Abstract
Impaired neuronal proteostasis is a salient feature of many neurodegenerative diseases, highlighting alterations in the function of the endoplasmic reticulum (ER). We previously reported that targeting the transcription factor XBP1, a key mediator of the ER stress response, delays disease progression and reduces protein aggregation in various models of neurodegeneration. To identify disease modifier genes that may explain the neuroprotective effects of XBP1 deficiency, we performed gene expression profiling of brain cortex and striatum of these animals and uncovered insulin-like growth factor 2 (Igf2) as the major upregulated gene. Here, we studied the impact of IGF2 signaling on protein aggregation in models of Huntington’s disease (HD) as proof of concept. Cell culture studies revealed that IGF2 treatment decreases the load of intracellular aggregates of mutant huntingtin and a polyglutamine peptide. These results were validated using induced pluripotent stem cells (iPSC)-derived medium spiny neurons from HD patients and spinocerebellar ataxia cases. The reduction in the levels of mutant huntingtin was associated with a decrease in the half-life of the intracellular protein. The decrease in the levels of abnormal protein aggregation triggered by IGF2 was independent of the activity of autophagy and the proteasome pathways, the two main routes for mutant huntingtin clearance. Conversely, IGF2 signaling enhanced the secretion of soluble mutant huntingtin species through exosomes and microvesicles involving changes in actin dynamics. Administration of IGF2 into the brain of HD mice using gene therapy led to a significant decrease in the levels of mutant huntingtin in three different animal models. Moreover, analysis of human postmortem brain tissue and blood samples from HD patients showed a reduction in IGF2 level. This study identifies IGF2 as a relevant factor deregulated in HD, operating as a disease modifier that buffers the accumulation of abnormal protein species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein misfolding and aggregation are considered one of the initial events resulting in neurodegeneration in a variety of brain diseases including amyotrophic lateral sclerosis (ALS), fronto-temporal dementia, Parkinson’s, Alzheimer’s and Huntington’s disease (HD), which are collectively classified as protein misfolding disorders (PMDs) [9, 83]. The proteostasis network is fundamental to maintain the stability of the proteome and prevent abnormal protein aggregation [6]. Chaperones resident in the cytosol and the endoplasmic reticulum (ER) ensure precise folding of newly synthesized native proteins. Quality control mechanisms identify misfolded proteins and facilitate their removal via the proteasome, lysosome and autophagy pathways [40]. Importantly, the buffering capacity of the proteostasis network is reduced during aging, which may increase the risk to accumulate protein aggregates and undergo neurodegeneration [40, 49]. One of the main nodes of the proteostasis network altered in neurodegenerative diseases involves the ER, the main site for protein folding and quality control in the cell [36, 76, 81]. ER stress instigates a rapid and coordinated signaling pathway known as the unfolded protein response (UPR) to enforce adaptive programs and restore proteostasis [103]. Conversantly, irremediable ER stress results in cell death by apoptosis [35].
The UPR is initiated by the activation of specialized stress transducers highlighting inositol-requiring transmembrane kinase/endonuclease (IRE1α) as the most conserved signaling branch [102]. IRE1α is an ER-located kinase and endoribonuclease that, upon activation, controls the processing of the mRNA encoding X-box-binding protein 1 (XBP1), leading to the expression of a stable and active transcription factor known as XBP1s [33]. XBP1s upregulates genes related to protein folding, quality control, ER translocation and ERAD, among other processes [1, 46, 78]. In C. elegans organisms, the expression of XBP1 in neurons controls organism aging [87], which may involve a crosstalk with classical signaling pathways regulating normal aging including the insulin-like growth factor (IGF)-1 and FOXO (DAF16) pathways [31]. We have previously studied the functional contribution of XBP1 to various neurodegenerative conditions. Despite initial expectations that targeting this central UPR mediator will exacerbate disease progression, we observed that genetic disruption of the IRE1α/XBP1 pathway during brain development led to neuroprotection in models of ALS [37], Parkinson’s disease [93], Alzheimer’s disease [19, 72] and HD [97]. These results were explained by hormetic adaptive changes that reduced the accumulation of abnormal protein aggregates. These findings illustrate the broader significance of XBP1 and the UPR to the progression of neurodegenerative diseases, highlighting the need to identify the mechanisms of action mediating neuroprotection.
To uncover novel regulatory elements that may underlie the beneficial effects of ablating XBP1 expression in the brain, here we performed a gene expression profile study in cortex and striatum of a XBP1 conditional knockout mice (XBP1cKO) and identified Igf2 as the major upregulated gene. We then investigated the significance of IGF2 to abnormal protein aggregation using HD models as proof of concept. HD is an autosomal disease, which belongs to a group of tandem repeat diseases caused by a CAG codon expansion known as polyglutamine disorders. HD is caused by a mutation in the huntingtin (Htt) gene, involving the generation of an abnormal tract of over 40 glutamine residues (polyQ) at the N-terminal region [7]. HD patients develop a combination of motor, cognitive and psychiatric symptoms, associated with progressive neurodegeneration of the striatum and cerebral cortex [17]. Mutant Htt (mHtt) often forms insoluble inclusions that alter cellular homeostasis including the function of the ER [96, 112]. Here, we provide evidence indicating that IGF2 signaling decreases the accumulation of intracellular mHtt and polyglutamine peptide aggregates in various cell culture models of HD. Unexpectedly, the decrease in mutant huntingtin and polyglutamine peptide aggregation was independent of the activity of the proteasome or autophagy pathways. Instead, IGF2 treatment enhanced the non-conventional disposal of soluble polyglutamine peptide species into the extracellular space through the secretion via microvesicles and exosomes. To validate our results in vivo, we developed a therapeutic approach to deliver IGF2 into the brain using recombinant viruses, which resulted in significant reduction in mHtt levels in three different preclinical models of HD. Furthermore, we observed a dramatic reduction in IGF2 levels in brain and blood samples derived from HD. We propose that IGF2 signaling operates as a disease modifier, where its deregulation enhances the abnormal accumulation of mHTT in HD. Overall, this study uncovered a previously unanticipated role of IGF2 in handling protein aggregates in HD.
Materials and methods
Animal care
XBP1flox/flox mice were crossed with mice expressing Cre recombinase under the control of the Nestin promoter to achieve the conditional deletion of XBP1 in the nervous system (XBP1cKO) [34]. We employed as HD models the R6/2 transgenic mice, expressing exon 1 of the human huntingtin gene carrying approximately 150 CAG repeat expansions [48], and the full-length human Htt transgenic mice with 128 CAG repetitions termed YAC128 [80], both obtained from The Jackson Laboratory. To generate experimental animals, XBP1cKO mice were crossed with the YAC128 model on FVB background and every generation breed to pure background XBP1cKO mice for four to six generations to obtain experimental animals. For proper comparison, all biochemical and behavioral analyses were performed on groups of littermates of the same breeding generation. Unless indicated, YAC128 and littermate control animals were used and maintained on a pure C57BL/6J background. Mice were maintained in a quiet, ventilated and temperature-controlled room (23 °C), with a standard 12 h light cycle, and monitored daily. Mice were housed in polystyrene solid bottom plastic cages fitted with a filtertop. Mice were fed with LabDiet pellets and drinking water ad libitum. For euthanasia, mice received CO2 narcosis. Animal care and experimental protocols were performed according to procedures approved by the “Guide for the Care and Use of Laboratory Animals” (Commission on Life Sciences. National Research Council. National Academy Press 1996), the Institutional Review Board’s Animal Care and Use Committee of the Harvard School of Public Health, the Bioethical Committee of the Faculty of Medicine, University of Chile (CBA 0670 FMUCH) and the Bioethical Committee of the Center for Integrative Biology, University Mayor (07-2017). We used XBP1cKO (n = 17) and litter matte control (n = 12) animals to gene expression analysis. For gene therapy analysis, we used 4–6 YAC128 mice per experimental group and 8 R6/2 mice per experimental group.
Microarray
XBP1cko animals were euthanized at 12–14 months of age and perfused with heparinized saline buffer. Brains were removed and placed in RNAlater. Brain cortex and striatum were dissected, and mouse whole-genome profiling was performed using the Illumina BeadChip® platform (Illumina; San Diego, CA). According to the manufacturer, probes on the Illumina MouseWG-6 v2.0 Expression BeadChip were derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database (Build 36, release 22), supplemented with probes derived from the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) set, as well as exemplar protein-coding sequences described in the RIKEN FANTOM2 database. The MouseWG-6 v2.0 BeadChip contains the full set of the MouseRef-8 Expression BeadChip probes with an additional 11,603 probes from the above databases. Raw data were quantile normalized and differentially expressed genes identified using the ArrayAnalysis software [20]. Genes that were significantly up- or downregulated were identified using significance analysis of microarrays (SAM) [92]. SAM assigns a score to each gene based on a change in gene expression relative to the standard deviation of repeated measurements. For genes with scores greater than an adjustable threshold, SAM uses permutations of the repeated measurements to estimate the percentage of genes identified by chance the false discovery rate (FDR). Analysis parameters were set to 800 permutations, and genes with q value = 0% were used for genomics analysis.
To identify functional connections between deregulated transcripts, both network and pathway analyses of the genes filtered by microarray were performed using Ingenuity Pathways Analysis (IPA; QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). The significance of networks was calculated by integrated Ingenuity algorithms. IPA computes a score for each gene network according to the fit of that network to the user-defined set of “focus genes.” The score is derived from a p value and indicates the likelihood of the focus genes in a network being found together due to random chance. Relevant pathways with p values less than 0.05 were considered. In addition, IPA compares the direction of change for the differentially expressed genes with expectations based on the literature (cured in the Ingenuity Knowledge Base) to predict an integrated direction of change for each function, using the z-score algorithm. For heatmap representation of deregulated transcripts, the software HeatmapGenerator was used [43].
RNA isolation, RT-PCR and real-time PCR
Total RNA was prepared from tissues or cells placed in cold PBS using Trizol following the manufacturer’s instructions (Life Technologies). The cDNA was synthesized with SuperScript III reverse transcriptase (Life Technologies) using random primers p(dN)6 (Roche). Quantitative PCR reactions were performed using standard protocols using EvaGreen™ in the Stratagene Mx3000P system (Agilent Technologies). The relative amounts of mRNAs were calculated from the values of comparative threshold cycle by using Actin as a control. PCR or RT-PCR were performed using the following primers: For mouse: Igf2 5′- GTCGCATGCTTGCCAAAGAG-3′ and 5′-GGTGGTAACACGATCAGGGG-3′, mouse Actin 5′- TACCACCATGTACCCAGGCA-3′ and 5′- CTCAGGAGGAG AATGATCTTGAT-3′, Igf1 5′- AAAGCAGCCCCGCTCTATCC-3′ and 5′- CTTCTGAGTCTTGGGCATGT A-3′, Acot1 5′- TGCAAAGCCCTCTGGTAGAC-3′ and 5′- CTCGCTCTTCCAGTTGTGGT-3′, insulin receptor 5′- TCAAGACCAGA CCGAAGATT-3′ and 5′- TCTCGAAGATAACCAGGGCATAG-3′, Igf1r 5′- CATGTGCTGGCAGTATAACCC-3′ and 5′- TCG GGA GGC TTG TTC TCC T-3′. For human IGF2 5′- GTGCTGTTTCCGCAGCTG-3′ and 5′- AGGGGTCGACACGTCCCTC-3′, Actin 5′- GCGAGAAGATGACCCAGATC-3′ and 5′- CCAGTGGTACGGCCAGAGG-3′, HA 5′- TAGACGTAATCTGGAACATCG-3′.
Reagents, plasmids and transfection
Lactacystin, MG-132, chloroquine, 3-methyladenine, bafilomycin A1, brefeldin A and recombinant insulin were purchased from Sigma. NSC23766 was purchased from Santa Cruz. GW4869 was purchased from Cayman Chemicals. Phalloidin was purchased from Molecular Probes, Invitrogen. Blocking antibody for IGF2R was purchased from Santa Cruz (sc-25462). Cell media and antibiotics were obtained from Invitrogen (MD, USA). Fetal calf serum was obtained from Hyclone and Sigma. All transfections for plasmids were performed using the Effectene reagent (Qiagen). DNA was purified with Qiagen kits. PolyQ79-EGFP is a 79 polyglutamine tract in-frame fused to EGFP in the N-terminal; GFP-mHttQ85 contains the first 171 amino acids of the first exon of the huntingtin gene with a tract of 85 glutamines fused to GFP in the N-terminal, was kindly provided by Dr. Paulson Henry (University of Michigan). Myc-tagged full-length huntingtin with a 103 polyglutamine region (FL Htt103Q-myc) in pcDNA3 was kindly provided by the Coriell Institute. pAAV-mHttQ85-mRFP contains the first 588 amino acids of the huntingtin gene with a tract of 85 glutamines, fused to mRFP [113]. IGF2 cDNA was obtained from pSPORT6 kindly provided by Dr. Oliver Bracko and subcloned with or without the HA epitope into a pAAV vector. SOD1G85R-EGFP was described before [91]. siRNA pool for IGFR1 and InsR were purchased from Origene, and transfections were made with Lipofectamine® RNAiMAX transfection Reagent from Invitrogene.
