Abstract
While many gene expression studies have focused on male pattern baldness (MPB), few studies have investigated the genetic differences between bald and non-bald hair follicles in female pattern hair loss (FPHL). This study aimed to identify molecular biomarkers associated with FPHL through genetic analysis of paired bald and non-bald hair follicles from 18 FPHL patients, using next-generation sequencing (NGS) techniques. RNA transcriptome analysis was performed to identify differentially expressed genes (DEGs) between bald and non-bald hair follicles in FPHL. The DEGs were validated using real-time PCR, and protein expression was confirmed through immunohistochemistry and western blot analysis. Our findings suggest that HOXB13, SFRP2, PTGDS, CXCR3, SFRP4, SOD3, and DCN are significantly upregulated in bald hair follicles compared to non-bald hair follicles in FPHL. SFRP2 and PTGDS were found to be consistently highly expressed in bald hair follicles in all 18 samples. Additionally, elevated protein levels of SFRP2 and PTGDS were confirmed through western blot and immunohistochemical analysis. Our study identified SFRP2 and PTGDS as potential biomarkers for FPHL and suggests that they may play a role in inducing hair loss in this condition. These findings provide a foundation for further research on the pathogenesis of FPHL and potential therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Female pattern hair loss (FPHL) is a prevalent hair loss disorder in women characterized by hair thinning from the mid-scalp to the vertex without the regression of the frontal hairline [6]. While FPHL differs from male pattern baldness (MPB) in its clinical features, hair follicle miniaturization is a common histological characteristic observed in both conditions. Androgens have been identified as the primary cause of hair loss in MPB [20], with dihydrotestosterone (DHT) resulting from testosterone conversion causing hair follicles to miniaturize [8, 13, 21]. However, the role of androgens in inducing hair loss in women has not been fully elucidated [3, 12, 14].
Although gene expression profiling studies to identify the genetic etiology of hair loss have largely focused on MPB, few genetic studies have been conducted to compare bald and non-bald hair follicles in FPHL. Previous research on FPHL has typically focused on androgenic factors and aromatase due to the established role of the androgen-dependent pathway in the etiology of MPB, with the assumption that FPHL shares the same underlying mechanism [17, 18, 22]. However, FPHL is influenced by multiple factors, including hormones, environmental factors, and genetics. Recent genome-wide association studies (GWAS) of FPHL have not found significant candidate genetic variants shared with MPB, highlighting the need to explore specific genetic biomarkers for FPHL [15, 16, 22].
This study aims to investigate critical genetic biomarkers for FPHL by conducting a whole-transcriptomic analysis of bald and non-bald hair follicles using high-throughput sequencing. Significant differentially expressed genes (DEGs) between balding and non-balding hair follicles in FPHL will be confirmed using real-time PCR and Western blotting.
Prostaglandins are highly expressed in male bald scalps and are associated with alopecia and miniaturization, although this has not been studied in FPHL. SFRP2 is functional role in regulating hair growth is controversial, as previous reports indicate high expression in the catagen phase until telogen in mouse hair follicles and highly expressed in vertex scalp in AGA1, while in in vitro study shows that it is induce trichogenicity.
Our findings suggest that SFRP2 and PTGDS are significantly upregulated in bald hair follicles compared to non-bald hair follicles in FPHL. SFRP2 and PTGDS were found to be consistently highly expressed in bald hair follicles in all 18 samples.
Materials and methods
Acquisition of scalp tissue
This study involved 18 women diagnosed with FPHL according to the Ludwig classification. Human scalp tissues were collected from the mid-scalp or vertex scalp area with miniaturized hairs, as well as from the occiput area with normal terminal hair follicles in the same participant. The Institutional Review Board of Dankook University Hospital approved this study (IRB No. DKUH 2015-02-014).
RNA isolation and sequencing with NGS technique
Total RNA was extracted from balding and non-balding hair follicles of 18 female individuals using Trizol reagent, and RNA quality was assessed by analyzing rRNA band integrity using an Agilent RNA 6000 Nano kit (Agilent Technologies, CA). mRNA sequencing libraries were constructed by enriching mRNA with oligo-dT attached to magnetic beads from total RNA and fragmenting it into short sizes. The fragmented mRNA was reverse-transcribed to cDNA, modified by end-repair, poly-A addition, and linked with 5´ and 3´ adaptors on both ends using the TruSeq RNA Sample Prep Kit (Illumina, CA) according to the manufacturer’s instructions. The modified mRNA fragments were separated on a BluePippin 2% agarose gel cassette (Sage Science, MA), and the suitable fragments were amplified as PCR templates for enrichment. The final products were quantified by an Agilent High Sensitivity DNA Kit (Agilent Technologies, CA) on an Agilent Bioanalyzer 2100 system. High-throughput RNA sequencing was then performed on an Illumina Hiseq2500 platform (Illumina, CA). All processes were carried out by the Theragen Bio Institute (Suwon, Korea).
