The dashed vertical line illustrates the selected enzyme concentration
Models for data analysis
Multiple mathematical models were developed to compute the probability of induction of SSB and DSB per dose from the electrophoresis results (Śmiałek 2012). A frequently used model was developed by Cowan et al. (1987). It was primarily developed to describe the transition between plasmid forms during restriction enzyme treatment, which corresponds to the dynamics after irradiation with dose (D). The full version of the model is difficult to implement due to its complexity and is rarely used. Most of the studies implemented simplified equations, where the induction of a linear form by coincidence of two independent events is neglected:
Parameters \( \mu \) and ϕ are the average numbers of single and double strand breaks per plasmid and Gy; μ 0 and ϕ 0 are initial parameters at zero dose.
Recently, a new model has been introduced by McMahon and Currell (2011), which is assumed to be more robust. The model uses the following equations in integrated form, where the factor \( \rho \) represents the probability that a DSB is formed by two or more SSB events on opposite strands within a certain distance, and S 0 and C 0 are, respectively, supercoiled and open-circular fractions at zero dose:
Both models were implemented in Origin 8.5 (OriginLab Corp, USA), with ρ = 10/4361 for McMahon model, and they were compared in order to verify which of them provides a better fit to the measured data. For that purpose, an artificial set of measured-like data was created using μ and ϕ parameters calculated from selected set of real measured data. The measured-like set was generated for each dose as normal distributed values with mean defined by the parameters and standard deviation given by experimental error. The generated data were normalized and fitted with both models.
Results
DNA damage yields
Proton-induced DNA damage yields (average of three experiments) were calculated with both tested models (Table 1). Excess strand breaks, observed after incubation with Nth or Fpg enzymes and compared to the samples not treated with enzymes, are marked as strand breaks with corresponding Nth or Fpg index. The yields calculated with McMahon model for gamma rays were 45.2 ± 6.6 (SSB), 72.9 ± 1.9 (SSBNth), 82.1 ± 16.3 (SSBFpg), 2.0 ± 0.8 (DSB), 5.3 ± 0.5 (DSBNth), 4.6 ± 2.4 (DSBFpg) 10−3 Mbp−1 Gy−1 for dry samples and 39.3 ± 6.6 (SSB), 68.6 ± 19.5 (SSBNth), 66.8 ± 13.4 (SSBFpg), 2.0 ± 0.5 (DSB), 3.0 ± 0.6 (DSBNth) and 5.7 ± 0.8 (DSBFpg) Mbp−1 Gy−1 for liquid samples.
Comparison of the models
The comparison was based on the data from liquid samples irradiated by 20 MeV protons. Averaged S, C, L relative fractions dependent on dose and quantified by AGE were fitted by Cowan and McMahon models (Fig. 5). The obtained parameters μ = 60.08 Mbp−1 Gy−1 and ϕ = 2.29 Mbp−1 Gy−1 were used for generation of 105 measured-like data.
Figure 6 shows the distributions of the ϕ parameter obtained with the models fitting the generated measured-like data. It can be seen that values for \( \phi \) are closely distributed around the true value in case of McMahon model, with standard deviation being 14 % smaller than for Cowan model. On the other hand, standard deviation of distribution of fitted parameter μ (the data not shown) was 2 % higher for McMahon model.
Discussion
Comparison of the models confirms that the McMahon model is more robust for DSB calculation. With a reasonable effort, the model also enables the implementation of clustering of two or more SSBs into DSBs. This probability is not negligible even for the proton radiation as it is clear from Table 1. Therefore, only the results calculated with the McMahon model are discussed further.
The low hydration of the dry samples (8 water molecules per nucleotide) is below the limit where hydroxyl radicals are created (10 water molecules per nucleotide); therefore, the yields of damage measured under such conditions can be considered as the result of direct events (Swarts et al. 1992). Irradiation of the plasmid DNA in solution and in dry form enables estimating the contribution of direct and indirect effects. The estimation has limited validity due to different energy loss of protons in the dry or liquid matter and corresponding differences in average LET. The LET change is manifested particularly for liquid samples irradiated by 20 MeV protons, while for the corresponding irradiation of the dry plasmid in thin layer, only minor LET changes are expected. Comparison of the DNA yields shows that the indirect effects in case of 20 and 30 MeV protons caused more than 99 % of the total damage.