Microscopy, western blot and filter trap analysis
Neuro2a and HEK293T cells were obtained from ATCC and maintained in Dulbecco’s modified Eagles medium supplemented with 5% fetal bovine serum and Penicillin/Streptomycin (Gibco). 3 × 105 cells were seeded in 6-well plate and maintained by indicated times in DMEM cell culture media supplemented with 5% bovine fetal serum and nonessential amino acids. Treatment with autophagy, proteasome or exocytosis inhibitors was generally performed for 16 h unless indicated.
We visualized the formation of intracellular polyQ79-EGFP, GFP-mHttQ85 and Htt588Q95-RFP inclusions in live cells after transient transfection using epifluorescent microscopy. We quantified the intracellular inclusion using automatized macros done in Image J software. Protein aggregation was evaluated by western blot in total cell extracts prepared in 1% Triton X-100 in PBS containing proteases and phosphatases inhibitors (Roche). Sample quantification was performed with the Pierce BCA Protein Assay Kit (Thermo Scientific).
For western blot analysis, cells were collected and homogenized in RIPA buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Triton X-100) containing protease and phosphatase inhibitors (Roche). After sonication, protein concentration was determined in all experiments by micro-BCA assay (Pierce), and 25–100 µg of total protein was loaded onto 8 to 15% SDS-PAGE minigels (Bio-Rad Laboratories, Hercules, CA) prior transfer onto PVDF membranes. Membranes were blocked using PBS, 0.1% Tween-20 (PBST) containing 5% milk for 60 min at room temperature and then probed overnight with primary antibodies in PBS, 0.02% Tween-20 (PBST) containing 5% skimmed milk. The following primary antibodies and dilutions were used: anti-GFP 1:1000 (Santa Cruz, Cat. no. SC-9996), anti-SOD1 (Cell signaling, Cat. no. 2770), anti-polyQ 1:1000 (Sigma, Cat. no. P1874), anti-HSP90 1:2000 (Santa Cruz, Cat. no. SC-13119), anti-GAPDH 1:2000 (Santa Cruz, Cat. no. SC-365062), anti-HA 1:500 (Santa Cruz, Cat. nºSC-805), anti-IGF2 1:1000 (Abcam, Cat. no. Ab9574), anti-LC3 1:1000 (Cell Signaling, Cat. no. 2775S), anti-p62 1:1000 (Abcam, Cat. no. Ab56416), anti-polyUbiquitin 1:5000 (Santa Cruz, Cat. no. SC-8017), anti-DARPP32 1:1000 (Millipore, Cat. no. ab10518), anti-tubulin 1:2000 (Oncogene, Cat. no. CP06) and Rac1 clone 23A8 1:1000 (Millipore, Cat. no. 05–389), and for cofilin and p-cofilin we used homemade antibody at a 1:1000 dilution (Dr. James Bamburg). Bound antibodies were detected with peroxidase-coupled secondary antibodies incubated for 2 h at room temperature and the ECL system.
For western blot analysis using FL Htt103Q-myc, cells were grown in 100-mm dishes and cell lysis was done in 100 µl of 1% SDS in PBS for 5 min at 95 ˚C. Samples were then diluted 1:5 with 2% Triton X100, pelleted in a microcentrifuge for 30 min at 4 ˚C and 50 µl of the supernatant was loaded on SDS–PAGE. After transfer to a nitrocellulose membrane, blocking was done in 5% low-fat milk and 0.1% Tween-20 in PBS for 1 h. Incubation with the primary antibody was performed overnight at 4 ˚C. The following primary antibodies and dilutions were used: anti-calnexin 1:1000 (Sigma-Aldrich, Cat. no. C4731), anti-Htt 1:1000 (Cell Signaling, cat nº2773S) and anti-myc 1:1000 (custom produced from 9E10 hybridoma). After three washes in 0.1% Tween-20 in PBS, incubation with the appropriate 1:10,000 secondary antibody was performed for 1 h at room temperature. After washing, ECL was performed and the membrane was exposed and quantified in a Bio-Rad ChemiDocXRS Imaging System (Bio-Rad, Hercules, CA).
For western blot analysis using HD MSNs protein extracts, MSNs were harvested in M-PER Mammalian Protein Extraction Reagent (Pierce) with cOmplete Mini EDTA-free protease inhibitor (1 tablet/10 mL) (Roche). Whole-cell lysates were sonicated with a 5 s pulse followed by a 5 s rest (X 5 times) at 40% amplitude. Samples were centrifuged at 14,000 rpm at 4 °C for 20 min, and supernatant was collected and stored at − 20 °C. Protein concentrations were estimated using the BCA assay (Pierce). Cell lysates were denatured under reducing conditions by boiling 10–20 μg total protein with 1 μL of 1 M DTT and 4X LDS sample buffer (Invitrogen) at 95 °C for 10 min. SDS-PAGE was performed using NuPage 4–12% Bis–Tris gels (Invitrogen) or 3–8% tris–acetate gels. Gels were run in 1X MOPS or 1X tris–acetate SDS running buffer containing 500 μL of antioxidant (Life technologies) for 90 min and transferred to 0.45-μm nitrocellulose membrane using 1X NuPage transfer buffer at 20 V for 14 h. Membranes were blocked with 5% non-fat milk in TBS with 0.1% Tween 20 (TBS-T) for 1 h at room temperature (RT) and incubated with primary antibody reconstituted in 5% non-fat milk overnight at 4 °C. The following primary antibodies and dilutions were used: anti-Htt 1:500 (Millipore, MAB2166) and anti-vinculin 1:500 (Sigma-Aldrich, Cat. no. V9131). Secondary antibodies were incubated for 2 h at RT. Blots were washed in TBS-T for 10 min (3×) and developed using Pierce ECL (Thermo Scientific). Vinculin served as loading control. Densitometry analysis was performed using ImageQuant TL v2005.
For western blots using SCA3 iPSCs-derived neurons, neuronal cells were washed in HBSS and scraped, and cells were immediately frozen in liquid N2 followed by lysis in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.2% Triton X-100) containing 25 mM EDTA. For fractionation, lysates containing 1–2 µg/µl total protein dissolved in 50 mM Tris, 150 mM NaCl, 0.2% Triton X-100, 25 mM EDTA (RIPA buffer) were centrifuged at 22,000g for 30 min at 4 °C. The pellet fractions were separated from supernatants (Triton X-100-soluble fraction) and homogenized by sonication in RIPA buffer containing 2% SDS (SDS fraction). β-mercaptoethanol (5%) was added in all the samples and subsequently incubated at 99 °C for 5 min. Gels were loaded with 10–20 µg of the Triton X-100 fraction and 40 µl of the SDS fraction. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane and then processed for western blotting. Membranes were subsequently incubated with HRP-conjugated secondary antibodies (Amersham) at 1:7000 dilution. Visualization was performed with enhanced chemiluminescence. Samples were probed with primary antibodies: Ataxin-3 (Aviva Systems Biology; Cat#ARP50507_P050), βIII-tubulin (Abcam; Cat#ab7751) and GAPDH (Fitzgerald; Cat#10R-G109A).
For filter trap assays, protein extracts were diluted into a final concentration of SDS 1% and were subjected to vacuum filtration through a 96-well dot blot apparatus (Bio-Rad Laboratories, Hercules, USA) containing a cellulose acetate membrane with pores with a 0.2 μm diameter (Whatman, GE Healthcare) as described in [89]. Membranes were then blocked using PBS, 0.1% Tween-20 (PBST) containing 5% milk and incubated with primary antibody at 4 °C overnight. Image quantification was done with the Image Lab software from BioRad.
Pulse-chase experiments
HEK293T cells were seeded in 6-well plates and transiently transfected using polyethylenimine (PEI) with pcDNA-GFP-HttQ43 and mRFP/IGF2 constructs (in a ratio of 1:9). We used these constructs instead of the polyQ79-EGFP peptide because this experiment was standardized using GFP-HttQ43. For pulse labeling of cells (35S incorporation), after 24 h of transfection cells were washed twice with pre-warmed pulse labeling medium (methionine- and cysteine-free DMEM (Gibco) containing 10% dialyzed FBS (Thermo Scientific), Glutamax (Life technologies) and penicillin/ streptomycin (Gibco) and kept for 45 min in pre-warmed pulse labeling medium to deplete the intracellular pool of cysteine and methionine. 0.1 mCi/mL 35S (EasyTag™ EXPRESS35S Protein Labeling Mix) were added in pre-warmed pulse labeling medium and pulsed for different time periods. Before harvesting the cells, the radioactive solution was removed, and cells were washed twice with 1 mL PBS containing 15 μM cycloheximide. Cells were scraped in 250 μL immunoprecipitation lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 0.5% NP-40, 0.5 mM MgCl2, 3% glycerol, 0.9 mM DTT) and protease inhibitor cocktail (complete Sigma-Aldrich) and lysed by repetitive passage (8 times) through a syringe with 26-gauge needle (0.45 mm). Samples were centrifuged (20.000 × g), and the supernatant was collected. 50 μL cell lysate was kept for input. 800μL TNE buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA) was added to rest of the sample, and the GFP-HttQ43 was immunoprecipitated using GFP-trap (Chormotek, gtma-100). GFP-trap beads were added to each sample and incubated overnight at 4ºC. Beads were collected on a magnetic rack and washed twice in immunoprecipitation wash buffer (0.2% NP40, 0.2% SDS, protease inhibitors), and twice in high salt buffer (TNE with 500 mM NaCl). Beads were boiled in sample buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue and 0.125 M tris HCl) and analyzed using SDS page and western blotting or autoradiography. To determine half-life, 0.1 mCi 35S was added to each well and incubated for 90 min. After washing, chase medium [DMEM containing 10% FCS, Glutamax, penicillin/streptomycin, 1 mg/mL methionine and 1 mg/mL cysteine (Gibco)] was added to cells and incubated for different time points. Sample preparations and immunoprecipitation of GFP-HttQ43 was performed as described for the pulse labeling experiments.
mHtt secretion and dot blot detection
PolyQ79-EGFP secretion was analyzed as described before [90]. In brief, to collect secreted proteins, Neuro2a cells were transiently transfected for a total period of 32 h, incubated in Optimem for the last 24 h. Cell culture media was collected and centrifuged for 5 min at 5000 rpm to eliminate cell debris and then applied to a 96-well dot blot apparatus (Bio-Rad Laboratories, Hercules, USA) on a PVDF membrane. Membranes were then blocked, and proteins were detected following the western blot protocol.
For the detection of FL Htt103Q-myc using dot blot, HEK293 cells were grown on 60-mm plates in 3-mL medium and transfected with plasmids for the expression of FL Htt103Q-myc together with IGF2 or empty vector. Cells and medium were collected 24 h post transfection. After washing with PBS, cells were lysed in 200 µl of 1% SDS in PBS with protease inhibitor cocktail. After boiling for 5 min, sample duplicates (50 µl for cell lysates and 100 µl for medium) were loaded on a nitrocellulose membrane (pre-wet with PBS) in a dot blotting apparatus. After two washes with PBS, the membrane was transferred into 5% milk blocking solution. Proteins were detected following the western blot protocol.
Vesicle isolation and nanoparticle tracking analysis
Extracellular vesicles were isolated from cell culture media. Briefly, Neuro2a cells were cultured for 24 h in Optimem. Collected medium was then depleted of cells and cellular debris at 2000×g for 10 min at 4 °C. Extracellular vesicles were isolated by centrifugation of the collected supernatant at 110,000×g for 70 min at 4 °C. The resultant pellet was resuspended in PBS 1× with proteases and phosphatases inhibitors cocktail. Vesicles were analyzed for size and concentration using nanoparticle tracking analysis (NanoSight, NS300/ZetaSizer instrument). Microvesicles and exosomes were isolated from cell culture media using Neuro2a. First, cell culture media was depleted of cells and cellular debris at 2000×g for 10 min at 4 °C. Then, microvesicles were pelleted by centrifugation of the collected supernatant at 11,000×g for 34 min at 4 °C. Finally, exosomes were isolated from the collected supernatant from microvesicles purification by centrifugation at 110,000×g for 70 min at 4 °C. The resultant pellet was then washed in PBS and centrifuged again for 70 min at 110,000×g at 4 °C. The resultant pellet was resuspended in PBS 1× with proteases and phosphatases inhibitors cocktail.
