Abstract
Understanding regulation of MAPT splicing is important to the etiology of many nerurodegenerative diseases, including Alzheimer disease (AD) and progressive supranuclear palsy (PSP), in which different tau isoforms accumulate in pathologic inclusions. MAPT, the gene encoding the tau protein, undergoes complex alternative pre-mRNA splicing to generate six isoforms. Tauopathies can be categorized by the presence of tau aggregates containing either 3 (3R) or 4 (4R) microtubule-binding domain repeats (determined by inclusion/exclusion of exon 10), but the role of the N-terminal domain of the protein, determined by inclusion/exclusion of exons 2 and 3 has been less well studied. Using a correlational screen in human brain tissue, we observed coordination of MAPT exons 2 and 10 splicing. Expressions of exon 2 splicing regulators and subsequently exon 2 inclusion are differentially disrupted in PSP and AD brain, resulting in the accumulation of 1N4R isoforms in PSP and 0N isoforms in AD temporal cortex. Furthermore, we identified different N-terminal isoforms of tau present in neurofibrillary tangles, dystrophic neurites and tufted astrocytes, indicating a role for differential N-terminal splicing in the development of disparate tau neuropathologies. We conclude that N-terminal splicing and combinatorial regulation with exon 10 inclusion/exclusion is likely to be important to our understanding of tauopathies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ninety-five percent of all human multi-exonic genes are subject to alternative pre-mRNA splicing [48, 60]. Correct regulation of this mechanism is essential for proteomic diversity by the production of multiple distinct isoforms from a single gene [46]. The microtubule-associated protein tau (MAPT) is a neuronally expressed gene consisting of 16 exons, many of which are differentially spliced within the central nervous system and peripheral tissues. Tau proteins are involved in axonal transport, synaptic plasticity, and stabilization of the microtubule network [26, 30, 44]. In the human brain, the splicing of MAPT exons 2, 3 and 10 results in the expression of six different isoforms, which can be parsed into two groups depending on their inclusion or exclusion of exon 10 (Fig. 1a).
Splicing factor and RNA-binding protein expression is differentially correlated with the inclusion of MAPT exons 2 and 10. a MAPT exons 2, 3 and 10 are alternatively spliced, resulting in the expression of 6 different isoforms. At the N-terminus, exons 2 and 3 may be included or excluded, although exon 3 inclusion requires the inclusion of exon 2. The absence of either exon results in 0N isoforms, exon 2 alone results in 1N isoforms, and exon 3 inclusion defines 2N isoforms. At the C-terminus, the inclusion of exon 10 (encoding the second microtubule-binding repeat domain) defines 4R isoforms, whereas its exclusion results in 3R isoforms. b MAPT exon expression (RPKM) in the AMP-AD ROSMAP cohort. Error bars ± SEM. c MAPT exons 2, 3 and 10 percent spliced in (PSI) across four Brodmann regions examined in the AMP-AD MSBB cohort. Error bars ± SEM. d PSI values for MAPT exons 2, 3 and 10 in cerebellum and temporal cortex in the AMP-AD MAYO cohort. Error bars ± SEM. e–f MAPT PSI values for exons 2, 3 and 10 between 17q21.31 H1 and H2 haplotypes in e MSBB and f MAYO AMP-AD datasets. Error bars ± SEM. g Pearson’s correlation coefficients with unsupervised hierarchical clustering between SF/RBP expression (x-axis) and MAPT exon 2/exon 10 PSI values in ROSMAP and MSBB data. Blue indicates a positive correlation (“includers”) and red indicates a negative correlation (“excluders”), while yellow denotes no association. All comparisons carried out using linear regression model. ***p < 0.001
The ratio of tau isoform expression changes during human brain development [25, 27] with only the shortest 0N3R isoform of tau being expressed in fetal brain [16, 25, 27, 45] Following birth, there is a sudden shift in the expression of both exons 2 and 10; exon 10 inclusion increases dramatically during the perinatal period [25] where it reaches a stable 3R:4R ratio of roughly 1:1 [3, 12, 59], whereas exon 2 expression increases gradually throughout the first decade of life [25]. The reason for this shift and the function of different MAPT isoforms is not fully understood. However, 4R tau has an increased affinity for binding microtubules that results in their increased stabilization [21]; therefore, shorter 3R isoforms may allow for greater neuroplasticity during brain development. It has been proposed that different N-terminal isoforms may also contribute to microtubule stabilization [14], and that the inclusion of exons 2 and 3 results in the extension of the acidic region of tau, lengthening its projection domain [9], which in turn may increase the distance between microtubules and increase bundling [33].
Understanding MAPT splicing is of critical importance to the etiology of tauopathies, which are characterized by the presence of neuronal and/or astroglial tau aggregates. Primary tauopathies include frontotemporal dementia (FTD), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), primary age-related tauopathy (PART) and Pick’s disease (PiD), while other tauopathies, such as AD, are secondary to amyloid-beta (Aß) deposition. In several primary tauopathies, the regulation of MAPT splicing is altered and mis-spliced isoforms are differentially incorporated into neurofibrillary tangles (NFTs) and pathogenic inclusions [13, 15]. The most striking evidence in support of the importance of regulated splicing is the numerous synonymous and intronic MAPT mutations, such as S305S [53, 55], IVS10+16 [28] and N296N [54], which result in increased exon 10 inclusion, and the subsequent development of autosomal dominant FTD. The ability for these mutations to induce tau pathology in the absence of an altered amino acid sequence is indicative of the relevance of MAPT splicing to disease pathogenesis, and the importance of maintaining the correct tau isoform ratio.
Alternative splicing of MAPT exon 10 in both healthy and diseased brain has been well characterized, although studies examining exon 10 expression in AD have yielded inconsistent results [3, 12, 19, 20, 29, 62]. In contrast, less is known about the regulation of exons 2 and 3, and there have been no studies directly assessing the contribution of N-terminal tau isoforms to primary tauopathies. To date, splicing of exon 10 has been associated with the function of several candidate splicing factors (SFs) and RNA-binding proteins (RBPs), the most comprehensively investigated of which are the serine and arginine-rich family of splicing factors (SRSFs). Multiple SRSFs have been associated with both exon 10 inclusion and exclusion [28,29,30,31], as have Tra2β [6, 13, 31, 57], FUS [57], RBM4 [34], NOVA1 [61] and hnRNPs E2 and E3 [5, 61]. However, there is limited evidence for many of these associations, and their impact on MAPT splicing lacks robust replication.
Here, we report a correlational screen for SFs/RBPs in human brain tissues that revealed novel genes associated with MAPT splicing, and uncovered coordinated regulation of MAPT exons 2 and 10. We found that the splicing factor RSRC1 bound directly to MAPT pre-mRNA and was associated with both exon 10 exclusion and exon 2 inclusion. Furthermore, RSRC1 was also differentially expressed in PSP and AD brain, suggesting a regulatory role for this SF in disease pathogenesis. Consistent with discordant RSRC1 expression, we observed increased expression of exon 2 and exon 10-containing transcripts in PSP brain, and increased expression of 0N transcripts in AD brain, which correlate with the accumulation of different N-terminal tau isoforms in different neuropathological features of AD and PSP. We therefore conclude that differential expression of MAPT splicing regulators in AD and PSP brain results in the loss of coordination between MAPT exon 10 and exon 2 splicing. In turn, this results in the expression and accumulation of different N-terminal isoforms in each disease, which may underlie the development of different neuropathological features characteristic of AD and PSP.
Methods
RNA-seq data analysis
Aligned BAM files and gene expression count data were downloaded from the AMP-AD consortium through Synapse (https://adknowledgeportal.synapse.org/). MAPT exon level counts were calculated using the FeatureCounts feature within the Subread package [39, 40], and percent spliced in (PSI) values were determined using the Mixture of Isoforms (MISO) package [35]. For statistical analysis, associations between MAPT PSI values and the expression of SFs/RBPs were carried out using a linear model in R with RNA integrity number (RIN), postmortem interval (PMI), sex, and age at death included in the model as covariates. The resulting p-values were Bonferroni-corrected for the number of comparisons. For heatmap plotting, correlation coefficients were generated using Pearson’s correlation as part of the cor function in R. For the comparison of SF/RBP expression in PSP and AD brain, the fold change expression of each SF/RBP in disease brain was calculated in comparison to controls and plotted in R using ComplexHeatmap [23], and statistically significant differences were determined by linear regression of expression values including the previously described covariates.
At the time of analysis, genotype data were unavailable for the MSBB cohort, so 17q21.31 haplotype was determined by Taqman genotyping. The relevant DNA was obtained from the NIH Neurobiobank. Taqman genotyping was carried out for H2 tag SNPs rs8070723 and rs1052553 using commercially available assays. Haplotypes were determined for the other cohorts using the same tag SNPs from genotype data downloaded from the AMP-AD knowledge portal (https://adknowledgeportal.synapse.org/).
Single-nuclei and single-soma sequencing analysis
Snuc-seq processed gene counts and covariates derived from AD and control entorhinal cortex [22] were downloaded from the Gene Expression Omnibus (GEO) (GSE138852). Data were further normalized and analyzed in Seurat 3.0 dev [7, 58]. Data from different individuals were integrated and scaled using SCTransform [24] while regressing out the percentage of mitochondrial genes, the number of genes per cell and the number of reads per cell. Principal components analysis (PCA) was carried out in Seurat using the top 3000 most variable genes, and data was reduced using UMAP [43]. Cell types present within each cluster were already annotated in the downloaded metadata. Differential gene expression analysis was carried out between AD cases and controls across the whole data set, or within the neuronal cluster only, using the MAST model applied to log normalized raw count data, including the percent of mitochondrial genes as a covariate.