Mapping and assembly of transcriptome data
To ensure exact and high-quality analysis, the total output reads underwent quality control and filtering for sequence quality score, GC content, and duplicated sequences before mapping and assembly. Raw quality-reads were those with more than 10% skipped bases, those including more than 40% of bases with less than Q20, and those with average quality scores less than Q20. The entire filtering process was performed using in-house scripts, and this step resulted in clean reads that were mapped to the human reference genome [7] using the STAR aligner v.2.3.0e [4]. The expression level of each gene was measured based on fragments per kilobase of exon per million mapped reads (FPKM), calculated by Cufflinks v2.1.1 [19] from the gene annotation database of Ensembl release 72.
Quantitative real-time PCR
One microgram of total RNA was used to synthesize cDNA following the M-MLV reverse transcription protocol (Promega, Madison, WI, USA). The mRNA expression levels of HOXB13, SFRP2, PTGDS, CXCR3, SFRP4, SOD3, and DCN were quantified by real-time RT-PCR using the Eco Real-time PCR System with Illumina. All reactions were performed in triplicate and analyzed by the delta-delta Ct method. The primer sequences used in this study are listed in Table 1, which were designed using the Primer3 software (V. 0.4.0).
Western blots for hair follicle
Two additional pairs of bald and non-bald HF samples were collected for Western blot analysis. The HF samples were homogenized using a lysis buffer containing 1mM PMSF and protease inhibitor, and the resulting lysates were centrifuged. The supernatants were collected, and the protein concentration was measured. The primary antibodies used were against SFRP2 (Abcam), PTGDS (Thermo Fisher scientific), and β-actin (Santacruz).
Immunohistochemistry in HF
Two additional pairs of bald and non-bald HF samples were collected for immunohistochemistry. The HF samples were embedded in paraffin, and 10-µm-thick histological sections were produced. The HF slides were incubated overnight at 4 °C with primary antibodies against SFRP2 or PTGDS. The following day, the slides were incubated with the secondary antibody for 1 h at room temperature. After removing the secondary antibody, hematoxylin staining was performed for nucleus staining. The stained HF were observed under a microscope (Olympus CX35, Tokyo, Japan).
Statistical analysis
Data are presented as mean ± standard deviation (SD) and were analyzed using the Student’s t-test. A p-value of less than 0.05 was considered statistically significant.
Result
Demographics
A total of 18 female patients with FPHL were included in this study, of whom 10 were under 50 years old (pre-menopause) and 8 were over 50 years old (menopause). The Ludwig classification revealed that 11 FPHL cases were L1, 5 were L2, and 1 was L3 type.
Differential gene expression analysis between bald and non-bald HF
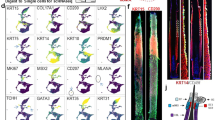
To identify the differentially expressed genes (DEGs) between bald and non-bald HF, we considered a fold change (FC) of ≥ 2 and a corrected p-value ≤ 0.05 as a cut-off value. We identified a total of 360 DEGs, among which 345 were up-regulated and 15 were down-regulated in bald HF. Figure 1 shows the hierarchical clustering of the differentially expressed genes. To further investigate FPHL-specific genes’ regulation, we selected hair growth-related genes from the 345 up-regulated genes based on previous reports. As a result, HOXB13, SFRP2, PTGDS, CXCR3, SFPR4, SOD3, and DCN were significantly upregulated in bald HF compared to non-bald HF. Individual DEG analysis showed that HOXB13, SFRP2, PTGDS, CXCR3, SFPR4, SOD3, and DCN expression levels were elevated in the balding area compared to the non-balding area in all 18 samples.