Quantitative proteomics
Neuro2a cells in 6-well plates were transiently transfected with IGF2 or empty expression vectors for 24 h. Lysates were prepared in RIPA buffer containing proteases and phosphatases inhibitors cocktail (Roche). After extracts sonication, protein concentration was determined with BCA (Thermo Fisher). For each sample, 20 μg of lysate was washed by chloroform/methanol precipitation. Samples for mass spectrometry analysis were prepared as described [63]. Air-dried pellets were resuspended in 1% RapiGest SF (Waters) and brought up in 100 mM HEPES (pH 8.0). Proteins were reduced with 5 mM Tris(2-carboxyethyl) phosphine hydrochloride (Thermo Fisher) for 30 min and alkylated with 10 mM iodoacetamide (Sigma Aldrich) for 30 min at ambient temperature and protected from light. Proteins were digested for 18 h at 37 °C with 0.5 μg trypsin (Promega). After digestion, the peptides from each sample were reacted for 1 h with the appropriate TMT-NHS isobaric reagent (Thermo Fisher) in 40% (v/v) anhydrous acetonitrile and quenched with 0.4% ammonium bicarbonate for 1 h. Samples with different TMT labels were pooled and acidified with 5% formic acid. Acetonitrile was evaporated on a SpeedVac, and debris was removed by centrifugation for 30 min at 18,000×g. MuDPIT microcolumns were prepared as described [71]. LCMS/MS analysis was performed using a Q Exactive mass spectrometer equipped with an EASY nLC 1000 (Thermo Fisher). MuDPIT experiments were performed by 5 min sequential injections of 0, 10, 20, 30, …, 100% buffer C (500 mM ammonium acetate in buffer A) and a final step of 90% buffer C/10% buffer B (20% water, 80% acetonitrile, 0.1% formic acid, v/v/v) and each step followed by a gradient from buffer A (95% water, 5% acetonitrile, 0.1% formic acid) to buffer B. Electrospray was performed directly from the analytical column by applying a voltage of 2.5 kV with an inlet capillary temperature of 275 °C. Data-dependent acquisition of MS/MS spectra was performed with the following settings: Eluted peptides were scanned from 400 to 1800 m/z with a resolution of 30,000 and the mass spectrometer in a data dependent acquisition mode. The top ten peaks for each full scan were fragmented by HCD using a normalized collision energy of 30%, a 100 ms activation time and a resolution of 7500 and scanned from 100 to 1800 m/z. Dynamic exclusion parameters were 1 repeat count, 30 ms repeat duration, 500 exclusion list size, 120 s exclusion duration and exclusion width between 0.51 and 1.51. Peptide identification and protein quantification was performed using the Integrated Proteomics Pipeline Suite (IP2, Integrated Proteomics Applications, Inc., San Diego, CA) as described previously [63].
Rac1 pull-down assay and actin dynamics
Neuro2a cells were cultured for 2 days in 100-mm-diameter plastic plates (107 cells/plate) and then stimulated with IGF2 conditioned medium for 5 min at 37 °C and 5% CO2. For Rac1 activation assays, the expression and purification of the CRIB domain (amino acids 67–150) of p21-activated kinase (Pak1) were performed as follows: BL21 (DE3) E. coli strains carrying pGEX-GST-CRIB were grown overnight at 37 °C in Luria broth media containing ampicillin. Cultures were diluted 1:100 and grown in fresh medium at 37 °C to an OD600 of 0.7. Next, IPTG was added to a final concentration of 1 mM, the cultures were grown for an additional 2 h, and then, samples were collected and sonicated in lysis buffer A [50 mM Tris–HCl, pH 8.0; 1% Triton X-100; 1 mM EDTA; 150 mM NaCl; 25 mM NaF; 0.5 mM PMSF and protease inhibitor complex (Roche)]. Cleared lysates were affinity purified with glutathione-Sepharose beads (Amersham). Loaded beads were washed ten times with lysis buffer B (lysis buffer A with 300 mM NaCl) at 4 °C. GST fusion protein was quantified and visualized in SDS–polyacrylamide gels stained with Coomassie brilliant blue. Purified loaded beads were incubated for 70 min at 4 °C with 1 mg of either Neuro2a lysates using fishing buffer (50 mM Tris–HCl, pH 7.5; 10% glycerol; 1% Triton X-100; 200 mM NaCl; 10 mM MgCl2; 25 mM NaF and protease inhibitor complex). The beads were washed three times with washing buffer (50 mM Tris–HCl, pH 7.5; 30 mM MgCl2; 40 mM NaCl) and then resuspended in SDS-PAGE sample buffer. Bound Rac1-GTP was subjected to western blot analysis.
To measure actin dynamics, Neuro2a cells were seeded onto fibronectin-coated 25-mm coverslips, transfected with EGFP-Lifeact using Lipofectamine® 2000 Transfection Reagent and imaged in HBSS medium supplemented with HEPES using a confocal microscope (Zeiss LSM 710) with a 63 × /1.4 NA oil-immersion objective at 37 °C. Images were acquired every 30 s for 5 min, and the number of cells showing actin clusters were quantified. For phalloidin staining, we used Phalloidin tetramethylrodamyne (Sigma) following the manufacturer’s instructions.
Adeno-associated viral vector injections
All adeno-associated vectors (AAV, serotype 2) were produced by triple transfection of HEK293T cells using a rep/cap plasmid and pHelper (Stratagene, La Jolla CA, USA) and purified by column affinity chromatography as previously described [57]. For stereotactic injections, mice were anesthetized using a 100 mg/kg Ketamine, 10 mg/kg Xylazine mixture (Vetcom), and affixed to a mouse stereotaxic frame (David Kopf Instruments). Wild-type C57BL/6 J male mice (3-month old) were co-injected in the right striatum with 4 µl of virus AAV-Htt588Q95-mRFP and AAV-IGF2 or AAV-Mock in a relation 3:2 as we described before [113]. The injection of AAVs suspension was performed at two points of the striatal region using a 5-µl Hamilton syringe (Hamilton) using the following coordinates: + 0.7 mm anterior, + 1.7 mm lateral and − 3 to 3.2 mm depth, with a 1 µl/min infusion rate. After 2 weeks, mice were euthanized for biochemistry analysis. YAC128 mice (3-month old) were injected bilaterally in a single point in the striatum with 2 µl of AAV-IGF2 or AAV-Mock. Stereotaxic coordinates with respect to the bregma used were: + 0.7 mm anterior, + 1.7 mm lateral and -3.1 mm depth, with a 1 µl/min infusion rate. After 1 month, mice were euthanized for biochemistry analysis. Four-week-old R6/2 mice were injected bilaterally in a single point in the striatum with 1.5 μl of AAV-IGF2 or AAV-Mock. Stereotaxis coordinates with respect to the bregma used were: + 0.8 mm anterior, + 1.8 mm lateral and 3.1 mm depth, with a 1 μl/min infusion rate. Four weeks after the injection, mice were euthanized for histochemical analysis.
Rotarod test
Disease progression was monitored once every two weeks, using the rotarod assay as previously reported [27]. In brief, a training period was performed consisting of a first day of conditioning in the rotarod, following by three trials per day (with 2 h in between trials) over the course of four consecutive days. The data collected were considered an accurate reflection of the animal's coordination. For each testing, mice were acclimated to the room for ∼15 min. The rotarod protocol consisted in (1) maximum speed, 40 rpm; (2) time to reach maximum rpm, 120 s; (3) time to “no fall”, 600 s; (4) starting speed, 4 r.p.m; and (5) trials per day, 3.
Tissue preparation and analysis
For biochemistry of the striatum, mice were euthanized by CO2 narcosis, and brains were rapidly removed, rinsed with ice-cold 0.1 M phosphate buffer saline pH 7.4 (PBS) and placed on a cold surface to dissect different brain areas. Tissue samples were homogenized in 100 µl of ice-cold PBS (pH 7.4), supplemented with proteases and phosphatases inhibitor cocktail (Roche). The homogenate was divided for further mRNA and protein extraction following standard purification and quantification protocols. Protein extraction was performed in RIPA buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate and 0.5% Triton X-100) containing proteases and phosphatases inhibitor cocktail (Roche).
For tissue immunofluorescence analysis, mice were anesthetized and subjected to transcardial perfusion with ice-cold PBS (pH 7.4). Brains were removed, and one hemisphere was fixed in 4% PFA for 24 h and then stored in a 30% sucrose solution. The other hemisphere was kept at − 80 °C. Fixed hemispheres were cut using a cryostat (Leica CM1520) and 20-μm-thick coronal slices were obtained from the striatum. Free-floating slices were preserved in PBS-Sodium azide 0.02% at 4 °C.
Tissue immunofluorescence analysis
Brain slices were washed three times with PBS (pH 7.4) and then blocked at room temperature for 1 h with blocking solution (BSA 0.5% and Triton 0.2% in PBS pH 7.4). Brain slices were incubated with EM48 1:1000 (Millipore, MAB5374) and HA 1:500 (Cell Signaling, cat no. C29F4) antibodies in blocking solution for 24 h at 4 °C. Afterward, slices were washed three times with PBS (pH 7.4) and then incubated for 2 h at room temperature with Alexa 488 goat anti-mouse 1:1000 (Jackson ImmunoResearch, cat no. 115-545-166), Alexa 594 goat anti-rabbit 1:1000 (Jackson ImmunoResearch, cat no. 111-545-144) and DAPI 1:10,000 (Merck, cat no. D9542). Finally, slices were washed three times with PBS (pH 7.4) and mounted in slides cover with gelatin using Fluoromount-G (Electron Microscopy Sciences, cat no. 17984-25).
Microscopy and quantification of brain slices
High-resolution images (330.49 × 330.49 μm) from brain slices were obtained using an inverted Leica DMi8 light microscope using a 40× objective. For the quantification of mutant huntingtin inclusions and DAPI-positive cells, 20–30 z-stacks were acquired in three different regions of the striatum of a single slice for each experiment (six-nine slices per animal analyzed). Images were deconvolved using the microscope software and mutant huntingtin inclusions and DAPI-positive cells were quantified using automatized macros from the Image J software.
Human samples
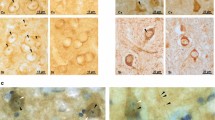
A total of 26 brain samples from the NIH NeuroBioBank were included in the study. Samples were obtained from 13 patients with clinical diagnosis of HD (grades 3 and 4 according to the neuronal loss observed) and 13 age-matched controls. Patients with HD met criteria proposed by the neuropathologist are responsible in the Harvard Brain Tissue Resource Center [18, 98, 99]. Samples of human adult peripheral blood (18 mL) were collected using BD vacutainer heparin tubes and processed in ≤ 4 h after extraction. Plasma and PBMC were obtained by Ficoll gradient. Sampling was done according to the ethics committee form by the advisory board of the director of the local health service and the San Jose Hospital, University of Chile. Patients were recruited in the Enroll-HD project at the Movement disorder center (CETRAM) in Santiago, Chile (https://www.enroll-hd.org). Enroll-HD is a worldwide observational study for Huntington’s disease families. It will monitor how the disease appears and changes over time in different people and is open to people who either have HD or are at-risk. This platform collects standardized data, in the same way and using the same methods around the world. This project develops a comprehensive database of patients followed by a team of neurologists. Totally, circulating IGF2 levels were measured in plasma from 49 individuals, using an ELISA IGF2 kit (CrystalChem, Cat. no. 80575).
NSC culture and MSN differentiation
Six-well plates were coated with 100 μg/mL poly-d-lysine (Sigma-Aldrich) followed by Matrigel (1:60) coating. HD and corrected C116 iPSC-derived neural stem cells (NSCs) were plated and cultured in Neural Proliferation Medium (NPM) (Neurobasal medium, 1× B-27 supplement (Life Technologies), 2 mM l-Glutamine, 100 U/ml penicillin, 100 μg/mL streptomycin, 10 ng/mL Leukemia inhibitory factor (LIF) (Peprotech, 300-05), 25 ng/mL basic Fibroblast Growth Factor (bFGF) (Peprotech, 100-18B) and 25 ng/mL Activin A (Peprotech, AF-120-14E) in humidified incubator under 37 °C, 5% CO2. For differentiation into medium spiny neurons (MSNs), NSCs (when at ~ 90% confluence) were treated with Synaptojuice A medium for 7 days followed by Synaptojuice B medium for 10 days at 37 °C, 5% CO2 [41]. Activin A of 25 ng/mL was added to both Synaptojuice A and Synaptojuice B media. Half media change was performed every 2 days. NSCs were treated with AAV2-IGF2 or AAV2-PGK-empty (1 × 1011 VG per well) on day 1 of Synaptojuice A treatment. Untransduced MSNs were used as controls.
Generation of iPSCs of SCA-3 patients with episomal vectors
Generation of iPSCs derived from patients with spinocerebellar ataxia class 3 was described previously [88]. Human dermal fibroblasts (HDFs) were cultured in Dulbecco’s modified Eagle media (DMEM, Gibco) containing 10% fetal bovine serum (FBS), 1 mM nonessential amino acids (NEAAs), 1× GlutaMAX and 100 units/mL of penicillin with 100 µg/mL streptomycin. The episomal iPSC reprogramming plasmids, pCXLE-hOCT3/4, pCXLE-hSK and pCXLE-hMLN were purchased from Addgene. The plasmids used in our experiments were mixed in a ratio of 1:1:1 for efficient reprogramming. 3 μg of expression plasmid mixtures was electroporated into 5 × 105 HDFs with Amaxa® Nucleofector Kit according to the manufacturer’s instructions. After nucleofection, cells were plated in DMEM containing 10% FCS and 1% penicillin/streptomycin until it reaches 70–80% confluence. The culture medium was replaced the next day by human embryonic stem cell medium (HESM) containing knock-out (KO) DMEM, 20% KO serum replacement (SR), 1 mM NEAAs, 1× GlutaMAX, 0.1 mM β-mercaptoethanol, 1% penicillin/streptomycin and 10 ng/mL bFGF (Invitrogen). Between 26 and 32 days after plating, colonies developed and colonies with a phenotype similar to human ESCs were selected for further cultivation and evaluation. Selected iPSC colonies were mechanically passaged on matrigel (BD, hES qualified matrigel)-coated plates containing mTeSR™1 (defined, feeder-free maintenance medium for human ESCs and iPSCs).