Protons and 60Co gamma rays are more effective in inducing SSB as compared to DSB, with slightly higher SSB/DSB ratio for the dry samples. The increase in the DNA damage complexity with hydration is expected. The reason lies in the lack of hydroxyl radical action under dry conditions and prevailing of single hit events (Adhikary et al. 2014, 2012). The yields for dry samples appear to be independent on LET. The same trend was found for proton- or alpha-induced yields measured in dry plasmid films and published in previous studies (Wyer et al. 2009; Urushibara et al. 2008; Ushigome et al. 2012); these are shown as half-full symbols in Fig. 7. The data were also compared to damage induced by 130 keV protons in dry plasmid film from experiments performed at the Université Paul Sabatier in Toulouse. The same experimental protocol as in this study was used, only with the following adaptations: 500 ng of pBR322 and 0.1× TE were used, the irradiation was performed in vacuum, and the enzymatic treatment was not applied. It should be emphasized that the results can differ due to density of the plasmid layer taken into account in dose and LET calculations. The SSB and DSB yields for liquid samples also do not seem to vary within the LET range used in this study. A comparison of the few existing studies performed with proton beams (Leloup et al. 2005; Sui et al. 2013; Pachnerová Brabcová et al. 2015) is problematic. The studies were performed with different scavengers and scavenging capacities; in other words, the indirect effect was suppressed to some extent that is difficult to estimate. Figure 7 includes the results with the following scavenging capacities: 21 s−1 due to residual TE buffer (this study); 1.5 × 103 s−1 due to residual TE buffer and 6.8 × 105–6.8 × 107 s−1 due to coumarin-3-carboxylic acid (Pachnerová Brabcová et al. 2015); 3.8 × 106 and 3.8 × 108 s−1 due to glycerol (Leloup et al. 2005); 3.0 × 108 s−1 due to TE buffer (Sui et al. 2013). The cellular environment has an estimated scavenging capacity of about 108 s−1 (Klimczak et al. 1993). Figure 7 illustrates suppressing of the indirect effect with increasing scavenging capacity. In addition to the outlined scavenging differences, an irradiation of liquid samples requires a careful consideration of how the geometry affects the delivered dose and LET, particularly for particles with short ranges. Differences in irradiation geometry bring a further uncertainty into the comparison of results. Considering these factors, the LET dependence cannot be confirmed within the compared interval.
Yields of SSB (a) and DSB (b) in dependence on LET calculated in dry DNA plasmid samples (full and half-full symbols) and DNA plasmid in solutions (empty symbols); the data include the results of the McMahon model (Table 1). Scavenging capacities of the solutions are marked by approximate values in s−1
Regarding the strand breaks detected after and without the enzymatic treatment, the used radiation sources are more effective in induction of (SSBNth + SSBFpg) as compared to (DSBNth + DSBFpg), also with slightly higher ratio for the dry samples. The data from this study do not show a LET dependence, neither for the liquid nor for the dry samples. However, comparison of the existing studies in wider LET ranges (Urushibara et al. 2008; Ushigome et al. 2012; Sui et al. 2013; Pachnerová Brabcová et al. 2015) suggests a decreasing trend for dry samples.
The question is what the excess of strand breaks, detected after enzymatic incubation over those detected without enzymes, actually represents. Many authors interpret it as isolated (excess of SSB) or clustered (excess of DSB) oxidized bases, where increase in strand breaks after Fpg treatment corresponds to detected purines, and, similarly, increase in strand breaks after Nth treatment corresponds to detected pyrimidines. However, the detection method distinguishes only vaguely between defined groups of plasmid damage sites: Type 1—modified base with possible tandem modified bases (on the same DNA strand); Type 2—SSB with possible tandem SSBs or modified bases; Type 3—SSB with bystanding modified base (on opposing strands) with possible tandem modified bases; Type 4—bystanding modified bases optionally complemented by other modified bases; Type 5—DSB with possible other modified bases or SSBs. It means that all groups can contain a complex cluster, which, by the current method, cannot be distinguished from a single lesion. Illustrations of the described damage sites in a simple and in a possible more complex configuration are shown in Fig. 8.