Aligned HDF5 feature barcode matrices for the single-soma sequencing data of AT8-positive and -negative neurons from AD PFC [47] were downloaded from GEO (GSE129308) and processed in Seurat [7, 58]. Data were filtered for cells expressing > 200 genes, < 2500 reads and < 10% mitochondrial genes. Data were integrated, transformed and reduced as described above. Differential gene expression analysis was carried out between AT8-positive and AT8-negative cells across the whole data set, using the MAST model applied to log normalized raw count data, including the percent of mitochondrial genes, age, RNA integrity number (RIN) and postmortem interval (PMI) as covariates.
Aligned HDF5 feature barcode matrices for PSP snuc-seq data [52] were kindly shared by Drs. Pereira and Crary, and were filtered, integrated, transformed and reduced in the same manner as the AD snuc-seq and AT8 soma-seq data described above. Individual clusters were identified in Seurat using the default resolution factor 0.5. Cell types within each cluster were defined using visualization of specific markers utilized in the AD snuc-seq data [22]; CD74 (microglia), AQP4 (astrocytes), MEGF11 (oligodendrocyte precursor cells (OPCs)), MOBP (oligodendrocytes), SYT1 (neurons) and FLT1 (endothelial cells). Differential gene expression analysis was carried out between PSP cases and controls across the whole data set, or within the neuronal cluster only, using the MAST model applied to log normalized raw count data, including the percent of mitochondrial genes and age as covariates.
Human brain tissue
Fresh frozen human control, AD and PSP temporal cortices were acquired from the Mount Sinai Neuropathology Core brain bank and the Harvard Brain Tissue Resource Center, University of Maryland Brain and Tissue Bank and Mount Sinai Brain Bank via the NIH Neurobiobank. Formalin-fixed paraffin-embedded sections from temporal cortex were acquired from the Mount Sinai Neuropathology Core brain bank, with neuropathological diagnosis being determined by Dr. John Crary. All post-mortem tissues were collected in accordance with the relevant guidelines and regulations of the respective institutions. A summary of tissues used in this project are described in Supplementary Table 1 (online resource).
Cell culture
All cell culture reagents were purchased from Thermo Fisher Scientific, unless otherwise stated. SH-SY5Y cells were grown in IMDM media supplemented with 1% penicillin/streptomycin and 10% FBS, and grown and maintained at 37 °C with 5% CO2 in a humid environment. Prior to transfection, cells were seeded into 6-well plates at a density of 1.6 × 105 cells per well. The next day, cells were transfected with 1.25 μg of LI9LI10 mini-gene and 1.25 μg of plasmid DNA of the SF/RBP of interest (all Origene) using Lipofectamine 3000. Cells were collected for RNA extraction and analysis 48 h later. For N-terminal antibody validation, N2a cells were transfected with 2.5 μg of either a 0N3R, 1N3R or 2N3R MAPT cDNA vector (Origene), and after 48 h were either fixed with 10% formalin (Sigma Aldrich) for 15 min at room temperature for immunofluorescence, or pelleted in PBS for protein extraction and western blotting.
RNA pull-down
The LI9LI10 mini-gene was digested by NotI (Cell Signaling Technologies) and SgfI (Promega) to excise the MAPT coding sequence and upstream T7 promoter from the PCI-Neo backbone. The resulting DNA was transcribed in vitro using the T7 Megascript kit (ThermoFisher Scientific), incubated at 37 °C for 4 h, followed by 15 min treatment with DNase to degrade any remaining DNA template. The resulting RNA was isolated by Lithium Chloride precipitation, and examined on a 1% agarose gel for the anticipated product size, compared against Lambda DNA digested with HindIII and EcoRI (both Cell Signaling Technologies). RNA was labelled using the 3′ end desthiobiotinylation RNA labelling kit (ThermoFisher), with 150 μg/25 nM RNA per reaction incubated at 16 °C overnight. Labelled RNA was then isolated by chloroform and ethanol precipitation. A negative control consisting of a scrambled RNA sequence was labelled at the same time. Labelling efficiency was measured by comparison of chemiluminescent signal from labelled sample RNA with a positive control using the ThermoFisher Scientific Chemiluminescent Nucleic Acid detection module. 25 nM of labelled RNA was then bound to nucleic acid-compatible streptavidin magnetic beads using the Pierce Magnetic RNA–Protein Pull-down kit (ThermoFisher Scientific) and incubated with 100ug protein lysate from human brain overnight at 4 °C. Bound protein was eluted from the beads and immediately subject to western blot analysis for SFs/RBPs of interest.
SDS-PAGE gel electrophoresis and western blot
Soluble protein was collected from cell pellets and human brain tissue by resuspension in Cell Lysis Buffer (Cell Signaling Technologies) supplemented with 10 µM PMSF on ice. Cells or tissue were then sonicated briefly on ice and spun at 13,000×g for 10 min at 4 °C to pellet debris. Protein concentration was determined by BCA assay (ThermoFisher Scientific). Prior to western blotting, human brain protein lysates were dephosphorylated to accurately determine Tau isoforms by size. Lysates were incubated with 100 units of Lambda protein phosphatase (LPP; Cell Signaling Technologies) per 10 μg total protein, supplemented with 1 × Protein MetalloPhosphatases buffer and 1 mM MnCl2, and incubated at 30 °C for 3 h before being analyzed by western blot.
For SDS-PAGE gel electrophoresis, 10–30 μg of protein was incubated with 1 × reducing agent and 1 × LDS sample buffer at 70 °C for 10 min before immediately being loaded onto a BOLT 4–16% Bis–Tris gel (ThermoFisher Scientific) in 1 × MES buffer. For splicing factor analyses, electrophoresis was carried out for 20 min at 200 V before blotting. For tau isoform analyses, electrophoresis was carried out for 60 min at 100 V before blotting. Gels were blotted onto nitrocellulose membranes using the iBlot system (ThermoFisher Scientific), and blocked for a minimum of 30 min in 5% milk in PBS-T. Primary antibodies were prepared at dilutions described in Supplementary Table 2 (online resource) in 5% milk in PBS-T and incubated with the membrane at 4 °C overnight. Membranes were washed 3 × in PBS-T, then incubated with either HRP Goat Anti-Rabbit or HRP Horse Anti-Mouse secondary antibodies (Vector laboratories) at a dilution of 1:20,000 in 5% milk in PBS-T for 2 h at room temperature. Following three additional washes, membranes were then incubated with WesternBright ECL HRP substrate (Advansta) for 3 min before imaging on a UVP ChemiDoc. For re-staining, blots were incubated in Restore PLUS stripping buffer (ThermoFisher Scientific) for 15 min at room temperature, followed by one wash in PBS and re-blocking.
qRTPCR
Cell pellets were collected by washing and scraping into ice-cold PBS. RNA was extracted from cell pellets using the Qiagen RNeasy mini RNA extraction kit, and reverse-transcribed using the high-capacity RNA-to-cDNA kit (ThermoFisher Scientific). qRTPCR for specific MAPT exons and isoforms was carried out using SybrGreen mastermix with the following primers: MAPT 4R Forward 5′-CGGGAAGGTGCAGATAATTAA-3′, Reverse 5′-GCCACCTCCTGGTTTATGATG-3′; MAPT 3R Forward 5′-AGGCGGGAAGGTGCAAATA-3′, Reverse 5′-GCCACCTCCTGGTTTATGATG-3′; MAPT 0N Forward 5′-TTTGAACCAGGATGGCTGAG-3′, Reverse 5′-ATGCCTGCTTCTTCAGCTTT-3′; MAPT Exon 2 Forward 5′-TTTGAACCAGGATGGCTGAG-3′, Reverse 5′-CTGCAGGGGAGATTCTTTCA-3′. SF/RBP overexpression was validated and quantified by qRTPCR using commercially available Taqman assays (ThermoFisher Scientific).
MAPT targeted iso-seq
RNA was extracted from human temporal cortex brain tissue as described above and submitted to the Icahn School of Medicine at Mount Sinai Genomics CoRE for single molecule real-time (SMRT) isoform sequencing (iso-seq) on the PacBio RS II platform using the following primers: Forward 5′-ATG GAA GAT CAC GCT GGG AC-3′, Reverse 5′-GAG GCA GAC ACC TCG TCA G-3′. Raw sequencing reads were passed through the ISOseq3 pipeline to detect full-length transcripts expressed in each sample. Beginning with raw subreads, single consensus sequences were generated for each MAPT amplicon with a SMRT adapter on both ends of the molecule. SMRT adapter sequences were then removed and MAPT-specific primer sequences were identified to orient the isoforms. Isoforms were subsequently trimmed of poly(A) tails and concatemers were identified and removed. Isoform consensus sequences were then predicted using a hierarchical alignment and iterative cluster merging algorithm to align incomplete reads to longer sequences. Finally, clustered isoform sequences were polished using the arrow model and binned into groups of isoforms with predicted accuracy of either ≥ 0.99 (high quality) or < 0.99 (low quality). The resulting isoforms were aligned to hg38 using the GMAP aligner [63] and isoform calling, collapsing and measurements of abundance were carried out using the Cupcake/ToFU pipeline (https://www.github/Magdoll/cDNA_Cupcake).