Generation of iPSC-derived neurons
IPSCs were dissociated manually and plated on a non-coated dish in human embryonic stem cell medium (HESM). After 4 days, embryoid bodies (EBs) were formed and transferred to neural differentiation medium containing DMEM/F12, 1 mM NEAAs, 1 × GlutaMAX, 1% penicillin/streptomycin and 1X N1 supplement (100×) for another 4 days. EBs were plated on matrigel-coated plates for neural rosette formation for 8–10 days with 0.01 mM retinoic acid. Neural rosettes were handpicked and cultured in neural stem cell medium containing DMEM/F12, 1 mM NEAAs, 1× GlutaMAX, 1% penicillin/streptomycin, 1× N1 supplement (100×), 20 ng/mL FGF2 (peprotech), 20 ng/mL EGF (peprotech) and 2 µl/mL B27 supplement (Invitrogen). Terminal neural differentiation was induced by dissociating the neural stem cells (NSCs) using accutase (Sigma) for 20 min at 37 °C and plating them on a matrigel-coated plate for attachment. The next day, the medium of these cell cultures was changed to neuronal differentiation medium containing DMEM/F12, 1 mM NEAAs, 1× GlutaMAX, 1% penicillin/streptomycin, 1× N1 supplement (100×), 20 ng/mL BDNF (Peprotech), 20 ng/mL GDNF (Peprotech), 1 mM dibutyryl-cAMP (Sigma) and 2 µl/mL B27 supplement (Invitrogen) for 80–90 days.
Excitatory stimulation of neurons
SCA3 neurons cultured in 6-well plates were washed three times with 2 mL HBSS (balanced salt solution) containing 25 mM Tris, 120 mM NaCl, 15 mM glucose, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2 and pH 7.4. After treatment with l-glutamate 0.1 mM (Sigma no. G8415) in HBSS for 30 min, cells were washed again three times and left them to recover for 30 min in differentiation media followed by a second 30 min l-glutamate treatment in HBSS and subsequently cultured in differentiation media or condition media (with or without IGF for 24 and 48 h until analyzed). For analysis of fragmentation and aggregation of Ataxin 3 by western blotting, extracts were analyzed either immediately after lysis or after fractionation.
Immunocytochemistry
IPSCs-derived neurons from SCA-3 patients were fixated with 4% paraformaldehyde for 20 min. Cells were blocked in 5% normal goat serum and 2% fetal calf serum; subsequently, samples were probed with primary antibodies: TRA-1-81 (Santa Cruz; sc-21706), TRA-2-54 (made by group Peter Andrews lab, The University of Sheffield), OCT-4 (Santa Cruz; sc-5279), SOX-2 (Cell Signaling; #4900S) and βIII-tubulin (Abcam; ab7751). Alexa 488 and Cy3-conjugated secondary antibodies were used in combination with Hoechst nuclear staining (1:1000). Images were obtained using a Leica TCS SP8 confocal microscope (Leica Microsystems).
Statistical analysis
The results were statistically compared using the Student’s t test which was performed for unpaired or paired groups. A p value of < 0.05 was considered significant (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Results
Igf2 is upregulated in the brain of XBP1 deficient mice
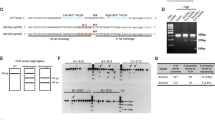
To define the regulatory network involved in the neuroprotective effects triggered by XBP1 deficiency in the brain, we generated XBP1cKO mice using the Nestin-Cre system [34]. Animals of 12–14-month old were euthanized, and brain tissue was dissected to obtain both cortex and striatum to perform a global gene expression profile analysis using a total of 29 animals. Surprisingly, global analysis indicated that the extent of gene expression changes was modest. A comparison between the two brain tissues indicated poor overlap in gene expression changes between brain cortex and striatum where only the expression of Igf2 was modified in both brain areas, showing higher expression in XBP1cKO mice (Fig. 1a).
XBP1 deficiency increases Igf2 expression levels. a Gene expression was determined in the brain cortex and striatum of XBP1cKO (n = 17) and litter mate control (n = 12) animals using Illumina BeadChip® platform and analyzed as described in materials and methods. Heatmaps of microarray analysis of cortex or striatum tissue of two experimental groups considering 1.5- or 1.6-fold change as cutoff, respectively. The normalized average signal of each group was displayed in the heatmap. Red color is used to indicate upregulated genes and green color for downregulation. b Network significantly enriched in genes differentially expressed in the brain cortex and striatum of XBP1cKO. This set of genes was grouped in the Ingenuity Top Network 2 “Cancer, Drug Metabolism, Endocrine System Development and Function.” Red: significantly upregulated gene; Green: significantly downregulated gene; Gray: nonsignificant differential expression; White: biological terms related to the network. c Igf2 mRNA levels were measured by real-time PCR in cDNA generated from cortex, striatum, substantia nigra and hippocampus region of 9-month old XBPWT and XBP1cKO mice. Data represent the average and SEM of the analysis of 3–6 animals per group. Statistically significant differences detected by one-tailed unpaired t test (***p < 0.001; *p < 0.05)
Although the top regulated genes in XBP1cKO mice displayed a consistent fold change of at least 1.5-fold, few genes including Igf2 showed a robust statistical significantly (q value = 0) (Supplementary table 1, online resource). Ingenuity pathway analysis (IPA) suggested a relation between Igf2 with key biological pathways related to neuronal dysfunction associated with Htt and the amyloid precursor protein (Fig. 1b). A second IPA hit found Htt as candidate of “upstream regulators” with several target genes deregulated (Igf2, Gfap, Mmp14, Mpeg1 and Serpine3) and lowest p-value (data no shown).
We then validated the changes in the expression levels of Igf2 in the brain of XBP1cKO mice using real-time PCR and detected a significant increase when compared to wild-type littermates (Fig. 1c). To determine whether the upregulation of Igf2 occurs in the context of neurodegenerative diseases when XBP1 expression is ablated, we analyzed brain tissue derived from a HD model (YAC128 transgenic mice) that was crossed with our XBP1cKO mice [97]. We measured mRNA levels in the striatum of XBP1cKO-YAC128 mice and confirmed the increase in the levels of Igf2 compared to control animals (Supplementary Fig. 1a, online resource). As control, we also monitored Igf1 levels and did not detect any significant differences between XBP1cKO-YAC128 and control mice (Supplementary Fig. 1b, online resource), indicating that Xbp1 deficiency in the nervous system results in the selective upregulation of Igf2 expression.
IGF2 reduces polyQ and mHtt aggregation
Although IGF2 is the less studied member of the insulin-like peptide family, which includes IGF1 and insulin, recent studies have uncovered the importance of IGF2 in brain physiology and neurodegeneration. For example, IGF2 overexpression restored memory function and decreased the accumulation of amyloid β in models of AD [52, 60]. In ALS, IGF2 is differentially expressed in resistant motoneurons and its overexpression extends the survival of mutant SOD1 mice [4]. However, the mechanisms involved in these protective activities need to be further defined. Since the accumulation of misfolded protein in neurons is a common feature of all of these diseases, we decided to explore the possible effects of IGF2.
We first performed cell culture studies to determine the consequences of IGF2 expression over abnormal protein accumulation. Neuro2a cells were transfected with expression vectors for polyQ79-EGFP together with IGF2 or empty vector (Mock) followed by the analysis of intracellular inclusions. Quantification of the number of cells containing GFP puncta by fluorescence microscopy revealed a strong reduction in the number of polyQ79-EGFP inclusions, when co-transfected with IGF2 vector (Fig. 2a). Then, we monitored the levels of polyQ79-EGFP using biochemical approaches. Again, IGF2 or IGF2-HA expression led to a near-complete reduction in the accumulation of both high molecular weight (HMW) and detergent insoluble species, in addition to monomeric forms of polyQ79-EGFP using standard detection by western blot (Fig. 2b and Supplementary Fig. 2a, online resource), whereas polyQ79-EGFP mRNA levels were equal in both conditions (Supplementary Fig. 2b, online resource). Similar results were obtained when protein extracts were analyzed by filter trap, an assay that detects protein aggregates by size using a 200-nm cellulose acetate filter (Fig. 2c and Supplementary Fig. 2c, online resource). Because similar results were obtained with both IGF2 and IGF2-HA constructs, the following experiments were performed using the tagged version.
IGF2 expression reduces the levels of polyQ79 aggregates and HMW species. a Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock). 24 or 48 h later, the number of polyQ79-EGFP inclusions were visualized by fluorescence microscopy (left panel). Quantification of inclusions was normalized by the number of GFP-positive cells (right panel; n = 100 to 350 cells per experiment). Scale bar, 20 µm. b PolyQ79-EGFP expression was analyzed in whole cell extracts by western blot analysis using an anti-GFP antibody. A non-tagged version of IGF2 expression vector was also included. The presence of high molecular weight (HMW) species is indicated. Hsp90 and IGF2 levels were also determined (left panel). PolyQ79-EGFP HMW species levels were quantified and normalized to Hsp90 levels (right panel). c Filter trap assay was performed in the same cell extracts analyzed in b (left panel) and quantified at 24 h (right panel). d Neuro2a cells were transiently co-transfected with polyQ79-EGFP together with plasmids to express IGF1, IGF2 or empty vector (Mock). The presence of polyQ79-EGFP HMW species was analyzed in whole cell extracts after 24 h by western blot analysis. Hsp90 expression was monitored as loading control. e Neuro2a cells were co-transfected with polyQ79-EGFP or SOD1G85R-GFP and IGF2 plasmid or empty vector (Mock). After 24 h, polyQ79 and SOD1 HMW species were measured in total cell extracts using western blot under non-reducing conditions (without DTT). Hsp90 expression was monitored as loading control. In all quantifications, values represent the mean and SEM of at least three independent experiments. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05)
To evaluate if the effects on aggregation were specific for IGF2, we measured the accumulation of polyQ79-EGFP in cells co-transfected with an IGF1 expression vector. Side-by-side comparison indicated that only IGF2 reduced the levels of HMW polyQ79-EGFP species (Fig. 2d). Since XBP1 deficiency also affects the accumulation of proteins associated with other diseases, we performed additional experiments to determine the effects of IGF2 over ALS-linked mutant SOD1G85R. Remarkably, the co-expression of mutant SOD1 with IGF2-HA also modified the levels of HMW species of the protein as assessed by western blot, in addition to reduce the levels of its monomeric form the levels of (Fig. 2e), suggesting broader implications to neurodegeneration.
To define if IGF2 exerts its function from the extracellular compartment, we treated cells expressing polyQ79-EGFP with conditioned media enriched in IGF2. In these experiments, we also included a construct that expresses a fragment of mutant huntingtin spanning exon 1 with 85 CAG repeats (GFP-mHttQ85). We transiently expressed polyQ79-EGFP or GFP-mHttQ85 in Neuro2a cells pre-treated with IGF2-enriched media generated from Neuro2a cells transfected with IGF2 or empty vector for 24 h. A clear reduction in the extent of protein aggregation/inclusions was observed in cells exposed to IGF2-enriched media using three different assays (Fig. 3a–c). These results suggest that IGF2 may exert its function in a paracrine manner, probably activating signaling pathways through the binding to a membrane receptor. Interestingly, treatment of cells with insulin did not reduce the levels of polyQ79-EGFP HMW species in our cellular model (Fig. 3d), similar to the results obtained with IGF1 overexpression.
IGF2 treatment reduces the accumulation of polyQ79 and mHttQ85 HMW species. a Neuro2a cells were transfected with polyQ79-EGFP expression vectors in the presence of IGF2-enriched cell culture media. 24 h later, polyQ79-EGFP and mHttQ85-GFP inclusions were visualized by microscopy (upper panel) and quantified (bottom panel; n = 100 to 350 cells per experiment). Scale bar 20 µm. b Neuro2a cells were transfected with polyQ79-EGFP or mHttQ85-GFP expression vectors in the presence of IGF2-enriched cell culture media. 24 h later, the HMW species levels of polyQ79-EGFP or mHttQ85-GFP were determined by western blot analysis using anti-GFP antibody. Hsp90 expression was monitored as loading control. c Filter trap assay was performed using the same protein extracts analyzed in b to quantify polyQ79-EGFP and mHttQ85-GFP aggregates. d Neuro2a cells were transfected with polyQ79-EGFP in the presence of 1 μM insulin. 24 h later, the HMW species levels of polyQ79-EGFP protein were measured in total cell extracts by western blot. Hsp90 expression was monitored as loading control. e Neuro2a cells were transfected with polyQ79-EGFP or mHttQ85-GFP expression vectors. After 24 h, cells were treated with IGF2-enriched cell culture media and 24 h later, HMW species levels were determined and quantified by western blot analysis using anti-GFP antibody. Hsp90 levels were monitored as loading control. f Filter trap assay was performed in the same cell extracts analyzed in (e) and polyQ79-EGFP, and mHttQ85-GFP aggregates were detected using anti-GFP antibody. In all quantifications, average and SEM of at least three independent experiments are shown. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05)
We then evaluated if IGF2 could reduce the content of pre-formed aggregates. Thus, we expressed polyQ79-EGFP or GFP-mHttQ85 for 24 h and then treated Neuro2a cells with IGF2-enriched or control media. In agreement with our previous results, exposure of cells to IGF2-conditioned media attenuated the load of misfolded polyQ79-EGFP or GFP-mHttQ85 species (Fig. 3e, f), suggesting that IGF2 may trigger the degradation or disaggregation of abnormal misfolded protein.