Validation of DEG expression levels by real-time PCR
To validate the expression levels of the selected genes (HOXB13, SFRP2, PTGDS, CXCR3, SFRP4, SOD3, and DCN), we performed real-time PCR on the 18 pairs of HF samples used for RNA sequencing. The results showed that the expression levels of these seven genes were significantly up-regulated in bald HF in real-time PCR analysis, consistent with NGS results (Fig. 2). Moreover, individual mRNA expression of these seven genes was analyzed in all 18 pairs of samples. The relative mRNA expression levels of HOXB13, SFRP2, PTGDS, CXCR3, SFPR4, SOD3, and DCN were higher in the balding area than in the non-balding area in 17, 18, 18, 15, 9, 11, and 17 samples, respectively, out of the total 18 FPHL samples (Table 1). In the RNA sequencing data, the FPKM values of HOXB13, SFRP2, PTGDS, CXCR3, SFPR4, SOD3, and DCN were higher in the balding area of all 18 samples.
Protein expression of SFRP2 and PTGDS in bald and non-bald scalp
Next, we analyzed the protein expression levels of SFRP2 and PTGDS, which were highly expressed in the mRNA of all 18 bald samples. First, immunohistochemistry staining showed that SFRP2 and PTGDS were strongly expressed in bald scalp than in non-bald scalp, particularly around the epidermis, including the hair shaft, rather than the dermis (Fig. 3A). Second, western blot analysis revealed that SFRP2 and PTGDS were up-regulated in the balding area compared to the non-balding area, consistent with the tissue staining results (Fig. 3B).
Discussion
Although MPB and FPHL share many common characteristics, their exact patho-mechanisms are different. The important genetic biomarkers such as androgen, 5 alpha reductase in MPB have not been validated in FPHL, highlighting the need to investigate the different genetic expression between balding and non-balding areas of FPHL. This study presents the first whole transcriptomic analysis of the balding and non-balding area of FPHL in Asians.
Differential gene expression (DEG) was screened using high output RNA sequencing based on NGS technology, which is a useful technique for investigating DEG between normal and pathological states quickly and widely. However, NGS results often need to be validated by real-time PCR. In this study, HOXB13, SFRP2, PTGDS, CXCR3, SFRP4, SOD3, and DCN were selected as important biomarkers in the DEG analysis using RNA sequencing by NGS technology. The mRNA expression of these genes was verified by real-time PCR, and the RNA expression patterns of these genes between the balding and non-balding areas were consistent with the real-time PCR analysis. However, individual analysis of mRNA expression by real-time PCR showed that only SFRP2 and PTGDS were significantly higher expressed in balding hair follicles (HFs) of all 18 FPHL samples, which was consistent with the RNA sequencing analysis. Although the individual pattern of mRNA expression in real-time PCR between the lesion and control was generally similar to the FPKM value in RNA sequencing analysis, the data was not identical and requires validation.
Based on the RNA sequencing data and real-time PCR results, the protein expression pattern of SFRP2 and PTGDS in the hair follicles of FPHL was investigated. SFRP2 is a representative Wnt inhibitor that plays an important role in various cellular regeneration and differentiation Its functional role in regulating hair growth is controversial, as previous reports indicate high expression in the catagen phase until telogen in mouse hair follicles and highly expressed in vertex scalp in AGA [9, 11], while in in vitro study shows that it is induce trichogenicity [10]. Considering our results, it can be known that SFRP2 is significantly upregulated in both M-AGA and FPHL vertex scalp although the functional role of SFRP2 in regulating hair growth still should be studied.
Prostaglandins are highly expressed in male bald scalps and are associated with alopecia and miniaturization [1], although this has not been studied in FPHL. In our study, PTGDS mRNA and PGD2 protein were elevated in bald compared to haired scalp, which is consistent with previous reports implicating the prostaglandin pathway in the pathogenesis of AGA in both males and females. Although PGE2 and PGF2a have been reported to be more generated in female than male scalps [2], contributing to hair growth induction, they were not identified in the DEGs analysis between balding and non-balding HFs. The recent Phase 2 clinical trial of the PTGDR2 inhibitor did not show efficacy on AGA, suggesting that further study on the PTGDS pathway and alopecia is needed [5].
In conclusion, this study identified the genetic expression pattern between balding and non-balding scalp of FPHL and found that SFRP2 and PTGDS are significantly involved in FPHL. These findings provide a foundation for further research on the pathogenesis of FPHL and potential therapeutic targets.
Data availability
No datasets were generated or analysed during the current study.