Immunohistochemistry
Immunohistochemical staining was carried out on formalin fixed paraffin-embedded brain sections by the Neuropathology Brain Bank and Research CoRE at the Icahn School of Medicine at Mount Sinai using the Ventana BenchMark autostainer. Slides were scanned on a Leica SCN400 at 40×. A list of antibodies used and their relevant dilutions can be found in Supplementary Table 2 (online resource).
OPAL multiplexed immunofluorescence
Multiplexed immunofluorescent staining was carried out using the Opal Polaris 7 color IHC detection kit (Akoya biosciences) according to manufacturer’s instructions. Briefly, slides were baked for 1 h at 65 °C, then deparaffinized with xylene and rehydrated with a graded series of ethanol concentrations. For epitope retrieval, slides were microwaved in AR buffer (provided with the OPAL IHC detection kit) for 45 s at 100% power, followed by an additional 15 min at 20% power. After cooling, slides were blocked for 10 min in blocking buffer then incubated with the first primary antibody at room temperature for 30 min. Slides were rinsed three times in TBS-T, then incubated with the secondary polymer HRP for 1 h at room temperature. After additional washes, the first Opal fluorophore was incubated with the slides for 10 min at room temperature, followed by further washes in TBS-T. This process was repeated from the microwave treatment step for each additional primary antibody, followed by one final repetition of the microwave treatment to strip the primary-secondary antibody complex from the tissue. Antibodies, concentrations and relevant Opal fluorophores can be found in Supplementary Table 2 (online resource). Once all primary antibodies had been introduced, slides were counterstained with DAPI for 5 min at room temperature, washed with TBS-T and coverslips were mounted using ProLong Diamond Antifade mounting reagent (ThermoFisher Scientific). Multispectral imaging was carried out using the Vectra Quantitative Pathology Imaging system, applying quantitative unmixing of fluorophores and removal of tissue autofluorescence. Images were visualized using the HALO image analysis platform (Indica Labs).
Statistical analysis
RNA-seq count and PSI data were analyzed as described above. Enrichment of SFs/RBPs in specific clusters was determined by Fisher’s exact test in R. Western blot protein bands were quantified by densitometry analysis in ImageJ and normalized to GAPDH for each sample, and the resulting values were subjected to unpaired Student’s t test. For tau isoform analysis, ratios between each isoform were calculated per sample prior to statistical analysis. For assessment of RNA pull-downs, the amount of eluted SF/RBP protein was normalized to the total amount of SF/RBP protein (Flow-through + Eluate) to determine the percentage of SF/RBP protein bound to the labelled RNA, and this value was used for statistical analysis by Student’s t test. For iso-seq data, the expression of each isoform was calculated as a proportion of all detected isoforms, and average expression between control, AD and PSP cases was calculated. Statistical difference in isoform expression and expression ratios were calculated by one-way ANOVA with Tukey post hoc testing. qRTPCR gene expression was analyzed using the ΔΔCt method, and expression was normalized to β-actin as endogenous control. Statistical significance was determined by the appropriate one-way ANOVA and Bonferroni post hoc testing. For cell culture experiments, all tests were conducted in triplicate in three independent experiments (total replicates = 9). For human brain analyses, tissue was acquired for 4–6 PSP cases, 4–6 AD cases and 4–6 healthy aged controls. Significant comparisons are labelled in figures as *p < 0.05, **p < 0.01 and ***p < 0.001.
Results
MAPT alternative splicing differs by brain region and 17q21.31 haplotype
MAPT is alternatively spliced at exons 2, 3 and 10 in the human brain (Fig. 1a), and expression of these exons across brain regions has previously been measured by microarray analyses [59]. However, accurate measurement of gene or exon expression by microarray may be impacted by the specificity of probe design and a narrow dynamic range for signal detection. We therefore chose to characterize the relative expression of MAPT exons in multiple human postmortem RNA-seq datasets from the AMP-AD consortium (Religious Orders Study Memory and Aging Project [ROSMAP; Synapse syn3219045] N = 450, Mount Sinai Brain Bank [MSBB; Synapse syn3159438] N = 230, and the Mayo Clinic [MAYO; Synapse syn3157268, syn5550404] N = 276) (Fig. 1b–f, Supplementary Fig. 1A-H (online resource)). We observed a pattern of exon expression consistent with that previously described [8, 59] and known isoforms expressed in brain (i.e., constitutive expression of exons 1, 4, 5, 7, 9, 11 and 12, little to no expression of exons 4a, 6 and 8, and variable levels of exon 2, 3 and 10), which was consistent across AMP-AD datasets and brain regions (Fig. 1b, Supplementary Fig. 1A-C (online resource)).
To assess proportional exon expression across different brain regions included in MSBB and MAYO data, we calculated percent spliced in (PSI) values for the alternatively spliced exons 2, 3 and 10 using MISO (Mixture of Isoforms) (Fig. 1c, d). While there were no differences in PSI values across MSBB Brodmann regions BM10 (frontal pole), BM22 (superior temporal gyrus), BM36 (fusiform gyrus) and BM44 (inferior frontal gyrus; Fig. 1c), there was significantly increased inclusion of all three exons in cerebellum compared to the temporal cortex in the MAYO cohort (Exon 2 p = 5.46 × 10–16; Exon 3 p = 2.74 × 10–8; Exon 10 p = 1.54 × 10–9; Fig. 1d). This is consistent with previous reports that suggest MAPT splicing in the cerebellum differs from the forebrain [3, 59].
We then compared PSI values between the major 17q21.31 MAPT H1 and H2 haplotypes, and observed increased exon 3 inclusion in H2 haplotype carriers compared to H1 across most AMP-AD datasets and brain regions (MSBB H1H2 p = 1.14 × 10–09, MSBB H2H2 p = 1.09 × 10–13; MAYO H1H2 p < 2 × 10–16, MAYO H2H2 p = 1.9 × 10–04, Fig. 1e, f, Supplementary Fig. 1D-E (online resource)), which has been previously reported in other RNA-seq and microarray datasets [2, 38, 59]. In comparison, we did not observe any difference in exon 2 or 10 inclusion between haplotypes (Fig. 1e, f, Supplementary Fig. 1D-E (online resource)). In contrast to previous reports [32], we did not find altered total MAPT expression between these haplotypes in any dataset or brain region (Supplementary Fig. 1F-H (online resource)).
MAPT exon 2 and 10 splicing is coordinated by SF/RBP expression
To identify novel MAPT splicing regulators in human prefrontal cortex, we carried out a correlational analysis between the expression (RPKM) of 294 known splicing factors (SFs) and RNA-binding proteins (RBP) described in Gerstberger et al. [18] (Supplementary Table 3 (online resource)) with MAPT exon 2, 3 and 10 PSI values, using ROSMAP and MSBB datasets (Fig. 1g, Supplementary Fig. 2A-G (online resource)). We did not observe any significant correlations that withstood Bonferroni multiple test correction between MAPT exon 3 and any SF/RBP (Supplementary Fig. 2A (online resource)), likely due to the very low expression of exon 3 in human brain. However, when we separated the data by H1/H2 haplotype, we observed stronger associations between exon 3 inclusion and SF/RBP expression in H2 homozygotes compared to H1 homozygotes in both datasets (Supplementary Fig. 2B-C (online resource)), although these still did not pass multiple test correction due to low frequency of the H2 haplotype in these datasets. While we did not have the statistical power to pursue analysis of exon 3 splicing regulators, these data indicate there may be additional regulation of exon 3 in the context of H2, consistent with its increased expression on this background.
In contrast, there were no significant differences in SF/RBP and MAPT exon 2/10 PSI value associations between H1/H2 haplotypes, which were highly correlated across both haplotypes (ROSMAP exon 2 R2 = 0.81, p < 0.001, exon 10 R2 = 0.89, p < 0.001; MSBB exon 2 R2 = 0.67 p < 0.001, exon 10 R2 = 0.66 p < 0.001) (Supplementary Fig. 2D-E (online resource)). Associations between PSI values and SF/RBP expression were also significantly correlated between AD cases and controls (ROSMAP exon 2 R2 = 0.97 p < 2.2 × 10–16, exon 10 R2 = 0.96 p < 2.2 × 10–16; MSBB exon 2 R2 = 0.89 p < 2.2 × 10–16, exon 10 R2 = 0.93 p < 2.2 × 10–16) (Supplementary Fig. 2F-G (online resource)), indicating no differences in the regulatory effects of SF/RBP expression on MAPT splicing in AD. We therefore focused our analyses on exons 2 and 10 using pooled AD/control and H1/H2 data.
Unsupervised hierarchical clustering of the resulting Pearson’s correlation coefficients between exon PSI values and SF/RBP expression revealed that exons 2 and 10 clustered separately from each other and had distinct patterns of association with SF/RBP expression (Fig. 1g). To identify robust MAPT splicing regulator candidates, we selected SF/RBPs with significant (Bonferroni-corrected p-value < 0.05) associations with MAPT exons 2 or 10 across the three most anatomically similar datasets; ROSMAP (prefrontal cortex), MSBB BM10 (frontal pole) and MSBB BM44 (inferior frontal gyrus). Many more SFs/RBPs were consistently associated with exon 10 exclusion (94 genes, 69.1% of all significant and replicated correlations, defined by a negative correlation; Supplementary Table 4 (online resource)) compared with its inclusion (5 genes replicated in 2/3 datasets, [3.7%], defined by a positive correlation; Supplementary Table 4 (online resource)), suggesting that more complex regulation may be required to promote removal of exon 10 from pre-mRNA transcripts in brain.