IGF2 reduces the aggregation of expanded polyglutamine proteins
To further validate our results, we determined the effects of IGF2 expression on a cellular model that expresses full-length mHtt. To this aim, HEK293 cells were transfected with expression vectors for a myc-tagged version of full-length mHtt with a repetition of 103 glutamines (FL HttQ103-myc) together with IGF2 or empty vector (Mock). Again, we observed that the expression of IGF2 caused a significant reduction in the levels of both monomeric and detergent insoluble species of the FL HttQ103-myc using western blot analysis (Fig. 4a). These results were also confirmed when cell lysates were analyzed by dot blot (Fig. 4b). Overall, IGF2 expression led to decrease levels of monomeric and HMW forms of full-length mHtt.
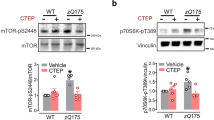
IGF2 overexpression reduced the levels of huntingtin in MSNs derived from HD patients. a HEK293T cells were co-transfected with full length (FL) HttQ103-myc and IGF2 or empty vector (Mock). 24 h later, FL HttQ103-myc levels were analyzed in whole cell extracts by western blot analysis using an anti-Htt antibody. The arrow indicates the migration of full-length huntingtin. b HEK293T cells were co-transfected with FL HttQ103-myc and IGF2 or empty vector (Mock). After 24 h, FL HttQ103-myc was detected in cell lysates by dot blot. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001). c Schematic representation of the differentiation process to generate mature medium spiny neurons (MSNs) from induced human pluripotent stem cells (iPSCs), showing cell morphology using contrast phase microscopy. d Confirmation of the expression of GABAergic (DARPP-32) and neuronal (MAP2) markers in control and HD patients MSNs using immunofluorescence. e MSNs from control subjects and HD patients were transduced with adeno-associated viral (AAV) vectors expressing IGF2 or empty vector (Mock). 17 days later, huntingtin protein levels were measured in total protein extracts by western blot using anti-Htt antibody. Vinculin expression was monitored as loading control (lower panel). Values represent the mean and SEM of three independent experiments. Statistically significant differences detected by one-way ANOVA (*p < 0.05). f Schematic representation of the generation of neuronal cultures generated from SCA-derived iPSCs. g Ataxin 3 levels were monitored in control and SCA3 patient fibroblasts using western blot analysis. “Asterisk” represents the mutant protein. h Differentiated SCA3 iPSC-derived neurons at 90 days post-differentiation were stained for βIII-tubulin neuronal marker. Scale bars: 50 µm. i Neurons were treated with 0.1 mM L-glutamate for 1 h. Then, detergent insoluble protein extracts were generated in triton X100 (TX-100) and pellets were resuspended in SDS to yield an SDS-soluble fraction (SDS) and SDS-insoluble fraction that was then dissolved in formic acid (FA). Ataxin 3 levels were monitored in all fractions using western blots analysis (*-expanded allele). Two independent experiments were performed. j SCA3 cells were stimulated with IGF2-conditioned media for indicated time points followed by stimulation with 0.1 mM L-glutamate followed by western blot analysis as described in (i). GAPDH levels were determined as loading control and βIII-tubulin as a neuronal marker
Next, to increase the relevance of the current study, we moved forward and determined the possible effects of IGF2 in a human model of HD that do not rely on overexpression using induced pluripotent stem cells (iPSCs) derived from patients [12, 55]. Thus, we generated human medium spiny neurons (MSNs) using iPSC-derived from HD patients and control subjects (Fig. 4c, d). To express IGF2, we packed its cDNA into an adeno-associated viral vector (AAV) using serotype 2, which has a high tropism for neurons. MSN cultures were transduced with AAV-IGF2 or AAV-Mock for 17 days followed by western blot analysis to determine the levels of mHtt. Remarkably, expression of IGF2 resulted in a decrease in the total levels of mHtt in HD-derived MSNs (Fig. 4e, right panel). Importantly, treatment of MSN cultures from control subjects did not modify the levels of wild-type Htt (Fig. 4e, left panel). Taken together, these results indicate that IGF2 expression reduces the content of mHtt in various models of HD.
The expansion of polyglutamine tracks is the underlying cause of different diseases, including six spinocerebellar ataxias (SCA), where SCA3 is the most common form of the disease and the second form of polyglutamine disease after HD [50]. SCA3 involves the expansion of the polyglutamine track in Ataxin-3, resulting in its abnormal protein aggregation. To validate the possible effects of IGF2 expression on the levels of other disease-related genes, we generated neuronal cultures using iPSC obtained from SCA3 patients (Fig. 4f–h) and exposed them to IGF2-enriched conditioned cell culture media. In this experimental setting, exposure of cells to glutamate results in the abnormal aggregation of mutant Ataxin-3, reflected in the accumulation of the mutant protein in the detergent insoluble fraction [45] (Fig. 4i). Exposure of cells to IGF2-enriched media led to reduced levels of mutant Ataxin-3 aggregates (Fig. 4j). Thus, IGF2 has broader effects in reducing the abnormal aggregation of polyglutamine-containing proteins.
IGF2 reduces the half-life of intracellular soluble mHtt
The reduction in the steady-state levels of mHtt aggregates induced by the exposure of cells to IGF2 may be explained by an attenuation in the synthesis rate or an increase its degradation. We performed pulse-label experiments in HEK293T cells co-transfected with a mHttQ43-GFP construct together with IGF2 or empty vector (Mock). To evaluate the translation rate, a pulse with S35 labeled methionine and cysteine was performed for increasing intervals on a total period of 60 min. These experiments indicated that the biosynthesis of mHtt was identical in cells expressing or not IGF2 (Fig. 5a). In addition, general protein synthesis was not affected by IGF2 expression (Supplementary Fig. 3a, online resource). Then, we monitored the decay in the levels of mHttQ43-GFP in cell extracts using pulse-chase to define its half-life. Remarkably, expression of IGF2 dramatically reduced the half-life of mHttQ43-GFP in the cell extract (Fig. 5b), without affecting the global stability of the proteome (Supplementary Fig. 3b, online resource). In these experiments, we did not observe the presence of HMW species of mHttQ43, suggesting that IGF2 was affecting the stability of the soluble or monomeric forms. Taken together, our data suggest that IGF2 signaling reduces the content of total mHtt associated with a shortening of the half-life of the protein inside the cell.
Stimulation of cells with IGF2 reduces the half-life of mHtt. a HEK293T cells were co-transfected with mHttQ43-GFP expression vector with IGF2 plasmid (middle panel) or empty vector (Mock) (upper panel) and then pulse labeled with 35S for indicated time points. Autoradiography (AR) indicated the 35S signal for each time point. Data were quantified and normalized to time point 0 h (lower panel). b Similar experiments as described in a were monitored for additional time points. Data were quantified and normalized to the time point 1 h (lower panel). c HEK293T cells were transfected with mHttQ43-GFP and IGF2 expression vectors. Pulse was performed 24 h after transfection. Cells were treated with 30 μM chloroquine (CQ) or 1 µM bortezomib (Bort) at the beginning of the chasing for additional 21 h (upper panel). Data were quantified and normalized to time point 1 h (lower panel). d Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock) for 8 h and then treated with 1 μM lactacystin (Lact) for additional 16 h (upper panel). Quantification of inclusions per GFP-positive cell was performed (lower panel; n = 100 to 350 cells per experiment). e Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock) for 8 h and then treated with 1 μM lactacystin (Lact) or 1 µM MG132 for additional 16 h. PolyQ79-EGFP HMW species were analyzed in whole cells extracts by western blot using anti-GFP antibody and quantified (lower panel). Hsp90 expression was analyzed as loading control (upper panel). f Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock) for 8 h and then treated with 30 μM chloroquine (CQ) for additional 16 h. Endogenous lipidated LC3-II levels were monitored by western blot using anti-LC3 antibody. Hsp90 expression was monitored as loading control (upper panel). LC3 II levels were quantified and normalized to Hsp90 (lower panel). g Neuro2a cells were co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock) for 8 h and then treated with 30 μM CQ for additional 16 h (upper panel). Quantification of inclusions per GFP-positive cell was performed (lower panel; n = 100 to 350 cells per experiment). h Neuro2a cells were co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock) for 8 h and then treated with 30 μM CQ for additional 16 h. PolyQ79-EGFP HMW species were analyzed in whole cell extracts by western blot using anti-GFP antibody and quantified (lower panel). Hsp90 expression was monitored as loading control (upper panel). i Filter trap was performed using the same cells extracts analyzed in (h). In all quantifications, average and SEM of at least three independent experiments are shown. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05)
The autophagy and proteasome pathways are not involved in the reduction of polyQ aggregates induced by IGF2
Two major protein degradation routes are well-established mediators of mHtt clearance, the proteasome and macroautophagy pathways [58, 75]. We monitored mHtt levels after inhibiting lysosomal function with chloroquine or the proteasome with bortezomib. To this end, HEK293T cells were co-transfected with mHttQ43-GFP and IGF2 expression vectors followed by a radioactive pulse 24 h later to perform a chase experiment in the presence of the inhibitors. Surprisingly, neither chloroquine nor bortezomib treatments reverted the effects of IGF2 on mHtt stability (Fig. 5c). As controls for the activity of the inhibitors, we monitored the levels of LC3 and p62 (markers of autophagy-mediated degradation) and the pattern of total ubiquitination (Supplementary Fig. 3c, online resource).
Since the pulse chase methodology demonstrated that IGF2 decreases mHtt intracellular content, we performed further experiments to determine if autophagy or the proteasome mediates the reduction in the load of polyQ aggregates. Although a slight increase in the number of intracellular inclusions was observed at the basal condition after treating cells with the proteasome inhibitors lactacystin or MG132, no changes were observed in the number of polyQ79-EGFP inclusions in IGF2 expressing cells in the presence of the inhibitors (Fig. 5d and Supplementary Fig. 4a, online resource). We confirmed these observations in Neuro2a cells by measuring polyQ79-EGFP HMW species and aggregates by western blot and filter trap analysis, respectively (Fig. 5e and Supplementary Fig. 4b, online resource; see positive controls of total ubiquitination in Supplementary fig. S4c, online resource). Again, the basal increase of polyQ HMW species formation after inhibiting the proteasome was lost in IGF2-treated cells. These results suggest that (1) IGF2 signaling may change the proportion between aggregate/soluble forms of polyQ inside the cell, or (2) it may redirect abnormal polyQ species toward a different pathway to mediate its clearance.
Based on our previous studies linking XBP1 deficiency with the upregulation of autophagy in neurons [97], we evaluated autophagy activation in cells overexpressing IGF2. Neuro2a cells were transfected with IGF2 in the presence or absence of polyQ79-EGFP expression vector. Then, autophagy was monitored after inhibiting lysosomal activity with chloroquine, followed by the analysis of LC3 and p62 levels [54]. We observed that IGF2 did not stimulate autophagy in Neuro2a cells, but slightly reduced LC3-II accumulation (Fig. 5f). A similar trend was observed when we measured p62 levels in cells expressing polyQ79-EGFP (Supplementary Fig. 4d, online resource). Likewise, treatment of cells with chloroquine did not recover protein aggregation of cells exposed to IGF2 as measured with three independent methods (Fig. 5g–i). As control, in these experiments we monitored the accumulation of LC3-II and p62 in cells exposed to chloroquine (Supplementary Fig. 4e, online resource). Similarly, inhibition of lysosomal activity with chloroquine did not recover the normal levels of polyQ79-EGFP HMW species formation in HEK293T cells stimulated with IGF2 (Supplementary Fig. 4f, g, see positive controls in Supplementary Fig. 4h, online resource), confirming our previous results. Finally, we obtained the similar results using the autophagy inhibitors bafilomycin A1 and 3-methyladenine (Supplementary Fig. 4i, j, respectively, online resource). Overall, these results suggest that the reduction in the load of polyQ79-EGFP aggregates induced by IGF2 treatment is independent of the proteasome and autophagy/lysosomal pathways. Our data suggest that IGF2 may reduce the levels of soluble and monomeric forms of mHtt or polyQ, reducing their aggregation levels by shifting the intracellular equilibrium toward a non-aggregated form. Alternatively, IGF2 signaling may reroute the clearance of abnormal protein species toward an alternative pathway that is independent of the activity of the proteasome and macroautophagy/lysosomes.
IGF2 signaling triggers polyQ secretion through extracellular vesicles
Since IGF2 administration bypassed the basal clearance of polyQ aggregates by the main degradation pathways, we explored the possibility that polyQ peptides were redirected to another compartment. mHtt has been found in the cerebrospinal fluid [104], as well as in neuronal allografts transplanted into the brain of HD patients [15] and can be secreted and transmitted from cell to cell. A cell-based study validated this concept, suggesting that mHtt can be exported to the extracellular space by non-conventional mechanisms [90]. To determine the impact of IGF2 treatment in mHtt secretion, we monitored its presence in the cell culture media. Remarkably, enhanced secretion of polyQ79-EGFP was observed in cells expressing IGF2 when culture media was analyzed using dot blot (Fig. 6a, left panel). This phenomenon was also observed in cells overexpressing FL HttQ103-myc in the present of IGF2 (Supplementary Fig. 4k, online resource). Similarly, analysis of SOD1G85R levels in the cell culture media indicated that IGF2 induces its secretion (Fig. 6a, middle panel), whereas no changes were observed in a cytosolic marker such as tubulin (Fig. 6a, right panel), suggesting no cell lysis or cytosol leaking. In agreement with this, analysis of cell viability using propidium iodide staining and FACS analysis in cells expressing polyQ79-EGFP in the presence or absence of IGF2 indicated no toxicity under our experimental conditions (Supplementary Fig. 4l, online resource). We then determined whether polyQ79-EGFP aggregates were also secreted upon IGF2 treatment. Although the signal was low, we detected the presence of large polyQ79-EGFP aggregates outside the cell at basal levels using filter trap, a phenomenon that was incremented with two-fold in cells expressing IGF2 (Fig. 6b). However, we cannot discard that these aggregated forms of polyQ79-EGFP were generated in the cell culture media after secretion or during the concentration process of the supernatants.