Groups of DNA damage sites detected with the current method, lines represent DNA backbone, and circles represent bases. Type 1: modified base with possible tandem modified bases; Type 2: SSB with possible tandem SSBs or modified bases; Type 3: SSB with bystanding modified base with possible tandem modified bases; Type 4: bystanding modified bases optionally complemented by other modified bases; Type 5: DSB with possible other modified bases or SSBs. The groups are illustrated in a simple (left) and a more complex configuration (right)
Figure 9 illustrates the link between the initial DNA damage groups as present in the irradiated plasmid DNA sample, plasmid forms as detected by agarose gel electrophoresis and the calculated μ and ϕ probability parameters.
The probability parameters are used to express yields of commonly used DNA damage categories as shown in Table 2. Note that particularly for the isolated oxidized bases, the yield is not coherent with an intuitive expectation and the yield is underestimated by the probability of supercoiled to open-circular form transition due to induction of Type 3 damage. This discrepancy could be decreased, assuming the same probabilities of SSB with possible tandem SSBs or modified bases (Type 2) and SSB with a bystanding modified base with possible tandem modified bases (Type 3).
Conclusions
Only few data on clustered DNA damage induced by protons are available. This is in sharp contrast with the importance of proton radiation as the dominant ionizing particle in space environment and with its increasing use in cancer treatment, as well as with severity of biological effects of the clustered damage.
Plasmid DNA in dry and liquid state was irradiated by 10, 20 and 30 MeV protons and 60Co, and the results were compared in terms of direct and indirect effect; the indirect effects were found to be dominant. Oxidized base damage was analyzed with base excision repair enzymes. The composition of the damage regarding its complexity was found slightly different when comparing the results of liquid and dry samples. This difference is due to processes dependent on DNA hydration level.
Two models used for calculation of strand break probabilities were compared, and their advantages were discussed. It was concluded that results strongly depend on irradiation geometry and the used model for DNA damage yield calculation; these factors should be carefully considered when interpreting the results.
The usual interpretation of strand breaks detected with base excision repair enzymes was reviewed. It was shown that the interpretation is not coherent with an intuitive expectation, and the yields of the damaged bases are underestimated.
This highlights the need for Monte Carlo models which can go beyond detection limits and close the gaps in current knowledge of DNA damage induced by ionizing radiation.
References
Adhikary A, Becker D, Palmer BJ, Heizer AN, Sevilla MD (2012) Direct formation of the C5′-radical in the sugar-phosphate backbone of DNA by high-energy radiation. J Phys Chem B 116(20):5900–5906
Adhikary A, Becker D, Sevilla MD (2014) Electron spin resonance of radicals in irradiated DNA. In: Lund A, Shiotani M (eds) Applications of EPR in radiation research. Springer, Berlin, pp 299–352
Asaithamby A, Hu B, Chen DJ (2011) Unrepaired clustered DNA lesions induce chromosome breakage in human cells. PNAS 108(20):8293–8298
Berger MJ, Coursey JS, Zucker MA, Chang J (2005) ESTAR, PSTAR, and ASTAR: computer programs for calculating stopping-power and range tables for electrons, protons, and helium ions (version 1.2.3). http://physics.nist.gov/Star. Accessed 30 Oct 2014
Bin JH, Allinger K, Assmann W, Dollinger G, Drexler GA, Friedl AA, Habs D, Hilz P, Hoerlein R, Humble N, Karsch S, Khrennikov K, Kiefer D, Krausz F, Ma WJ, Michalski D, Molls M, Raith S, Reinhardt S, Röper B, Schmid TE, Tajima T, Wenz J, Zlobinskaya O, Schreiber J, Wilkens JJ (2012) A laser-driven nanosecond proton source for radiobiological studies. Appl Phys Lett 101:243701–243704
Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2(2):95–113
Bolton PR, Borghesi M, Brenner C, Carroll DC, De Martinis C, Flacco A, Floquet V, Fuchs J, Gallegos J, Dn Giove, Green JS, Green S, Jones B, Kirby D, McKenna P, Neely D, Nuesslin F, Prasad R, Reinhardt S, Roth M, Schramm U, Scott GG, Ter-Avetisyan S, Tolley M, Turchetti G, Wilkens JJ (2014) Instrumentation for diagnostics and control of laser-accelerated proton (ion) beams. Phys Med 30(3):255–270
Cowan R, Collis CM, Grigg GW (1987) Breakage of double-stranded DNA due to single-stranded nicking. J Theor Biol 127:229–245
Do TT, Tang VJ, Konigsfeld K, Aguilera JA, Perry CC, Milligan JR (2012) Damage clusters after gamma irradiation of a nanoparticulate plasmid DNA peptide condensate. Radiat Environ Biophys 51(1):43–52
Durante M, Cucinotta FA (2008) Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer 8:465–472
Gebicki S, Gebicki JM (1999) Crosslinking of DNA and proteins induced by protein hydroperodixes. Biochem J 338:629–636
Georgakilas AG, O’Neill P, Stewart RD (2013) Induction and repair of clustered DNA lesions: What do we know so far? Radiat Res 180(1):100–109
Girdhani S, Sachs R, Hlatky L (2013) Biological effects of proton radiation: What we know and don’t know. Radiat Res 179(3):257–272
Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P (2002) Clustered DNA damage induced by γ radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acid Res 30(15):3464–3472
Hada M, Georgakilas AG (2008) Formation of clustered DNA damage after high-LET irradiation: a review. J Radiat Res 49(3):203–210
Hall EJ, Hei TK (2003) Genomic instability and bystander effects induced by high-LET radiation. Oncogene 22(45):7034–7042
ICRU Report 16 (1970) Linear energy transfer. International Commission on Radiation Units and Measurements
Klimczak U, Ludwig DC, Mark F, Rettberg P, Schulte-Frohlinde D (1993) Irradiation of plasmid and phage DNA in water-alcohol mixtures: strand breaks and lethal damage as a function of scavenger concentration. Int J Radiat Biol 64(5):497–510
Kraft SD, Richter C, Zeil K, Baumann M, Beyreuther E, Bock S, Bussmann M, Cowan TE, Dammene Y, Enghardt W, Helbig U, Karsch L, Kluge T, Laschinsky L, Lessmann E, Metzkes J, Naumburger D, Sauerbrey R, Schurer M, Sobiella M, Woithe J, Schramm U, Pawelke J (2010) Dose-dependent biological damage of tumour cells by laser-accelerated proton beams. New J Phys 12:085003
Leloup C, Garty G, Assaf G, Cristovão A, Breskin A, Chechik R, Shchemelinin S, Paz-Elizur T, Livneh Z, Schulte RW, Bashkirov V, Milligan JR, Grosswendt B (2005) Evaluation of lesion clustering in irradiated plasmid DNA. Int J Radiat Biol 81(1):41–54
Malka V, Faure J, Gauduel YA, Lefebvre E, Rousse A, Phuoc KT (2008) Principles and applications of compact laser-plasma accelerators. Nat Phys 4(6):447–453
McMahon SJ, Currell FJ (2011) A robust curve-fitting procedure for the analysis of plasmid DNA strand break data from gel electrophoresis. Radiat Res 175(6):797–805
Milligan JR, Aguilera JA, Nguyen TTD, Paglinawan RA, Ward JF (2000) DNA strand-break yields after post-irradiation incubation with base excision repair endonucleases implicate hydroxyl radical pairs in double-strand break formation. Int J Radiat Biol 76(11):1475–1483
Pachnerová Brabcová K, Sihver L, Yasuda N, Matuo Y, Štěpán V, Davídková M (2014) Clustered DNA damage on subcellular level: effect of scavengers. Radiat Environ Biophys 53(4):705–712
Pachnerová Brabcová K, Štěpán V, Karamitros M, Karabín M, Dostálek P, Incerti S, Davídková M, Sihver L (2015) Contribution of indirect effects to clustered damage in DNA irradiated with protons. Radiat Prot Dosim. doi:10.1093/rpd/ncv15
Prise KM, Pullar CHL, Michael BD (1999) A study of endonuclease III-sensitive sites in irradiated DNA: detection of α-particle-induced oxidative damage. Carcinogenesis 20(5):905–909
Semenenko VA, Stewart RD (2006) Fast Monte Carlo simulation of DNA damage formed by electrons and light ions. Phys Med Biol 51(7):1693–1706