Fewer SFs/RBPs were consistently associated with exon 2 splicing (7 [5.1%] excluders, 16 [11.8%] includers). However, a proportion of SFs/RBPs were significantly associated with both exon 2 and exon 10 PSI values in opposing directions (Table 1). Specifically, 14 SF/RBPs (10.3%) were significantly correlated with both exon 10 exclusion and exon 2 inclusion, suggesting a coordinated regulation of MAPT N- and C-terminal splicing that has not been previously characterized. We therefore chose to focus on this subset of genes (Table 1).
RSRC1 and RBM11 directly bind MAPT pre-mRNA and alter MAPT splicing in vitro
To prioritize SFs/RBPs for validation, we identified genes known to be expressed in brain (queried through the GTex portal; www.gtexportal.org) and neurons (Barres RNA-seq browser; www.brainrnaseq.org), as we predicted these would be most relevant to the regulation of MAPT, which is primarily neuronally expressed. This resulted in a panel of seven exon 2 includers/exon 10 excluders (Table 1, bold text).
To validate the association of candidate SF/RBPs with MAPT splicing, we first assessed whether they were able to directly interact with MAPT pre-mRNA in the context of human brain tissue. We carried out RNA pull-downs using desthiobiotinylated in vitro reverse-transcribed RNA generated from a mini-gene containing MAPT exons 9-11, with full intervening intronic sequences (LI9LI10) [64] and probed protein lysates derived from postmortem human prefrontal cortical tissue. The resulting eluates were examined by western blot for SFs/RBPs of interest. LI9LI10-derived MAPT pre-mRNA pulled down significant proportions of RSRC1 (41.62%, p < 0.001) and RBM11 (11.11%, p < 0.01) proteins from human brain lysates (Fig. 2a, b), as well as a minimal, but not significant proportion of THOC3 (5.77%) and SNRNP25 (4.77%), suggesting that these factors may regulate MAPT exon 10 splicing via a direct interaction with its pre-mRNA.
RBM11 and RSRC1 directly bind to MAPT pre-mRNA and regulate splicing. a MAPT pre-mRNA pull-downs from human brain tissue protein lysates for each target SF/RBP using the LI9LI10 minigene sequence or scrambled (Scr) RNA sequence as bait. FT, flow through fraction not bound to MAPT pre-mRNA or non-specific control; E, eluate fraction bound to target RNA. b Quantification of RNA pull-down western blots in a. Fraction of protein pulled down in E normalized to total protein present in FT and E fractions combined. N = 3, each SF/RBP compared to scrambled control using t test. **p < 0.01, ***p < 0.001, ns not significant. c Fold change (FC) expression of 4R:3R and Ex2:0N ratios in SH-SY5Y cells by qRTPCR following overexpression of either RBM11 or RSRC1. Asterisks denote significantly different expression compared to empty vector control, represented by grey line intersecting plot at FC = 1. One-way ANOVA and post-hoc Bonferroni tests for multiple comparisons. N = 3 independent experiments with 3 replicates each. **p < 0.01, ***p < 0.001
To functionally validate whether RSRC1 and RBM11 could alter MAPT splicing in vitro, we overexpressed these SF/RBPs in human neuroblastoma SH-SY5Y cells as an immortalized cell line proxy to neuronal cells (Supplementary Fig. 3A-B (online resource)). As expression of MAPT exon 10 was very low in this cell line, we co-expressed the LI9L10 minigene with each SF/RBP to facilitate measurement of exon 10 exclusion. Consistent with their putative roles as exon 10 excluders, overexpression of either RBM11 or RSRC1 significantly reduced the 4R:3R ratio, as measured by qRTPCR (RBM11 p < 0.001, RSRC1 p < 0.01) (Fig. 2c). While PPIH and SNRPB did not directly bind MAPT pre-mRNA, their overexpression resulted in a significantly reduced 4R:3R ratio in SH-SY5Y cells (SNRPB p < 0.01, PPIH p < 0.001) (Supplementary Fig. 3C (online resource)). There was no significant effect of THOC7, THOC3 or SNRNP25 (Supplementary Fig. 3C (online resource)), possibly due to poor overexpression efficiency. We then measured the ability for candidate SF/RBPs to alter N-terminal splicing of endogenous MAPT, and found that both RBM11 and RSRC1 overexpression significantly increased the Exon 2/0N ratio in SH-SY5Y cells (RBM11 p < 0.001, RSRC1 p < 0.01) (Fig. 2C), consistent with their hypothesized role as exon 2 includers. In contrast, we did not see any effect of other candidate SF/RBPs on exon 2 inclusion (Supplementary Fig. 3D (online resource)). We therefore conclude that RBM11 and RSRC1 may be important regulators of combinatorial N- and C-terminal MAPT splicing.
Regulators of MAPT N-terminal splicing and MAPT exon 2 are differentially expressed in PSP and AD brain
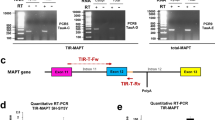
Altered MAPT C-terminal splicing is a characteristic of several tauopathies [15], including PSP19 and AD, for which there is inconsistent data [3, 12, 19, 20, 29, 62]. We therefore queried the AMP-AD MAYO temporal cortex RNA-seq dataset, which includes both AD and PSP cases, to investigate whether MAPT splicing regulation may be altered in tauopathy brain. We calculated the fold change (FC) expression of every significant SF/RBP from our initial correlational analysis (Supplementary Table 4 (online resource)) in PSP and AD compared to controls (Fig. 3a). While PSP and AD shared largely similar patterns of SF/RBP expression dysregulation compared to controls, there were many genes that exhibited differential patterns of expression in either disease (Fig. 3a). The group of SFs/RBPs that exhibited increased expression in PSP and reduced expression in AD comprised 4/7 of our candidate SF/RBPs, including both RBM11 and RSRC1, which were associated with both exon 2 inclusion and exon 10 exclusion. In contrast, SFs/RBPs that were increased in AD and reduced in expression in PSP were significantly enriched for exon 2 excluders (Fisher’s exact test p = 0.0005) (Fig. 3a). Furthermore, the net fold change (FC) expression of all exon 2 includers was significantly higher in PSP temporal cortex compared to AD (PSP average FC = 0.024, AD average FC = − 0.036, p < 0.01) (Fig. 3b), suggesting differential regulation of MAPT exon 2 splicing between diseases. To confirm this hypothesis, we compared MAPT exon 2, 3 and 10 PSI values in control, PSP and AD brain from the same dataset (Fig. 3d–f). Consistent with the observed patterns of exon 2 splicing regulator expression in PSP and AD, we observed significantly different exon 2 inclusion across cases compared to controls (F(2,134) = 4.01, p = 0.02), with significantly increased exon 2 PSI in PSP brain compared to controls (Tukey HSD p = 0.02), and a trend towards reduced exon 2 PSI in AD brain compared to controls (Fig. 3d). In contrast, there was no significant difference in exon 3 or exon 10 PSI values between either disease and controls (Fig. 3e, f). Taken together, this suggests there is differential dysregulation of MAPT N-terminal splicing regulators between AD and PSP brain, which results in altered expression of MAPT exon 2.
MAPT exon 2 expression and regulation is differentially altered in PSP and AD brain. a Fold change expression of each significant SF/RBP from the analysis in Fig. 1g in PSP and AD brain compared to controls (AMP-AD MAYO temporal cortex), with unsupervised hierarchical clustering. Red indicates increased expression compared to controls, blue indicates reduced expression compared to controls. The direction of association of each SF/RBP with MAPT exon 2 and exon 10 splicing is indicated to the right of the figure. Clusters containing target SF/RBPs of interest from prior analyses and MAPT exon 2 excluders (purple) are indicated. b Sum of the fold change expression of all exon 2 includers in PSP and AD brain compared to controls (AMP-AD MAYO temporal cortex). Statistical comparison between sum expression in PSP compared to AD brain, t test, Error bars ± SEM. **p < 0.01. c Single nuclei and single-soma RSRC1 expression in AD entorhinal cortex, AT8-positive/negative neurons and PSP subthalamic nucleus. FC, fold change expression disease/AT8-positive compared to controls/AT8-negative. Depth of color indicates scaled average expression, size of dot indicates proportion of cells expressing RSRC1, also denoted by percentage value above each dot. MAST linear model with Bonferroni correction, **p < 0.01, ***p < 0.001. d–f PSI values for exons 2 (d), 3 (e) and 10 (f) in control, AD and PSP brain (AMP-AD MAYO temporal cortex). Red dashed line indicates mean PSI value in controls for each exon. One-way ANOVA with post hoc Tukey tests. Error bars ± SEM. *p < 0.05
RSRC1 is differentially expressed in AD and PSP neurons in disease-relevant brain regions
While MAYO temporal cortex bulk RNA-seq data indicated altered expression of exon 2 splicing regulators in PSP and AD brain, these data may be influenced by altered cell type proportions in the disease context. Therefore, to validate neuronal and disease-specific patterns of expression of exon 2 splicing regulators, we assessed single nuclei sequencing (snuc-seq) from AD entorhinal cortex [22] and PSP subthalamic nucleus [52], as well as single-soma sequencing of hyperphosphorylated tau (AT8)-positive neurons from AD prefrontal cortex [47]. We found that while RBM11 was not detectable in AD snuc-seq data, and was expressed at very low levels in the other datasets, RSRC1 was consistently detected and more highly expressed across data sets. While RSRC1 expression was highest in microglia in both entorhinal cortex and subthalamic nucleus, we confirmed that it was also expressed in neurons in these regions (Supplementary Fig. 4A-B (online resource)). Consistent with data from bulk temporal cortex tissue, we observed significantly lower RSRC1 expression in AD tissue compared to controls (FC = 0.07, p < 0.01), as well as a lower proportion of RSRC1-expressing cells in AD compared to controls (8.9% vs 12.5%, respectively) (Fig. 3c). We observed the same fold change expression and reduction in RSRC1-expressing cells when assessing neuronal populations specifically (Fig. 3c), although due to the small proportion of neurons present in the data (Supplementary Fig. 4C (online resource), this was not significant. Interestingly, RSRC1 expression was also higher in neurons derived from AD prefrontal cortex that were negative for hyperphosphorylated tau (AT8-), compared to AT8 + neurons (FC = 0.13, p = 3.33 × 10–33) (Fig. 3c, Supplementary Fig. 4D (online resource)), indicating that there may be an interaction between RSRC1 expression, MAPT splicing regulation and the formation of tau pathology.