IGF2 enhances the extracellular release of polyQ79 and mHttQ85 through unconventional secretion. a Neuro2a cells were co-transfected with polyQ79-EGFP or SOD1G85R plasmid and IGF2 plasmid or empty vector (Mock). After 16 h, cell culture media was replaced for Optimem and then collected after 24 h for dot blot or b filter trap analysis. c Neuro2a cells were transfected with siRNAs against InsR or IGF1R mRNAs. After 24 h, cells were co-transfected with polyQ79-EGFP and IGF2 expression vector or empty vector (Mock). Then, cell culture media was replaced for Optimem for 24 h. In addition, an anti-IGF2R was added when indicated. The presence of polyQ79-EGFP in the cell culture media analyzed by dot blot using an anti-GFP antibody and quantified. The knockdown of indicated proteins was confirmed in cell extracts using semiquantitative PCR (right panel). d Neuro2a cells were co-transfected with polyQ79-EGFP and IGF2 plasmid or empty vector (Mock). Then, cell culture media was replaced for Optimem for 8 h and treated with 2 μM brefeldin A (Bref A) or vehicle for additional 16 h. Cell culture media was collected and analyzed by dot blot using an anti-GFP antibody (upper panel) and quantified (lower panel). Image was cropped from the same membrane and film exposure. e Cells described in d were analyzed by western blot using anti-GFP antibody. Hsp90 expression was monitored as loading control. PolyQ79-EGFP levels were quantified and normalized to Hsp90 (lower panel). f Extracellular vesicles from Neuro2a were analyzed by NanoSight nanotracking analysis and plotted by size (upper panel) and total concentration (lower panel) in cells expressing or not with IGF2. g Extracellular vesicles from Neuro2a cells co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock) were concentrated followed by western blot analysis. h Isolated microvesicles (MV) and exosomes, together with total cell extracts from Neuro2a cells co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock), were analyzed by western blot. In all quantifications, average and SEM of at least three independent experiments are shown. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05)
Given that IGF2 is a soluble factor, we evaluated which receptors mediated the enhancement of polyQ secretion. Thus, we used siRNAs to interfere insulin and IGF1 receptors (InsR and IGF1R), in addition to a blocking antibody to target the IGF2 receptor (IGF2R). Remarkably, antagonizing IGF2R decreased almost in half the levels of polyQ79-EGFP secretion, whereas the knockdown of InsR or IGF1R did not have any effect (Fig. 6c). Taken together, these results indicate that IGF2 signaling reduces the load of polyQ79-EGFP in the cell through the engagement of IGF2R, resulting in the extracellular disposal of polyQ proteins.
IGF2 stimulates polyQ secretion through exosomes and microvesicles
While the precise mechanisms of mHtt secretion are not completely understood, several possibilities have been suggested, including synaptic vesicle release [62], vesicular transport [5], exosomes/extracellular vesicles release [39, 109], exophers [51] and secretory lysosomes [90]. Since the initial link connecting IGF2 with mHtt was through the UPR, we evaluated if the polyQ peptide was secreted by the conventional secretory pathway. Blocking ER to Golgi trafficking with brefeldin A was unable to ablate polyQ79-EGFP secretion in IGF2 stimulated cells (Fig. 6d, e). We then explored the presence of polyQ79-EGFP in extracellular vesicles. First, we used nanoparticle tracking analysis system to determine the size distribution and concentration of vesicles in the cell culture media of cells expressing IGF2. We found enhanced release of vesicles in IGF2-expressing cells, showing an average diameter between 30 and 150 nm, whereas a minor portion (< 1%) had a larger diameter (Fig. 6f). Interestingly, we observed that the population of vesicles increased under IGF2 expression picked around 100 nm, suggesting an enhancement of exosome release [82]. Thus, we purified extracellular vesicles followed by western blot analysis and observed higher content of polyQ79-EGFP in vesicles obtained from IGF2-expressing cells (Fig. 6g).
We then isolated microvesicles and exosomes-enriched fractions through sequential centrifugation. Remarkably, increased levels of polyQ79-EGFP were detected in both fractions in cells expressing IGF2 using western blot analysis (Fig. 6h, see controls of purification in Supplementary Fig. 4m, n, online resource). Importantly, mostly monomeric and soluble forms of polyQ79-EGFP were secreted through microvesicles and exosomes, and no detergent insoluble HMW species were observed in this analysis. Thus, IGF2 signaling enhances the secretion of polyQ79-EGFP through exosomes and microvesicles, reducing the levels of polyQ79-EGFP inside the cell.
Cytoskeleton remodeling contributes to polyQ secretion induced by IGF2
To define possible downstream effectors mediating the effects of IGF2 signaling in abnormal protein aggregation, we employed an unbiased approach to screen for global changes in protein expression. To characterize proteome remodeling upon stimulation with IGF2, we applied quantitative proteomics using Tandem Mass Tag-Multi-Dimensional Protein Identification Technology (MuDPIT) [63]. This approach, we identified a total of 199 proteins that changed their expression levels with a p value < 0.05 and a minimum log2 > 0.1-fold change (Fig. 7a and Supplementary table 2, online resource). IGF2 expression did not result in major proteomic changes. However, functional enrichment analysis revealed that IGF2 expression triggered fluctuations in proteins related to macromolecular complex disassembly, protein transport and cytoskeleton reorganization (Fig. 7b). A cluster of proteins modified by IGF2 was related to actin cytoskeleton dynamics and regulation, including several actin binding proteins, Rho GTPases, vimentin, dynactin and dyneins, among other factors (Supplementary table 3, online resource).
IGF2 signaling induces the release of polyQ79 through cytoskeleton remodeling. a Quantitative proteomics was performed in protein extracts derived from Neuro2a cells transiently transfected with IGF2 or empty vector (Mock) for 24 h. Data were analyzed and plotted in a volcano graph as fold change. Vertical dashed lines indicate a log2 > 0.1 of fold change. Proteins related to actin cytoskeleton function are highlighted in green. Dots in blue represent associated proteins to actin cytoskeleton with a log2 < 0.1 fold change. b Functional enrichment analysis. Blue scale refers to the number of proteins that are known to be involved in each pathway. Red scale refers to the number of proteins altered in IGF2 overexpressing cells for each pathway. c Neuro2a cells were transfected with a plasmid encoding Life-Actin to monitor actin cytoskeleton dynamics. Cells were plated onto fibronectin-coated plates and recorded by time-lapse confocal microscopy every 40 s for 5 min. Time-lapse microscopy was performed after treatment with IGF2-enriched media (left panel). Quantification of cortical and internal actin clusters is shown (right panel). d Phalloidin-rhodamine staining of MEF cells after 5 min of treatment with IGF2-enriched media. Scale bar, 50 µm (right panel). Quantification of actin clusters is presented (left panel). e Neuro2a cells were treated with IGF2-enriched media or control media derived from Mock transfected cells. After 5 min, cells were lysed and cell extracts prepared to measure Rac1-GTP levels by pull-down assay. f Neuro2a cells were transfected with RacN17 or RacV12 or empty vector (Mock). 24 h later, cells were co-transfected with expression vectors for polyQ79-EGFP and IGF2 or empty vector (Mock) and then incubated with Optimem. The presence of polyQ79-EGFP in the cell culture media was determined using dot blot (upper panel) and quantified (lower panel). g Neuro2a cells were co-transfected with expression vectors for polyQ79-EGFP and IGF2 or empty vector (Mock). Then, cell culture media was replaced for Optimem for 8 h and treated with 100 μM NSC23766 (NSC) for additional 16 h. Cell culture media was collected and analyzed by dot blot using an anti-GFP antibody (upper panel) and quantified (lower panel). Image was cropped from the same membrane and film exposure. h Neuro2a cells were co-transfected with polyQ79-EGFP plasmid and IGF2 plasmid or empty vector (Mock) for 8 h and then treated with 100 μM NSC23766 (NSC) or vehicle for additional 16 h. PolyQ79-EGFP HMW species were analyzed in cell lysates by western blot using anti-GFP antibody. Hsp90 expression was monitored as loading control (left panel). PolyQ79-EGFP levels were quantified and normalized to Hsp90 levels (right panel). In all quantifications, average and SEM of at least three independent experiments are shown. Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; *p < 0.05)
Since actin cytoskeleton is key to modulate protein secretion and vesicular trafficking [64], we decided to explore if IGF2 had any impact in the regulation of actin cytoskeleton dynamics using Lifeact, a fluorescent peptide designed to visualize polymerized actin in living cells [69]. Neuro2a cells were plated on coverslips and recorded using time-lapse confocal microscopy. Unexpectedly, IGF2 treatment generated very fast changes in actin dynamics and cell morphology within minutes, reflected in increased development of filopodia and the appearance of actin clusters in the cytosol of the cell (Fig. 7c, see Supplementary videos 1 and 2, online resource). These results were confirmed in murine embryonic fibroblasts by visualizing the actin cytoskeleton in fixed cells stained with phalloidin-rhodamine (Fig. 7d).
Actin cytoskeleton dynamics are dependent on the activity of small GTPases from the Rho family [68]. Among the small GTPases of the Rho family, Cdc42, Rac and Rho are recognized as the most important regulators of actin assembly, controlling the formation of filopodia, lamellipodia and stress fibers [22]. Considering the central role of Rac1 in cytoskeleton dynamics, we evaluated Rac activity upon IGF2 stimulation. Neuro2a cells were treated for 5 min with IGF2-enriched media, and Rac1 activity was measured using pull-down assays using p21-activated kinase that binds specifically to the Rac1-GTP but not to the inactive form of Rac1 (Rac1-GDP) [32], followed by western blot analysis. These results demonstrate that the amount of active Rac1 coupled to GTP was increased very quickly after IGF2 treatment (Fig. 7e).
We tested the functional contribution of actin dynamics to the release of polyQ79-EGFP peptide into the extracellular space. Neuro2a cells were transiently transfected with both a dominant negative (Rac117N) and a constitutive active (Rac1V12) forms of Rac1. Remarkably, Rac117N was able to block IGF2-enhanced secretion of polyQ79-EGFP (Fig. 7f). In contrast, Rac1V12 expression facilitated polyQ79-EGFP secretion at basal levels, whereas no differences were observed in IGF2-stimulated cells, suggesting that the pathway was already fully activated (Fig. 7f). In agreement with these findings, inhibition of Rac1-GTPase activity with NSC23766 significantly decreased the secretion of polyQ79-EGFP (Fig. 7g). These effects paralleled with an increase in the levels of intracellular HMW species of polyQ79-EGFP in cells treated with IGF2 in the presence of NSC23766 (Fig. 7h). Taken together, these results suggest that IGF2 signaling triggers rapid changes in cytoskeleton dynamics that result in the routing of polyQ79-EGFP peptide into the extracellular space.
IGF2 reduces mHtt aggregation in vivo
Based on the significant reduction in mHtt aggregation observed in cells overexpressing IGF2, we moved forward to develop a therapeutic strategy to deliver IGF2 into the brain of HD mice using gene therapy. This, we stereotaxically injected AAVs to then assess the impact on mHtt levels. We previously developed an animal model of HD to monitor mHtt aggregation based on the local delivery into the striatum of a large fragment of mHtt of 588 amino acids containing 95 glutamine repetitions fused to monomeric RFP (Htt588Q95-RFP) [113]. To validate the effects of IGF2 in the aggregation of this mHtt construct, we first performed co-expression experiments in Neuro2a cells followed by fluorescent microscopy and western blot analysis. IGF2 expression decreased mHtt aggregation in both experimental settings (Supplementary Fig. 5a, online resource).
To determine the possible impact of IGF2 in mHtt aggregation levels in vivo, we performed unilateral stereotaxic injections of a mixture of AAVs to deliver Htt588Q95-RFP together with IGF2 or empty vector (Mock) into the striatum (Fig. 8a, left panel). Two weeks after AAV delivery, mice were euthanized, and the striatum was dissected for biochemical analysis. We confirmed the overexpression of IGF2 in the brain using PCR (Supplementary Fig. 5b, online resource). Local expression of IGF2 in adult mice resulted in a marked decrease in mHtt aggregation of near 90% when evaluated by western blot analysis (Fig. 8a, middle and right panel). Consistent with our cell culture experiments, monomeric forms of mHtt were also reduced in the brain of HD mice injected with AAV-IGF2 (Fig. 8a, middle and right panel). To assess the impact of overexpressing IGF2 on neuronal survival, we measured DARPP-32 levels as a marker of medium spiny neuron viability [97]. Significantly higher levels of DARPP-32 were detected in AAV-IGF2-treated animals when compared to control virus (Fig. 8a, middle and right panel).