References
Chovarda E, Sotiriou E, Lazaridou E et al (2021) The role of prostaglandins in androgenetic alopecia. Int J Dermatol 60(6):730–735
Colombe L, Vindrios A, Michelet JF et al (2007) Prostaglandin metabolism in human hair follicle. Exp Dermatol 16(9):762–769
Cousen P, Messenger A (2010) Female pattern hair loss in complete androgen insensitivity syndrome. Br J Dermatol 162(5):1135–1137
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
DuBois J, Bruce S, Stewart D et al (2021) Setipiprant for Androgenetic Alopecia in males: results from a Randomized, Double-Blind, placebo-controlled phase 2a trial. Clin Cosmet Invest Dermatology 14:1507–1517
Fabbrocini G, Cantelli M, Masara A et al (2018) Female pattern hair loss: a clinical, pathophysiologic, and therapeutic review. Int J Women’s Dermatology 4(4):203–211
Flicek P, Ahmed I, Amode MR et al (2013) Ensembl 2013. Nucleic Acids Res 41:D48–D55
Fu D, Huang J, Li K et al (2021) Dihydrotestosterone-induced hair regrowth inhibition by activating androgen receptor in C57BL6 mice simulates androgenetic alopecia. Biomed Pharmacother 137:111247
Kim BK, Yoon SK (2014) Expression of sfrp2 is increased in catagen of hair follicles and inhibits keratinocyte proliferation. Ann Dermatol 26(1):79–87
Kwack MH, Ahn JS, Jang JH et al (2016) SFRP2 augments Wnt/beta-catenin signalling in cultured dermal papilla cells. Exp Dermatol 25(10):813–815
Liu Q, Tang Y, Huang Y et al (2022) Insights into male androgenetic alopecia using comparative transcriptome profiling: hypoxia-inducible factor-1 and Wnt/beta-catenin signalling pathways. Br J Dermatol 187(4):936–947
Orme S, Cullen DR, Messenger AG (1999) Diffuse female hair loss: are androgens necessary? Br J Dermatol 141(3):521–523
Paus R, Muller-Rover S, Botchkarev VA (1999) Chronobiology of the hair follicle: Hunting the hair cycle clock. Journal of Investigative Dermatology Symposium Proceedings, 4(3);338–345
Price VH, Roberts JL, Hordinsky M et al (2000) Lack of efficacy of finasteride in postmenopausal women with androgenetic alopecia. J Am Acad Dermatol 43(5):768–776
Redler S, Brockschmidt FF, Tazi-Ahnini R et al (2012) Investigation of the male pattern baldness major genetic susceptibility loci AR/EDA2R and 20p11 in female pattern hair loss. Br J Dermatol 166(6):1314–1318
Rui W, Sheng Y, Hu R et al (2016) Polymorphic CAG repeat numbers in the androgen receptor gene of female pattern hair loss in a Han Chinese Population. Dermatology 232(4):464–467
Sanchez P, Serrano-Falcon C, Torres JM et al (2018) 5alpha-Reductase isozymes and aromatase mRNA levels in plucked hair from young women with female pattern hair loss. Arch Dermatol Res 310(1):77–83
Sawaya ME, Price VH (1997) Different levels of 5alpha-reductase type I and II, aromatase, and androgen receptor in hair follicles of women and men with androgenetic alopecia. J Invest Dermatology 109(3):296–300
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Trueb RM (2002) Molecular mechanisms of androgenetic alopecia. Exp Gerontol 37(7–8):981–990
Urysiak-Czubatka I, Kmiec ML, Broniarczyk-Dyla G (2014) Assessment of the usefulness of dihydrotestosterone in the diagnostics of patients with androgenetic alopecia. Postepy Dermatologii i Alergologii 31(4):207–215
Yip L, Zaloumis S, Irwin D et al (2009) Gene-wide association study between the aromatase gene (CYP19A1) and female pattern hair loss. Br J Dermatol 161(2):289–294
Acknowledgements
This study was supported by a research grant of the Amorepacific co. (2016). This research was supported by Basic Science Research Capacity Enhancement Project through Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2019R1A6C1010033).
Funding
This work was supported by the research grant of the Amorepacific co. (2016). This research was supported by Basic Science Research Capacity Enhancement Project through Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2019R1A6C1010033).
Author information
Authors and Affiliations
Contributions
S.K. and Y.K. wrote the main manuscript text and J.K., S.K, W.P. and S.K. made contrbutions to the analysis and interpretation of the study. J.C. approved the version to be published and M.C. and M.K. revised the work for important intellectual content. B.P. was in charge of the whole process of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The patients in this manuscript have given written informed consent to publication of their case details.
Competing interests
The authors has no conflict of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, S.R., Kim, Y.J., Kim, JH. et al. Comprehensive transcriptome profiling between balding and non-balding scalp of female pattern hair loss in Asian. Arch Dermatol Res 316, 360 (2024). https://doi.org/10.1007/s00403-024-03114-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00403-024-03114-w