In contrast, we found significantly higher RSRC1 expression in PSP subthalamic nucleus cells compared to controls (FC = 0.04, p < 0.01), as well as a higher proportion of RSRC1-expressing cells in PSP (10% compared to 7% controls) (Fig. 3c). These differences were exacerbated when assessing neurons alone (FC = 0.08, 17% PSP neurons vs 10% control neurons) (Fig. 3c), but similar to the AD data, the small proportion of neurons in this dataset (Supplementary Fig. 4E (online resource)) precluded these data from reaching statistical significance. Interestingly, we were able to detect the opposite pattern of effect for the exon 2 excluders QKI and PRPF38B in AD and PSP neurons by snuc-seq: consistent with the temporal cortex bulk data, expression of QKI and PRPF38B were higher in AD neurons (QKI FC = 0.239, PRPF38B FC = 0.04) and lower in PSP neurons compared to controls (QKI FC = − 0.03, 47.8% PSP vs 44.4% control neurons, PRPF38B FC = − 0.125, 15.7% PSP vs 12.5% control) (Supplementary Fig. 4F (online resource)). These data therefore support our assertion of differential expression of MAPT exon 2 regulators between AD and PSP brain. Furthermore, this demonstrates that SF/RBP expression is altered in neurons in disease-relevant brain regions.
MAPT N-terminal isoforms are expressed at different ratios in AD and PSP brain
As we found evidence of coordinated splicing between MAPT exons 2 and 10, but observed differential expression of only exon 2 and exon 2 regulators in AD and PSP brain, we hypothesized that there may be loss of coordinated combinatorial splicing regulation between N- and C-terminal MAPT in tauopathy brain. We therefore carried out targeted MAPT isoform (iso)-seq on temporal cortex tissue from 6 × control, 6 × AD and 6 × PSP cases to assess the expression of full-length transcripts (Fig. 4a–c). From control and PSP brain, we detected ~ 150,000 reads mapping to the 6 main MAPT isoforms (control = 157,561; PSP = 143,820), whereas only 24,237 reads were mapped in AD brain tissue, possibly due to longer post-mortem intervals and more severe degeneration of neurons in these cases. We observed similar ratios of expression for each isoform as previously described by western blot analyses [59]: 1N3R and 0N3R were the two most highly expressed isoforms (~ 30% and 25%, respectively), followed by 0N4R (~ 21%) 1N4R (~ 15%) and finally very low expression of both 2N isoforms 2N3R (~ 1.4%) and 2N4R (~ 0.8%) (Fig. 4a). This pattern of expression was largely similar in AD and PSP cases; however, there was a trend towards increased expression of both 0N isoforms in AD brain compared to controls (AD 0N3R 42.5% ± 12.7% vs 25% ± 5% controls, AD 0N4R 31.6% ± 3% vs 21% ± 4.6% controls), although due to high variability and lower read mapping, these differences did not reach statistical significance (Fig. 4a). We then compared the 4R:3R ratios for each N-terminal isoform, and found that despite increased expression of 0N isoforms in AD, there was no difference for either AD or PSP in the 0N4R:0N3R ratio (Fig. 4b). In contrast, we observed a trend towards an increase in exon 2-containing 4R:3R ratios in PSP cases compared to controls (1N4R:1N3R PSP = 1.1 ± 0.38 vs 0.53 ± 0.07 controls, 2N4R:2N3R PSP = 0.8 ± 0.13 vs 0.55 ± 4.9 controls) (Fig. 4b), consistent with both our observation of increased exon 2 inclusion in temporal cortex (Fig. 3d) and the known pathological accumulation of 4R tau isoforms in PSP pathology. Lastly, when comparing N-terminal isoform expression, we found no differences in PSP brain, but a trend towards reduced 1N/0N and 2N/0N ratios in AD brain specifically (1N/0N AD = 0.43 ± 0.13 vs 1.2 ± 0.33 controls, 2N/0N AD = 0.02 ± 0.01 vs 0.05 ± 0.01 controls) (Fig. 4c), likely due to the increase in expression of 0N isoforms (Fig. 4a). Interestingly, we also observe an increase in the 2N/1N ratio in AD brain (0.1 ± 0.03 vs 0.05 ± 0.005 controls) (Fig. 4c), which may be due to slightly decreased 1N expression or increased 2N3R expression we observe in AD compared to controls (Fig. 4a).
Coordination of N- and C-terminal splicing regulation is altered in AD and PSP brain, and is associated with tau pathology. a–c Proportion of each full-length MAPT isoform (a), 4R:3R ratio for each N-terminal isoform (b) and N-terminal isoform ratios (c) as detected by targeted MAPT iso-seq in human control, AD and PSP temporal cortex. One-way ANOVA with post hoc Tukey tests, all comparisons did not reach statistical significance. N = 6. Error bars ± SEM. d Dephosphorylated tau isoform expression in PSP temporal cortex compared to tau ladder detected by specific N-terminal antibodies against 0N, 1N and 2N. Anti-1N tau showed poor detection for 1N4R isoforms, so anti-4R tau antibody RD4 was used for detection of this isoform. GAPDH used as loading control. e Quantification and 4R:3R ratio of N-terminal isoforms in d. Each point denotes a different individual brain lysate, N = 6. Error bars ± SEM. Student’s t test **p < 0.01. f Dephosphorylated tau isoform expression in AD temporal cortex compared to tau ladder with same antibody detection as in d. GAPDH used as loading control. g Quantification and 4R:3R ratio of N-terminal isoforms in f. Each point denotes a different individual brain lysate, N = 6. Error bars ± SEM. Student’s t test *p < 0.05, **p < 0.01. h Representative images of labeling of control, AD and PSP temporal cortex with marker of hyperphosphorylated tau, AT8, indicating different tau pathologies present across cases. Sections counterstained with hematoxylin and eosin. N = 3–4. Scale bar = 100 µm. i Representative images of tau pathology labeling in control, AD and PSP temporal cortex with anti-0N, 1N and 2N tau antibodies from several individuals. Sections counterstained with hematoxylin and eosin. N = 3–4. Scale bar = 100 µm
To examine whether these transcriptional changes were apparent at the protein level, we carried out western blot analyses in the same tissues from the same individuals (Fig. 4d–g). Consistent with the direction of the trends observed in the iso-seq data, there was a significantly increased 1N4R:1N3R ratio in PSP brain compared to controls (control 1N4R:1N3R ratio = 0.52 (SEM = 0.1), PSP 1N4R:1N3R ratio = 1.7 (SEM = 0.4), p = 0.009), which was largely driven by a reduction in 1N3R tau (Fig. 4d, e). In contrast, there was no difference in either 0N or 2N 4R:3R ratios in PSP (Fig. 4d, e). In AD brain, there was an accumulation of soluble 0N and 2N isoforms by western blot, consistent with the iso-seq data (Fig. 4c, f, g). When examining 4R:3R ratios for each N-terminal isoform, we observed significantly increased 0N4R:0N3R (control ratio = 0.83 (SEM = 0.05), AD ratio = 1.49 (SEM = 0.15), p = 0.002) and 2N4R:2N3R ratios (control ratio = 0.83 (SEM = 0.05), AD ratio = 1.96 (SEM = 0.46), p = 0.03) in AD brain compared to controls (Fig. 4f–g), indicating that while there were no transcriptional differences in the 4R:3R ratio for these isoforms, there may be impaired degradation of 0N4R and 2N4R tau in AD resulting from their increased expression.
Pathologically aggregated tau in PSP and AD brain are associated with different N-terminal isoforms
While western blot analysis of AD and PSP temporal cortex revealed differences in the accumulation of soluble N-terminal tau isoforms, we wanted to determine whether the formation of different neuropathological features between tauopathies may be due to the insoluble aggregation of different N-terminal tau isoforms. After validating the specificity of each N-terminal antibody by overexpressing different tau isoforms in N2a cells (Supplementary Fig. 5A-B (online resource)), we carried out immunohistochemistry (IHC) on control, AD and PSP brain sections from the temporal cortex for hyperphosphorylated pathogenic tau (AT8; Fig. 4h)) and each N-terminal isoform (Fig. 4i). We observed the anticipated hyperphosphorylated tau neuropathology in both PSP and AD brain (Fig. 4h), specifically the widespread presence of neurofibrillary tangles and neuropil threads throughout the AD cortex, and sparse glial plaques and neurofibrillary tangles in PSP tissue. In comparison, there was little to no signal in control brain (Fig. 4h).