Gene therapy to deliver IGF2 into the striatum reduces mHtt levels in HD mouse models. a 3-month-old wild-type mice were co-injected with adeno-associated viral (AAV) vectors expressing mHtt588Q95-RFP and IGF2 or empty vector (Mock). Two weeks later, the striatum was dissected and mHtt levels were analyzed in total protein extracts by western blot analysis using anti-polyQ antibody. DARPP-32 levels were also determined in the same samples. Hsp90 and tubulin expression were monitored as loading control. The presence of the monomer is indicated with an arrowhead. mHtt588Q95-RFP aggregates and monomers or DARPP-32 levels were quantified and normalized to Hsp90 levels (lower panel) (AAV-Mock: n = 5; AAV-IGF2: n = 6). b 3-month-old YAC128 transgenic mice were injected with AAV-IGF2 or AAV-Mock into the striatum using bilateral stereotaxis. Four weeks later, the striatum was dissected and mHtt levels monitored by western blot using anti-polyQ antibody. Tubulin expression was monitored as loading control (upper panel). mHtt HMW species levels were quantified and normalized to tubulin levels (lower panel) (AAV-Mock: n = 6; AAV-IGF2: n = 5). c YAC128 mice were injected at postnatal stage (P1-P2) into the ventricle with AAV-IGF2 or AAV-Mock. 3 months later, the striatum was dissected and mHtt levels were analyzed by western blot using anti-polyQ antibody. Hsp90 levels were determined as loading control (left panel). mHtt HMW species were quantified and normalized to Hsp90 levels (right panel) (AAV-Mock: n = 5; AAV-IGF2: n = 4). d YAC128 and littermate control mice were injected with AAVs as indicated in (c). Motor performance was monitored using rotarod once every two weeks from 3- to 8-month old. The analysis shows the average of the group at each time point (AAV-Mock: n = 11; AAV-IGF2: n = 9). Data for time course progression of rotarod performance are presented in Supplementary Fig. 5d. Statistically significant differences detected by two-tailed unpaired t test (**p < 0.001; *p < 0.05). e R6/2 mice were injected at 4 weeks of age with AAV-IGF2 of AAV-Mock into the striatum using bilateral stereotaxis (left panel). Four weeks after the injection, mHtt was detected using the EM48 antibody by immunohistochemistry (upper right panel). High-magnification images of cortex and striatum region. Scale bar: 100 μm (low magnification) and 20 μm (high magnification). CC: corpus callosum; Cx: cortex; St: striatum. Using fluorescence microscopy mHtt and HA-epitope were detected using the EM48 and anti-HA antibodies, respectively (bottom right panel). Nuclei were stained using DAPI (bottom right panel). f High-resolution images of the slices were obtained, and quantification of the number of mHtt inclusions was performed by total area (upper panel) and number of DAPI-positive cells (lower panel). Statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05)
Considering our positive results obtained using the viral HD model, we then evaluated our gene therapy strategy to deliver IGF2 in the YAC128 transgenic model on a heterozygous condition. This mouse model expresses full-length mHtt with 128 tandem glutamines using an artificial chromosome that contains all endogenous regulatory elements [80]. First, we performed bilateral stereotaxic injections of AAV-IGF2 or empty vector into the striatum of 3-month-old YAC128 mice followed by biochemical analysis of brain extracts four weeks later (Fig. 8b, left panel). This strategy led to a significant reduction in full-length mHtt expression in the striatum, reaching near 80% decrease on average (Fig. 8b, right panel). Because stereotaxic injections only transduce a restricted area of the striatum restricting the analysis on motor function, we employed a second route of AAV delivery to generate a global spreading of the viral particles through the brain to assess the impact of IGF2 on mHtt levels and motor control. The delivery of AAVs into the ventricle of new-born pups has been reported to result in an efficient spreading of the virus throughout the nervous system [61]. We injected AAV-IGF2 or control vector in postnatal day 1 or 2 (P1-P2) YAC128 mice and dissected the striatum 3 months after (Fig. 8c, left panel). Western blot analysis demonstrated that IGF2 expression significantly reduced the total levels of mHtt in the brain of YAC128 animals (Fig. 8c, right panel; see controls for IGF2 expression in Supplementary Fig. 5c, online resource). Finally, we determined the functional consequences of administrating our IGF2-based gene therapy on the clinical progression of experimental HD. YAC128 mice develop a variety of functional deficits, including motor learning deficits on rotarod beginning at 2 months [95] and motor impairment starting at 4 months of age [28, 94]. We monitored the motor performance of postnatal-treated YAC128 mice with control or AAV-IGF2 using the rotarod assay. We reported that this model shows a sustained impairment of motor control when assessed using the rotarod assay, with constant values for several weeks [97] (Supplementary Fig. 5d, online resource). Remarkably, delivery of IGF2 into the nervous system improved the average motor performance over time of HD transgenic mice (Fig. 8d).
To complement our in vivo validation, we took advantage of a third model, the R6/2 mice, a transgenic HD model that expresses exon 1 of human huntingtin containing ~ 150 CAG repeats [67], which allows the visualization of intracellular mHtt inclusions. Thus, we evaluated the effects of IGF2 administration in the brain of R6/2 mice by performing bilateral stereotaxic injections of AAV-IGF2 or AAV-Mock into the striatum of 4-week-old R6/2 mice (Fig. 8e, left panel), followed by tissue immunohistochemistry and immunofluorescence analysis of the brains four weeks later (Fig. 8e, upper right and bottom right panel, respectively). A strong reduction in the content of mHtt-positive inclusions was observed upon AAV-IGF2 administration, analyzed by immunohistochemistry and immunofluorescence (Fig. 8e, upper right and bottom right panel, respectively). Quantification of these experiments indicated a significant reduction of near 60% in the brain of R6/2 mice treated with AAV-IGF2 (Fig. 8f). Taken together, these results indicate that the artificial enforcement of IGF2 expression in the brain reduces abnormal protein aggregation in different HD models.
IGF2 levels are reduced in the striatum and blood samples of HD patients
Due to the dramatic effects of IGF2 on intracellular mHtt levels, we moved forward to explore the possible alterations on IGF2 expression in HD patient samples. We monitored the levels of IGF2 in human caudate-putamen derived from HD patients and age-matched control subjects obtained from the Harvard Brain Bank (Supplementary table s4, online resource). Western blot analysis of protein extracts revealed a near 66% reduction on average of IGF2 levels in the brain of HD patients when compared to the control group (Fig. 9a).
IGF2 levels are downregulated in the brain and blood of HD patients. a IGF2 protein levels were measured by western blot in human post mortem samples containing the caudate-putamen region from HD patients in stages 3 and 4. GADPH was monitored as loading control (left panel). IGF2 levels were quantified and normalized to GADPH levels (right panel). b Total protein extracts were generated from freshly isolated peripheral blood mononuclear cells (PBMCs) from HD patients and control subjects. IGF2 expression levels were determined using western blot analysis. GADPH was determined as loading control (left panel). IGF2 levels were quantified and normalized to GADPH levels (right panel). c Igf2 mRNA levels were measured by real-time PCR in PBMCs obtained from HD patients and control subjects. d IGF2 content in plasma from blood obtained from HD patients and control subjects was measured by ELISA. In all quantifications, statistically significant differences detected by two-tailed unpaired t test (***p < 0.001; **p < 0.01; *p < 0.05). e Proposed model: IGF2 reduces mHtt levels through unconventional secretion. In HD patients, the downregulation of IGF2 expression enhances the content of mHtt (left panel). When IGF2 is upregulated using gene therapy, IGF2R signaling enhances the secretion of mHtt into the extracellular space involving actin cytoskeleton remodeling and possibly microvesicles and exosomes (right panel)
Considering the unexpected decrease of IGF2 levels in HD postmortem brain tissue, we evaluated the presence of IGF2 in peripheral blood mononuclear cells (PBMCs) from HD patients. We obtained blood samples from a cohort of 24 Chilean patients recruited in the Enroll-HD international platform (Supplementary table 5, online resource). Although control PBMCs presented a clear expression of IGF2, HD-derived cells showed reduced IGF2 levels, observing an almost 80% decrease on its protein levels using western blot analysis (Fig. 9b). These results were confirmed when mRNA levels of IGF2 were measured in the same samples, observing a near 90% decreased in its levels (Fig. 9c). Since IGF2 is a soluble secreted factor and its plasma levels have been suggested as a possible biomarker of cancer [38, 108], we decided to measure the quantity of IGF2 in plasma from HD patients using ELISA. This analysis revealed a slight but significant decrease in the amount of circulating IGF2 present in the plasma samples derived from HD patients when compared to control subjects (Fig. 9d), which may be related to the different contribution of tissues and cell types to plasmatic IGF2 levels. Taken together, these results suggest that IGF2 levels are drastically reduced in the brain and blood cells of HD patients.
Discussion
Proteostasis impairment is observed in a variety of neurodegenerative diseases and is a central hallmark of aging [107]. Our previous studies uncovered an unexpected connection between the UPR and autophagy, two central nodes of the proteostasis network, where a dynamic balance between both pathways sustains cellular function [37, 97]. However, it remained to be defined if other mechanisms may underlie the beneficial effects targeting the UPR in neurons. To identify novel disease modifiers, we performed a gene expression profile analysis of striatal and cortical areas of XBP1 ablated animals and uncovered the upregulation of Igf2 as the major hit. This prompted us to investigate the consequences of manipulating IGF2 in the context of PMDs using HD as a proof of concept. Based in our findings, and other studies, we speculate that growth factor signaling may play an important role in boosting relevant pro-survival effectors that fine-tune the proteostasis network [26].
IGF1 and IGF2 are mitogenic polypeptides with structural homology to insulin. IGF2 binds with higher affinity to IGF2R but can also associate with lower affinity to IGF1R and the insulin receptor [29, 47]. IGF2 regulates cell proliferation, growth, differentiation and survival [8]. In general, the biological effects of IGF2 have been historically mapped to the IGF1R and to a lower extent to the insulin receptor, impacting cell survival and proliferation [24]. Unlike insulin and IGF1 receptors, IGF2R does not have intrinsic tyrosine kinase activity and has higher affinity for IGF2 than IGF1 and does not bind insulin [30]. IGF2R controls extracellular IGF2 levels as it promotes its endocytosis toward lysosomal-mediated degradation [30]. However, IGF2R can initiate specific responses affecting various cellular processes, possibly involving the activation of heteromeric G proteins and downstream calcium signaling, in addition to the activation of PKC and MAP kinases (reviewed in [30]). Intracellular IGF2R also controls the uptake and trafficking of lysosomal enzymes from the trans-Golgi network to the lysosomes [30]. While fetal IGF2 is abundant, its levels are decreased after birth and are also further reduced during aging [60]. In humans, altered dosage of the IGF2 gene can result in developmental problems and its deregulation has been widely correlated with cancer [42]. In adults, IGF2 is almost exclusively expressed in the brain, especially in the choroid plexus, the brain vasculature and meninges [25]. Initial assessment of the significance of IGF2 expression to animal physiology was provided by the observation that full knockout animals develop growth defects [16], whereas it overexpression generates embryonic lethality [66, 105].
Recent findings highlight the physiological function of IGF2 in cognition, neuronal differentiation and survival. Several studies have demonstrated the relevance of IGF2 to memory consolidation [3, 13, 77], in addition to memory extinction [2], cognitive and social learning [84, 85] and brain stem cells proliferation in adults related to learning, memory and anxiety [111]. At the molecular level, IGF2 regulates the formation of dendritic spines [60] and synapses [77], in addition to control adult neurogenesis in the hippocampus [10]. At the dentate gyrus, the autocrine action of IGF2 may explain the proliferative effects on neural stem cells [10]. Importantly, the effects of IGF2 in learning, memory and neurogenesis have been mapped to the stimulation of IGF2R. Thus, it might be feasible that IGF2 contributes to neuroprotection in HD through additional mechanisms, improving synaptic plasticity, connectivity, neuronal survival and neuronal function. We are currently addressing this hypothesis by assessing the possible impact of IGF2 in the clinical progression of HD using gain- and loss-of-function. Our current study proposes that IGF2 has a relevant role in controlling cellular proteostasis by fine-tuning the load of misfolded proteins through a novel mechanism involving the extracellular disposal of these misfolded proteins through extracellular vesicles (Fig. 9e). The precise relationship between mHtt inclusion bodies and neuronal pathology in HD remains controversial since the neurotoxic species (oligomers, small fibrils versus large protein inclusions) remain to be defined [14]. Large aggregates may even represent a mechanism to preserve neuronal function by reducing the intracellular concentration of oligomers. Thus, as suggested in most protein misfolding associated with neurodegenerative diseases [83], the amyloid cascade is highly complex, where different abnormal protein species are generated over time which may alter neuronal function at multiple levels [14, 100]. We consider monitoring mHtt inclusions as an endpoint that reflects the occurrence of a molecular cascade that triggers neurodegeneration in HD. Here, we speculate that the reduction in the levels of soluble non-aggregated forms of mHtt and polyQ may shift the equilibrium in the amyloid cascade, reducing the proportion of abnormal protein aggregates inside the cell.