Shikazono N, Noguchi M, Fujii K, Urushibara A, Yokoya A (2009) The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J Radiat Res 50(1):27–36
Śmiałek MA (2012) Early models of DNA damage formation. J Phys Conf Ser 373(1):012013
Śmiałek MA, Jones NC, Hoffmann SV, Mason NJ (2013) Measuring the density of DNA films using ultraviolet-visible interferometry. Phys Rev E 87:060701
Stefančíková L, Porcel E, Eustache P, Li S, Salado D, Marco S, Guerquin-Kern JL, Réfrégiers M, Tillement O, Lux F, Lacombe S (2014) Cell localisation of gadolinium-based nanoparticles and related radiosensitising efficacy in glioblastoma cells. Cancer Nanotechnol. doi:10.1186/s12645-014-0006-6
Sui L, Wang Y, Wang X, Kong FQ, Liu JC, Zhou PK (2013) Clustered DNA damage induced by protons radiation in plasmid DNA. Chin Sci Bull 58(26):3217–3223
Sutherland BM, Bennett PV, Saparbaev M, Sutherland JC, Laval J (2001) Clustered DNA damages as dosemeters for ionising radiation exposure and biological responses. Radiat Prot Dosim 97(1):33–38
Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT (1992) Radiation-induced DNA damage as a function of hydration: I. Release of unaltered bases. Radiat Res 129(3):333–344
Terato H, Ide H (2004) Clustered DNA damage induced by heavy ion particles. Biol Sci Space 18(4):206–215
Urushibara A, Shikazono N, O’Neill P, Fujii K, Wada S, Yokoya A (2008) LET dependence of the yield of single-, double-strand breaks and base lesions in fully hydrated plasmid DNA films by 4He2+ ion irradiation. Int J Radiat Biol 84(1):23–33
Ushigome T, Shikazono N, Fujii K, Watanabe R, Suzuki M, Tsuruoka C, Tauchi H, Yokoya A (2012) Yield of single- and double-strand breaks and nucleobase lesions in fully hydrated plasmid DNA films irradiated with high-LET charged particles. Radiat Res 177(5):614–627
von Sonntag C (2006) Free-radical-induced DNA damage and its repair, a chemical perspective. Springer, Berlin
Watanabe M (2007) The first target of radiation carcinogenesis is not DNA. Int Congr Ser 1299:21–24
Willers H, Dahm-Daphi J, Powel SN (2004) Repair of radiation damage to DNA. Brit J Cancer 90:1297–1301
Wyer JA, Butterworth KT, Hirst DG, Latimer CJ, Montenegro EC, Shah MB, Currell FJ (2009) Fragmentation and plasmid strand breaks in pure and gold-doped DNA irradiated by beams of fast hydrogen atoms. Phys Med Biol 54(15):4705–4721
Yogo A, Sato K, Nishikino M, Maeda T, Sakaki H, Hori T, Ogura K, Nishiuchi M, Teshima T, Nishimura H, Kondo K, Bolton PR, Kawanishi S (2011) Measurement of DNA double-strand break yield in human cancer cells by high-current, short-duration bunches of laser-accelerated protons. Jpn J Appl Phys 50(10):106401
Ziegler JF, Ziegler MD, Biersack JP (2010) SRIM—The stopping and range of ions in matter (2010). Nucl Instrum Methods B 268(11–12):1818–1823
Zlobinskaya O, Siebenwirth C, Greubel C, Hable V, Hertenberger R, Humble N, Reinhardt S, Michalski D, Röper B, Multhoff G, Dollinger G, Wilkens JJ, Schmid TE (2014) The effects of ultra-high dose rate proton irradiation on growth delay in the treatment of human tumour xenofrafts in nude mice. Radiat Res 181(2):177–183
Acknowledgments
This work has been supported by ESA Project No. ESTEC/ITT AO/1-7146/12/NL/GLC “Radiation Biological End Effects Models and Interfaces to Physics Models” and MEYS CR Project No. LD12008. L. Vyšín and L. Juha appreciate a partial financial support provided by the Czech Science Foundation under the Grant No. 13-28721S. M. Vlk is grateful for a partial financial support by MEYS CR Project No. LK21310. The irradiation has been realized at the CANAM (Center of Accelerators and Nuclear Analytical Methods) supported by the MEYS CR Project No. LM 2011019. The authors also appreciate valuable comments of Dr. Amitava Adhikary.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vyšín, L., Pachnerová Brabcová, K., Štěpán, V. et al. Proton-induced direct and indirect damage of plasmid DNA. Radiat Environ Biophys 54, 343–352 (2015). https://doi.org/10.1007/s00411-015-0605-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00411-015-0605-6