There was little signal for 0N tau in PSP brain (Fig. 4i), although in one case we observed sparse labeling of neuropil threads or possible glial involvement (Fig. 4i). In AD brain the 0N antibody did not label neurofibrillary tangles, but we did observe labeling of neuropil threads and dystrophic neurites surrounding amyloid plaques (Fig. 4i). Interestingly, in one case, the 0N antibody labelled thorny astrocytes consistent with an age-related tau astrogliopathy (ARTAG) pathology that was not visible with the AT8 antibody (Fig. 4h, i). While it is surprising that there was little immunolabeling of 0N Tau in human brain, given the high levels of 0N transcripts, this pattern was replicated with a second antibody against 0N Tau (Supplementary Fig. 5C (online resource)).
1N Tau immunostaining was primarily present in neurofibrillary tangles and pre-tangles (Fig. 4i) throughout the cortex in all three PSP cases, as well as neuropil granules and threads. In AD, this isoform was also the most prevalent in neurofibrillary tangles (Fig. 4i), but was primarily present in dystrophic neurites surrounding amyloid plaques (Fig. 4i). Finally, while 2N Tau was observed in some neurofibrillary tangles in PSP and AD brain (Fig. 4i), these were less common than the 1N labelled neurons. 2N Tau was also observed in dystrophic neurites surrounding amyloid plaques in AD brain, but this labelling was far less dense than the 1N Tau and less tightly co-localized with plaques (Fig. 4i). A second 2N antibody revealed a similar labeling of neurofibrillary tangles in AD and PSP brain, but less involvement in dystrophic neurites (Supplementary Fig. 5C (online resource)).
To directly compare the accumulation of different tau N-terminal isoforms in pathogenic inclusions, we carried out Opal multiplex labelling of adjacent brain sections from the same individuals using all three N-terminal antibodies in conjunction with AT8 and β-amyloid staining (Fig. 5a, b, Supplementary Fig. 6A-B (online resource)). Consistent with the IHC staining, we observed primarily 1N and 2N tau in AT8-positive neurofibrillary tangles in PSP temporal cortex, but all three N-terminal isoforms were present in AD-associated tangles (Fig. 5a, b, Supplementary Fig. 6A-B). Dystrophic neurites surrounding amyloid plaques were primarily associated with 0N and 1N accumulation, with less 2N involvement (Fig. 5a, Supplementary Fig. 6A (online resource)). In contrast, amyloid plaques present in either controls or PSP cases were not associated with any tau staining (Supplementary Fig. 6B (online resource)). Interestingly, 2N tau was absent in glial pathology observed in both AD and PSP cases, suggesting that 2N tau is unable to accumulate in glia, while astrocytic tufts in PSP brain were labelled primarily by 1N tau (Fig. 4i), consistent with the increased accumulation of 1N4R tau isoforms we observed by western blot and transcriptomic analyses.
N-terminal isoforms accumulate differently in AD and PSP neuronal and glial pathologies. a Representative images of multiplex immunofluorescent labeling of AD temporal cortex with AT8 (red), β-amyloid (green), 2N tau (blue), 1N tau (yellow) and 0N tau (orange), and overlay of all three N-terminal tau antibodies in 4 different individuals. Examples of tau accumulation in dystrophic neurites can be found in AD1 and AD2, thorny astrocytes present in AD3, and an example of a neurofibrillary tangle shown in AD4. N = 4, scale bar = 50 µm. b Representative images of multiplex immunofluorescent labeling of PSP temporal cortex from 3 individuals as in a. Examples of early and pre-tangles shown in PSP1 and PSP3, while examples of astrocytic tufts are shown in PSP2. N = 3, scale bar = 50 µm
Discussion
To date, the contribution of N-terminal MAPT splicing to disease pathogenesis has largely been overlooked compared to evaluation of exon 10 splicing and 4R tau expression. However, N-terminal tau is relevant to disease pathogenesis: for example, N-terminal fragments are prominent in AD cerebrospinal fluid (CSF), and their secretion from cells can be inhibited by the presence of exon 2 [14, 36]. In contrast, 0N Tau is more readily cleaved and released from human neuroblastoma cells [36]. This is consistent with our IHC data, where we observe little 0N tau accumulation in neurons, but prominent 1N and 2N tau in neurofibrillary tangles. The regulation of tau release by N-terminal inserts also has implications for our understanding of tau spread and seeding, as different isoforms may be available extracellularly, and exhibit different seeding competencies. Furthermore, aberrant folding of N-terminal tau is one of the earliest pathological changes identified in tauopathies [11], thus supporting the assertion that N-terminal splicing is a relevant consideration when modeling and investigating tauopathy.
N-terminal tau splicing may play a role in modifying tau subcellular localization and aggregation propensity. The N-terminal contains a plasma-interacting domain [4] that interacts with synaptic proteins and Annexin A6 [17, 56], which results in retention of tau in the axonal compartment. While exons 2 and 3 are not within this domain, it is possible that they modify these interactions and impact the subcellular localization of tau, resulting in the increased propensity for certain isoforms to accumulate in these regions. Indeed, murine 0N tau has been found to localize in axons, whereas 1N was enriched in dendrites and 2N tau was depleted from cytoskeletal structures [41], indicating that N-terminal splicing likely modifies tau function. 1N tau has also been found to more readily accumulate and aggregate: the presence of exon 2 promotes the fibrillary extension of tau filaments in vitro [65] and exon 2-containing proteins more readily polymerize than 0N tau [37], therefore increased exon 2 inclusion would be likely to worsen tau aggregation. This is consistent with our observation of strong 1N tau immunostaining in PSP astroglia and tangles in both PSP and AD neurons. Despite this, we observe increased 0N expression in AD brain. However, the resulting protein accumulation was soluble, and associated with dystrophic neurites surrounding amyloid plaques rather than in neurofibrillary tangles. N-terminal tau splicing has also been found to be required for specific interactions with proteins associated with synaptic signaling and the plasma membrane [42], supporting the assertion that different N-terminal isoforms likely facilitate distinct cellular functions. Characterizing the disruption of N-terminal splicing is therefore important for understanding tau biology and the mechanisms underlying disease pathogenesis.
We have identified coordinated regulation of MAPT exons 2 and 10, indicating that N- and C-terminal splicing of either region likely does not occur fully independently of the other. This phenomenon has been reported once previously using polony-based exon profiling [12]. Disrupting this coordination will therefore lead to imbalanced expression of different tau isoforms. Interestingly, while the expression of numerous exon 10 splicing regulators were altered between AD, PSP and control brains, we observed enrichment of exon 2 regulators with opposing patterns of differential expression between PSP and AD. As anticipated from this pattern of expression, we observed increased exon 2 inclusion in PSP and reduced exon 2 in AD. However, assessment of full-length isoforms by iso-seq and western blot revealed that shifts in N-terminal splicing appear to be coupled to alterations in the 4R:3R ratio, supporting our hypothesis that loss of coordinated regulation between N- and C-terminal splicing contributes to tauopathy pathogenesis. The change in the 4R:3R ratio for specific N-terminal isoforms may explain why we did not observe an increase in exon 10 inclusion in PSP by short-read bulk RNA-seq; the most highly expressed 0N isoforms did not exhibit a change in the 4R:3R ratio in PSP brain, therefore the lack of change in 0N4R may have diluted out increases in 1N or 2N4R. Curiously, the increased 1N4R:1N3R ratio observed in PSP appeared to be largely due to a loss of 1N3R rather than an increase in 1N4R tau, thus raising the possibility that in combination with 1N4R tau accumulation, losing 1N3R tau expression and function could also be detrimental to neuronal health.
Aberrant regulation of splicing is a known phenotype of some forms of inherited tauopathy [1], which is hypothesized to be due to the sequestration and mislocalization of SF/RBPs to the cytosol and into stress granules [1]. SF/RBP dysfunction is therefore a likely mechanism underlying aberrant MAPT splicing in non-familial AD and PSP. While we do not characterize the mechanism of SF/RBP disruption in AD and PSP brain here, we do identify differential expression of numerous SF/RBPs between AD and PSP compared to controls, which is suggestive of separate downstream effects of splicing dysregulation that ultimately contribute to the pathogenesis of either disease.
We found that MAPT splicing is likely regulated by numerous splicing factors, of which RSRC1 and RBM11 may be of particular interest. However, it is likely that there are many other modifiers that were not identified in our screen. Several of our SF/RBPs of interest, including SNRPB, SNRNP25, THOC7 and THOC3, are known to be components of protein complexes that regulate splicing, and therefore would be unlikely to be isolated by the RNA pull-down assay. Indeed, overexpression of some of these SF/RBPs was able to significantly shift the 4R:3R ratio in human neuroblastoma cell lines. While expressed in brain and in neurons, many of these SF/RBPs are ubiquitously expressed in many different tissues and cell types. Indeed, we observed the highest expression of RSRC1 in microglia in human brain snuc-seq data. Therefore, while their dysregulation may be impacting MAPT splicing in neurons, there will likely be wider effects of altered expression and splicing in other neural cell types that may also be relevant for disease pathogenesis.
RSRC1 is a member of the serine- and arginine-rich-related protein family, which are highly conserved regulators of alternative splicing, but how RSRC1 regulates this process is unknown. It is hypothesized that RSRC1 plays a role in 3′ splice site selection by interaction with splicing regulators U2AF and SRSF2 [10], although it also possible that it may interact directly with pre-mRNA via its RS (arginine–serine) domain. However, there is no specific RNA motif that has been characterized as binding RS domains; thus, the region in which RSRC1 may be binding MAPT pre-mRNA is currently unknown. Mutations within this gene have been associated with intellectual disability [50, 51], whereas the silencing of RSRC1 in SH-SY5Y cells has been associated with downregulation of genes associated with schizophrenia, Alzheimer’s disease and dementia [50], thus indicating the relevance of this gene for brain function.
RBM11 is a brain-specific splicing factor that exhibits fluctuating expression with brain development, with high expression throughout embryogenesis, peaking at perinatal days 0–3 [49], after which MAPT exon 10 expression increases [25], consistent with our observations that RBM11 expression may promote exon 10 exclusion. RBM11 is associated with the choice of 5’ splice sites and may antagonize the activity of other SF/RBPs such as SRSF1 [49], potentially regulating MAPT splicing by direct interaction with pre-mRNA and by inhibiting binding of competing SFs/RBPs. RBM11 is downregulated in the PS19 mouse model of tauopathy [1], consistent with the direction of effect we observe in AD brain. However, this mouse model expresses only 1N4R tau and as such the effect of RBM11 disruption on MAPT splicing was not measurable in this model. It should be noted that we were unable to validate RBM11 expression in snuc-seq data as its expression was very low; therefore, the relevance it may have to tauopathy and MAPT splicing requires additional investigation.
We were unable to identify regulators of MAPT exon 3 splicing from these data, or to compare its regulation between 17q21.31 H1 and H2 haplotypes due to the low frequency of H2 carriers. Increased Exon 3 inclusion is consistently identified on the H2 background [38, 59], an effect we also observe in these data. However, the mechanism underlying increased exon 3 inclusion, and whether its expression is protective against tauopathy is currently unknown, although it may reduce the fibrillization of tau [65]. It should be noted that exon 3, and subsequently 2N tau expression, is very low in adult human brain (accounting for less than 3% of all transcripts in our targeted iso-seq data); therefore, the extent to which it may contribute to disease is unclear. Nevertheless, we observe some alterations in 2N expression and accumulation in AD and PSP, the most striking of which was the absence of 2N tau in glial pathology, indicating that 2N tau is either not released from neurons or is not internalized or aggregated by astrocytes. These data have implications for the design of experimental models of tauopathy where a single tau isoform is expressed to ensure the most disease and pathology appropriate isoform is utilized.
In conclusion, we propose that changes in SF/RBP expression result in differential splicing of the MAPT N-terminus between AD and PSP, resulting in the expression of isoforms with different aggregation properties and subcellular localizations, thus explaining the distinct neuropathological phenotypes of each disease. It would therefore be of great interest to investigate the role of N-terminal splicing in other primary tauopathies associated with different pathologies, such as Pick’s disease (PiD), primary age-related tauopathy (PART), age-related tau astrogliopathy (ARTAG) and chronic traumatic encephalopathy (CTE) to determine whether these diverse disorders also exhibit loss of MAPT exon 2 and 10 splicing coordination. These data indicate that it is unlikely that exon 10 splicing is alone in underlying and regulating disease pathogenesis and tau neuropathology in either AD or PSP, but rather the combinatorial expression of specific and N- and C-terminal MAPT isoforms is relevant for understanding the development of tauopathy. While alterations in the 4R:3R ratio are undoubtedly important and relevant to our understanding of tau pathogenesis, N-terminal splicing is likely to be an important modifier of disease and pathology, and should be carefully assessed.
References
Apicco DJ, Zhang C, Maziuk B, Jiang L, Ballance HI, Boudeau S et al (2019) Dysregulation of RNA splicing in tauopathies. Cell Rep 29:4377-4388.e4. https://doi.org/10.1016/j.celrep.2019.11.093
Beevers JE, Lai MC, Collins E, Booth HDE, Zambon F, Parkkinen L et al (2017) MAPT genetic variation and neuronal maturity alter isoform expression affecting axonal transport in iPSC-derived dopamine neurons. Stem Cell Rep 9:587–599. https://doi.org/10.1016/j.stemcr.2017.06.005
Boutajangout A, Boom A, Leroy K, Brion JP (2004) Expression of tau mRNA and soluble tau isoforms in affected and non-affected brain areas in Alzheimer’s disease. FEBS Lett 576:183–189. https://doi.org/10.1016/j.febslet.2004.09.011
Brandt R, Léger J, Lee G (1995) Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J Cell Biol 131:1327–1340. https://doi.org/10.1083/jcb.131.5.1327
Broderick J, Wang J, Andreadis A (2004) Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene 331:107–114. https://doi.org/10.1016/j.gene.2004.02.005
Bruch J, Xu H, De Andrade A, Höglinger G (2014) Mitochondrial complex 1 inhibition increases 4-repeat isoform tau by SRSF2 upregulation. PLoS ONE 9:e113070. https://doi.org/10.1371/journal.pone.0113070
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. https://doi.org/10.1038/nbt.4096
Caillet-Boudin M-L, Buée L, Sergeant N, Lefebvre B (2015) Regulation of human MAPT gene expression. Mol Neurodegener 10:28. https://doi.org/10.1186/s13024-015-0025-8
Caillet-Boudin ML, Fernandez-Gomez FJ, Tran H, Dhaenens CM, Buee L, Sergeant N (2014) Brain pathology in myotonic dystrophy: when tauopathy meets spliceopathy and RNAopathy. Front Mol Neurosci 6:1–20. https://doi.org/10.3389/fnmol.2013.00057
Cazalla D, Newton K, Cáceres JF (2005) A novel SR-related protein is required for the second step of pre-mRNA splicing. Mol Cell Biol 25:2969–2980. https://doi.org/10.1128/mcb.25.8.2969-2980.2005
Combs B, Hamel C, Kanaan NM (2016) Pathological conformations involving the amino terminus of tau occur early in Alzheimer’s disease and are differentially detected by monoclonal antibodies. Neurobiol Dis 94:18–31. https://doi.org/10.1016/j.nbd.2016.05.016
Conrad C, Zhu J, Conrad C, Schoenfeld D, Fang Z, Ingelsson M et al (2007) Single molecule profiling of tau gene expression in Alzheimer’s disease. J Neurochem 103:1228–1236. https://doi.org/10.1111/j.1471-4159.2007.04857.x
D’Souza I, Schellenberg GD (2005) Regulation of tau isoform expression and dementia. Biochim Biophys Acta Mol Basis Dis 1739:104–115. https://doi.org/10.1016/j.bbadis.2004.08.009
Derisbourg M, Leghay C, Chiappetta G, Fernandez-Gomez FJ, Laurent C, Demeyer D et al (2015) Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci Rep 5:1–10. https://doi.org/10.1038/srep09659
Espinoza M, de Silva R, Dickson DW, Davies P (2008) Differential incorporation of tau isoforms in Alzheimer’s disease. J Alzheimers Dis 14:1–16
Gao QS, Memmott J, Lafyatis R, Stamm S, Screaton G, Andreadis A (2000) Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J Neurochem 74:490–500. https://doi.org/10.1046/j.1471-4159.2000.740490.x
Gauthier-Kemper A, Alonso MS, Sündermann F, Niewidok B, Fernandez MP, Bakota L et al (2018) Annexins A2 and A6 interact with the extreme N terminus of tau and thereby contribute to tau’s axonal localization. J Biol Chem 293:8065–8076. https://doi.org/10.1074/jbc.RA117.000490
Gerstberger S, Hafner M, Tuschl T (2014) A census of human RNA-binding proteins. Nat Rev Genet 15:829–845. https://doi.org/10.1038/nrg3813
Ginsberg SD, Che S, Counts SE, Mufson EJ (2006) Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Neurochem 96:1401–1408. https://doi.org/10.1111/j.1471-4159.2005.03641.x
Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F et al (2006) The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer’s disease. J Neurochem 96:635–644. https://doi.org/10.1111/j.1471-4159.2005.03552.x
Goode BL, Feinstein SC (1994) Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol 124:769–781. https://doi.org/10.1083/jcb.124.5.769
Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C et al (2019) A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat Neurosci 22:2087–2097. https://doi.org/10.1038/s41593-019-0539-4
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. https://doi.org/10.1093/bioinformatics/btw313
Hafemeister C, Satija R (2019) Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20:1–15. https://doi.org/10.1101/576827
Hefti MM, Farrell K, Kim SH, Bowles KR, Fowkes ME, Raj T et al (2018) High-resolution temporal and regional mapping of MAPT expression and splicing in human brain development. PLoS ONE 13:1–14. https://doi.org/10.1371/journal.pone.0195771
Hernández F, Merchán-Rubira J, Vallés-Saiz L, Rodríguez-Matellán A, Avila J (2020) Differences between human and murine tau at the N-terminal end. Front Aging Neurosci 12:1–6. https://doi.org/10.3389/fnagi.2020.00011
Hernández F, Pérez M, de Barreda EG, Goñi-Oliver P, Avila J (2008) Tau as a molecular marker of development, aging and neurodegenerative disorders. Curr Aging Sci 1:56–61. https://doi.org/10.2174/1874609810801010056
Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H et al (1998) Association of missense and 5’ -splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998:393
Ingelsson M, Ramasamy K, Cantuti-Castelvetri I, Skoglund L, Matsui T, Orne J et al (2006) No alteration in tau exon 10 alternative splicing in tangle-bearing neurons of the Alzheimer’s disease brain. Acta Neuropathol 112:439–449. https://doi.org/10.1007/s00401-006-0095-3
Iqbal K, Liu F, Gong C-X, Grundke-Iqbal I (2010) Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res 7:656–664. https://doi.org/10.2174/156720510793611592
Jiang Z, Tang H, Havlioglu N, Zhang X, Stamm S, Yan R et al (2003) Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2β. J Biol Chem 278:18997–19007. https://doi.org/10.1074/jbc.M301800200
DeJong S, Chepelev I, Janson E, Strengman E, Van Den Berg LH, Veldink JH et al (2012) Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics 13:1. https://doi.org/10.1186/1471-2164-13-458
Kanai Y, Chen J, Hirokawa N (1992) Microtubule bundling by tau proteins in vivo: analysis of functional domains. EMBO J 11:3953–3961. https://doi.org/10.1002/j.1460-2075.1992.tb05489.x
Kar A, Havlioglu N, Tarn WY, Wu JY (2006) RBM4 interacts with an intronic element and stimulates tau exon 10 inclusion. J Biol Chem 281:24479–24488. https://doi.org/10.1074/jbc.M603971200
Katz Y, Wang ET, Airoldi EM, Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015. https://doi.org/10.1038/nmeth.1528
Kim WH, Lee S, Hall GF (2010) Secretion of human tau fragments resembling CSF-tau in Alzheimer’s disease is modulated by the presence of the exon 2 insert. FEBS Lett 584:3085–3088. https://doi.org/10.1016/j.febslet.2010.05.042
King ME, Gamblin TC, Kuret J, Binder LI (2000) Differential assembly of human tau isoforms in the presence of arachidonic acid. J Neurochem 74:1749–1757. https://doi.org/10.1046/j.1471-4159.2000.0741749.x
Lai MC, Bechy AL, Denk F, Collins E, Gavriliouk M, Zaugg JB et al (2017) Haplotype-specific MAPT exon 3 expression regulated by common intronic polymorphisms associated with Parkinsonian disorders. Mol Neurodegener 12:1–16. https://doi.org/10.1186/s13024-017-0224-6
Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt214
Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Liu C, Götz J (2013) Profiling murine tau with 0N, 1N and 2N isoform-specific antibodies in brain and peripheral organs reveals distinct subcellular localization, with the 1N isoform being enriched in the nucleus. PLoS ONE 8:1–18. https://doi.org/10.1371/journal.pone.0084849
Liu C, Song X, Nisbet R, Götz J (2016) Co-immunoprecipitation with Tau isoform-specific antibodies reveals distinct protein interactions and highlights a putative role for 2N Tau in disease. J Biol Chem 291:8173–8188. https://doi.org/10.1074/jbc.M115.641902
McInnes L, Healy J, Melville J (2018) UMAP: Uniform manifold approximation and projection for dimension reduction. J Open Source Softw 3:861. https://doi.org/10.21105/joss.00861
Morfini GA, Burns M, Binder L, Kanaan NM, Bosco DA, Junior RHB et al (2009) Minisymposium: axonal transport defects in neurodegenerative diseases. J Neurosci 29:12776–12786. https://doi.org/10.1523/JNEUROSCI.3463-09.2009.Minisymposium
Neve RL, Harris P, Kosik KS, Kurnit DM, Donlon TA (1986) Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Mol Brain Res 1:271–280. https://doi.org/10.1016/0169-328X(86)90033-1
Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463. https://doi.org/10.1038/nature08909
Otero-Garcia M, Xue T-Q, Shakouri T, Deng Y, Morabito S, Allison T et al (2020) Single-soma transcriptomics of tangle-bearing neurons in Alzheimer’s disease reveals the signatures of tau-associated synaptic dysfunction. BioRxiv. https://doi.org/10.1101/2020.05.11.088591
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415. https://doi.org/10.1038/ng.259
Pedrotti S, Busà R, Compagnucci C, Sette C (2012) The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res 40:1021–1032. https://doi.org/10.1093/nar/gkr819
Perez Y, Menascu S, Cohen I, Kadir R, Basha O, Shorer Z et al (2018) RSRC1 mutation affects intellect and behaviour through aberrant splicing and transcription, downregulating IGFBP3. Brain 141:961–970. https://doi.org/10.1093/brain/awy045
Scala M, Mojarrad M, Riazuddin S, Brigatti KW, Ammous Z, Cohen JS et al (2020) RSRC1 loss-of-function variants cause mild to moderate autosomal recessive intellectual disability. Brain. https://doi.org/10.1093/brain/awaa070
Sharma A, Song W-M, Farrell K, Whitney K, Zhang B, Crary JF et al (2021) Single-cell atlas of progressive supranuclear palsy reveals a distinct hybrid glial cell population. BioRxiv. https://doi.org/10.1101/2021.04.11.439393
Skoglund L, Viitanen M, Kalimo H, Lannfelt L, Jönhagen ME, Ingelsson M et al (2008) The tau S305S mutation causes frontotemporal dementia with parkinsonism. Eur J Neurol 15:156–161. https://doi.org/10.1111/j.1468-1331.2007.02017.x
Spillantini MG, Yoshida H, Rizzini C, Lantos PL, Khan N, Rossor MN et al (2000) A novel tau mutation (N296N) in familial dementia with swollen achromatic neurons and corticobasal inclusion. Ann Neurol 48:939–943. https://doi.org/10.1002/1531-8249(200012)48:6%3c932::AID-ANA15%3e3.0.CO;2-E
Stanford PM, Halliday GM, Brooks WS, Kwok JBJ, Storey CE, Creasey H et al (2000) Progressive supranuclear palsy pathology caused by a novel silent mutation in exon 10 of the tau gene expansion of the disease phenotype caused by tau gene mutations. Brain 2000:880–893
Stefanoska K, Volkerling A, Bertz J, Poljak A, Ke YD, Ittner LM et al (2018) An N-terminal motif unique to primate tau enables differential protein–protein interactions. J Biol Chem 293:3710–3719. https://doi.org/10.1074/jbc.RA118.001784
Storbeck M, Hupperich K, Gaspar JA, Meganathan K, Carrera LM, Wirth R et al (2014) Neuronal-specific deficiency of the splicing factor Tra2b causes apoptosis in neurogenic areas of the developing mouse brain. PLoS ONE. https://doi.org/10.1371/journal.pone.0089020
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM et al (2019) Comprehensive integration of single-cell data. Cell 177:1888-1902.e21. https://doi.org/10.1016/j.cell.2019.05.031
Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C et al (2012) MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet 21:4094–4103. https://doi.org/10.1093/hmg/dds238
Wang F, Zhao Y, Hao Y, Tan Z (2008) Identification of low-abundance alternatively spliced mRNA variants by exon exclusive reverse transcriptase polymerase chain reaction. Anal Biochem 383:307–310. https://doi.org/10.1016/j.ab.2008.09.002
Wang Y, Gao L, Tse S-W, Andreadis A (2010) Heterogeneous nuclear ribonucleoprotein E3 modestly activates splicing of tau exon 10 via its proximal downstream intron, a hotspot for frontotemporal dementia mutations. Gene. https://doi.org/10.1038/jid.2014.371
Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P et al (2020) Tau PTM profiles identify patient heterogeneity and stages of alzheimer’s disease. Cell 183:1699-1713.e13. https://doi.org/10.1016/j.cell.2020.10.029
Wu TD, Watanabe CK (2005) Sequence analysis GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. https://doi.org/10.1093/bioinformatics/bti310
Yu Q, Guo J, Zhou J (2004) A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J Neurochem 90:164–172. https://doi.org/10.1111/j.1471-4159.2004.02477.x
Zhong Q, Congdon EE, Nagaraja HN, Kuret J (2012) Tau isoform composition influences rate and extent of filament formation. J Biol Chem 287:20711–20719. https://doi.org/10.1074/jbc.M112.364067
Acknowledgements
This work was supported by funding from the BrightFocus Foundation (KRB), Association for Frontotemporal Degeneration (KRB), CurePSP (KRB), the Rainwater Charitable Foundation (AMG, KRB), and Alzheimer’s Association AARF-17-529888 (JDC). We thank the NIH Neurobiobanks at the University of Maryland, Harvard and Mount Sinai for supplying tissues for analyses. We also thank the Neuropathology brain bank and research CoRE and the Genetics CoRE at the Icahn School of Medicine at Mt. Sinai for supplying tissues and conducting immunohistochemical staining, and for carrying out targeted iso-seq and data analysis. We are grateful to the study participants and their families for their contributions to research.
Author information
Authors and Affiliations
Contributions
Conceptualization: KRB, AMG. Methodology: KRB, DAP, LMO, BMJ, KF, KW, AS, JDC. Validation: KRB, DAP, LMO. Formal analysis: KRB, KF. Investigation: KRB, DAP, LMO. Resources: JDC, TR, KF, KW, JFC, AP, AMG. Data curation: KRB. Writing—original draft: KRB. Writing—review and editing: KRB, DAP, LMO, KF, KW, JDC, JFC, AP, AMG. Visualization: KRB. Supervision: AMG. Funding acquisition: KRB, JFC, AMG.
Corresponding author
Ethics declarations
Conflict of interests
AMG: Scientific advisory board (SAB) for Denali Therapeutics (2015-2018), SAB for Pfizer (2019), SAB for Genentech, consultant for GSK, AbbVie, Biogen and Eisai. All other authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bowles, K.R., Pugh, D.A., Oja, LM. et al. Dysregulated coordination of MAPT exon 2 and exon 10 splicing underlies different tau pathologies in PSP and AD. Acta Neuropathol 143, 225–243 (2022). https://doi.org/10.1007/s00401-021-02392-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-021-02392-2