Our results suggest that the upregulation of IGF2 in animals with disrupted UPR may serve as a backup mechanism to sustain neuronal function when the adaptive capacity of the proteostasis network is severely compromised. We are currently investigating the mechanisms explaining the upregulation of IGF2 in XBP1 deficient brains. IGF2 expression is controlled by four different promoters and the presence of several mRNA binding proteins [21], in addition to the regulation by genomic imprinting according to a parental origin [16, 23, 53]. IGF2 is also modulated in various pathological contexts including brain injury [101], schizophrenia [59] and Alzheimer’s disease [60, 70], in addition to normal aging [44, 60]. Recent studies also indicated that the administration of IGF2 through gene therapy or the infusion of recombinant proteins into the brain ameliorates AD pathogenesis, improving synaptic dysfunction [52, 60]. Similarly, treatment of SOD1 transgenic mice with AAVs encoding for IGF2 delayed disease progression, improving motor neuron survival [4]. These findings are consistent with the pro-survival role of IGF2 in motor neurons [79], in addition to its ability to induce axonal sprouting [11, 56]. All these reports highlight the neuroprotective potential of IGF2 in various diseases affecting the nervous system.
Although several mechanisms have been proposed to be involved in the secretion of mHtt [5, 39, 62, 109], it was reported that secretory lysosomes are the major route for mHtt release into the extracellular space [90]. Our results suggest that IGF2 signaling leads to a re-routing of mHtt, mostly the soluble forms, toward the secretion in extracellular vesicles. As a limitation of our study, we cannot exclude the possibility that IGF2 administration will enhance the propagation of abnormal mHtt conformations because our experimental settings were not designed to study disease spreading throughout the brain. Based on our results, we propose that soluble species of mHtt are secreted upon stimulation with IGF2, a form of the protein that is expected to lack the capacity to seed aggregation and propagate the protein misfolding process. It is most likely that the reduced aggregation observed in our experiments is due to a restricted availability of the soluble mHtt substrate, shifting the balance toward the accumulation of non-pathogenic mHtt species and reducing the overall intracellular levels of the protein. In addition, microglia has been shown to express IGF2R and IGF1R [86], opening the possibility that IGF2 may impact other cell types affected in HD. In fact, based on the data available in AD models [52, 60], we speculate that IGF2 administration may also enhance the extracellular clearance of mHtt by glial cells or by the action of extracellular proteases. Besides, recent studies suggested that mHtt is cleared faster in astrocytes than in neurons [110].
Our findings in human HD-derived tissue suggest that the downregulation of IGF2 during the progression of the disease may enhance or accelerate the accumulation of abnormal mHtt species. In contrast, increased plasmatic levels of IGF1 in HD patients predict a worst prognosis, involving a faster and stronger cognitive decline [73, 74]. These effects were not observed when insulin levels were monitored in longitudinal studies [74]. Moreover, analysis of YAC128 mice also revealed an increase in IGF1 expression in the brain and blood of symptomatic animals [65, 106]. Since IGF2 levels were dramatically downregulated in human brain tissue derived from HD patients, we speculate that the delivery of IGF2 into the brain using gene therapy may serve as a strategy to restore its normal levels in the brain of HD patients.
Overall, our study places IGF2 as an interesting disease modifier agent and as an interesting candidate molecule for the treatment of HD. Since IGF2 is a soluble factor, the development of gene transfer strategies to augment IGF2 levels in the brain may emerge as an attractive approach for future therapeutic development. Based on the available literature and our current study, we speculate that IGF2 administration to HD patients may have important beneficial consequences beyond proteostasis control and mHtt aggregation, including multiple points of action such as improvement in neuronal connectivity and synaptic plasticity, in addition to enhance axonal regeneration, neuronal survival, neurogenesis/tissue repair and improvement in motor control.
References
Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C et al (2007) XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27:53–66. https://doi.org/10.1016/j.molcel.2007.06.011
Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S et al (2011) A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J 30:4071–4083. https://doi.org/10.1038/emboj.2011.293
Alberini CM, Chen DY (2012) Memory enhancement: consolidation, reconsolidation and insulin-like growth factor 2. Trends Neurosci 35:274–283. https://doi.org/10.1016/j.tins.2011.12.007
Allodi I, Comley L, Nichterwitz S, Nizzardo M, Simone C, Benitez JA et al (2016) Differential neuronal vulnerability identifies IGF-2 as a protective factor in ALS. Sci Rep 6:25960. https://doi.org/10.1038/srep25960
Babcock DT, Ganetzky B (2015) Transcellular spreading of huntingtin aggregates in the Drosophila brain. Proc Natl Acad Sci USA 112:E5427–5433. https://doi.org/10.1073/pnas.1516217112
Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319:916–919. https://doi.org/10.1126/science.1141448
Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR et al (2015) Huntington disease. Nat Rev Dis Primers 1:15005. https://doi.org/10.1038/nrdp.2015.5
Bergman D, Halje M, Nordin M, Engstrom W (2013) Insulin-like growth factor 2 in development and disease: a mini-review. Gerontology 59:240–249. https://doi.org/10.1159/000343995
Bertram L, Tanzi RE (2005) The genetic epidemiology of neurodegenerative disease. J Clin Invest 115:1449–1457. https://doi.org/10.1172/JCI24761
Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J et al (2012) Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J Neurosci 32:3376–3387. https://doi.org/10.1523/JNEUROSCI.4248-11.2012
Caroni P, Grandes P (1990) Nerve sprouting in innervated adult skeletal muscle induced by exposure to elevated levels of insulin-like growth factors. J Cell Biol 110:1307–1317. https://doi.org/10.1083/jcb.110.4.1307
Chamberlain SJ (2016) Disease modelling using human iPSCs. Hum Mol Genet 25:R173–R181. https://doi.org/10.1093/hmg/ddw209
Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, Bambah-Mukku D et al (2011) A critical role for IGF-II in memory consolidation and enhancement. Nature 469:491–497. https://doi.org/10.1038/nature09667
Chiti F, Dobson CM (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem 86:27–68. https://doi.org/10.1146/annurev-biochem-061516-045115
Cicchetti F, Saporta S, Hauser RA, Parent M, Saint-Pierre M, Sanberg PR et al (2009) Neural transplants in patients with Huntington's disease undergo disease-like neuronal degeneration. Proc Natl Acad Sci USA 106:12483–12488. https://doi.org/10.1073/pnas.0904239106
DeChiara TM, Robertson EJ, Efstratiadis A (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859. https://doi.org/10.1016/0092-8674(91)90513-x
Di Prospero NA, Fischbeck KH (2005) Therapeutics development for triplet repeat expansion diseases. Nat Rev Genet 6:756–765. https://doi.org/10.1038/nrg1690
Dickson DW, Weller RO (2011) Neurodegeneration: the molecular pathology of dementia and movement disorders. https://doi.org/10.1002/9781444341256
Duran-Aniotz C, Cornejo VH, Espinoza S, Ardiles AO, Medinas DB, Salazar C et al (2017) IRE1 signaling exacerbates Alzheimer's disease pathogenesis. Acta Neuropathol 134:489–506. https://doi.org/10.1007/s00401-017-1694-x
Eijssen LM, Goelela VS, Kelder T, Adriaens ME, Evelo CT, Radonjic M (2015) A user-friendly workflow for analysis of Illumina gene expression bead array data available at the arrayanalysisorg portal. BMC Genom 16:482. https://doi.org/10.1186/s12864-015-1689-8
Engstrom W, Shokrai A, Otte K, Granerus M, Gessbo A, Bierke P et al (1998) Transcriptional regulation and biological significance of the insulin like growth factor II gene. Cell Prolif 31:173–189. https://doi.org/10.1111/j.1365-2184.1998.tb01196.x
Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420:629–635. https://doi.org/10.1038/nature01148
Feil R, Walter J, Allen ND, Reik W (1994) Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 120:2933–2943
Fernandez AM, Torres-Aleman I (2012) The many faces of insulin-like peptide signalling in the brain. Nat Rev Neurosci 13:225–239. https://doi.org/10.1038/nrn3209
Ferron SR, Radford EJ, Domingo-Muelas A, Kleine I, Ramme A, Gray D et al (2015) Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat Commun 6:8265. https://doi.org/10.1038/ncomms9265
Garcia-Huerta P, Troncoso-Escudero P, Jerez C, Hetz C, Vidal RL (2016) The intersection between growth factors, autophagy and ER stress: a new target to treat neurodegenerative diseases? Brain Res 1649:173–180. https://doi.org/10.1016/j.brainres.2016.02.052
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G et al (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125:1179–1191. https://doi.org/10.1016/j.cell.2006.04.026
Graham RK, Slow EJ, Deng Y, Bissada N, Lu G, Pearson J et al (2006) Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis 21:444–455. https://doi.org/10.1016/j.nbd.2005.08.007
Hawkes C, Jhamandas JH, Harris KH, Fu W, MacDonald RG, Kar S (2006) Single transmembrane domain insulin-like growth factor-II/mannose-6-phosphate receptor regulates central cholinergic function by activating a G-protein-sensitive, protein kinase C-dependent pathway. J Neurosci 26:585–596. https://doi.org/10.1523/JNEUROSCI.2730-05.2006
Hawkes C, Kar S (2004) The insulin-like growth factor-II/mannose-6-phosphate receptor: structure, distribution and function in the central nervous system. Brain Res Brain Res Rev 44:117–140. https://doi.org/10.1016/j.brainresrev.2003.11.002
Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M et al (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA 107:9730–9735. https://doi.org/10.1073/pnas.1002575107
Henriquez DR, Bodaleo FJ, Montenegro-Venegas C, Gonzalez-Billault C (2012) The light chain 1 subunit of the microtubule-associated protein 1B (MAP1B) is responsible for Tiam1 binding and Rac1 activation in neuronal cells. PLoS ONE 7:e53123. https://doi.org/10.1371/journal.pone.0053123
Hetz C (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. https://doi.org/10.1038/nrm3270
Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C et al (2008) Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci USA 105:757–762. https://doi.org/10.1073/pnas.0711094105
Hetz C, Papa FR (2018) The unfolded protein response and cell fate control. Mol Cell 69:169–181. https://doi.org/10.1016/j.molcel.2017.06.017
Hetz C, Saxena S (2017) ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 13:477–491. https://doi.org/10.1038/nrneurol.2017.99
Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R et al (2009) XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev 23:2294–2306. https://doi.org/10.1101/gad.1830709
Huang GS, Brouwer-Visser J, Ramirez MJ, Kim CH, Hebert TM, Lin J et al (2010) Insulin-like growth factor 2 expression modulates Taxol resistance and is a candidate biomarker for reduced disease-free survival in ovarian cancer. Clin Cancer Res 16:2999–3010. https://doi.org/10.1158/1078-0432.CCR-09-3233
Jeon I, Cicchetti F, Cisbani G, Lee S, Li E, Bae J et al (2016) Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol 132:577–592. https://doi.org/10.1007/s00401-016-1582-9
Kaushik S, Cuervo AM (2015) Proteostasis and aging. Nat Med 21:1406–1415. https://doi.org/10.1038/nm.4001
Kemp PJ, Rushton DJ, Yarova PL, Schnell C, Geater C, Hancock JM et al (2016) Improving and accelerating the differentiation and functional maturation of human stem cell-derived neurons: role of extracellular calcium and GABA. J Physiol 594:6583–6594. https://doi.org/10.1113/JP270655
Kessler SM, Haybaeck J, Kiemer AK (2016) Insulin-like growth factor 2—the oncogene and its accomplices. Curr Pharm Des 22:5948–5961. https://doi.org/10.2174/1381612822666160713100235
Khomtchouk BB, Van Booven DJ, Wahlestedt C (2014) HeatmapGenerator: high performance RNAseq and microarray visualization software suite to examine differential gene expression levels using an R and C++ hybrid computational pipeline. Source Code Biol Med 9:30. https://doi.org/10.1186/s13029-014-0030-2
Kitraki E, Bozas E, Philippidis H, Stylianopoulou F (1993) Aging-related changes in IGF-II and c-fos gene expression in the rat brain. Int J Dev Neurosci 11:1–9. https://doi.org/10.1016/0736-5748(93)90029-d
Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D et al (2011) Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature 480:543–546. https://doi.org/10.1038/nature10671
Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. https://doi.org/10.1128/mcb.23.21.7448-7459.2003
Li R, Pourpak A, Morris SW (2009) Inhibition of the insulin-like growth factor-1 receptor (IGF1R) tyrosine kinase as a novel cancer therapy approach. J Med Chem 52:4981–5004. https://doi.org/10.1021/jm9002395
Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C et al (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506. https://doi.org/10.1016/s0092-8674(00)81369-0
Martinez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C (2017) Endoplasmic reticulum proteostasis impairment in aging. Aging Cell 16:615–623. https://doi.org/10.1111/acel.12599
McLoughlin HS, Moore LR, Paulson HL (2020) Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol Dis 134:104635. https://doi.org/10.1016/j.nbd.2019.104635
Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC et al (2017) C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542:367–371. https://doi.org/10.1038/nature21362
Mellott TJ, Pender SM, Burke RM, Langley EA, Blusztajn JK (2014) IGF2 ameliorates amyloidosis, increases cholinergic marker expression and raises BMP9 and neurotrophin levels in the hippocampus of the APPswePS1dE9 Alzheimer's disease model mice. PLoS ONE 9:e94287. https://doi.org/10.1371/journal.pone.0094287
Mikaelsson MA, Constancia M, Dent CL, Wilkinson LS, Humby T (2013) Placental programming of anxiety in adulthood revealed by Igf2-null models. Nat Commun 4:2311. https://doi.org/10.1038/ncomms3311
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326. https://doi.org/10.1016/j.cell.2010.01.028
Naphade S, Tshilenge KT, Ellerby LM (2019) Modeling polyglutamine expansion diseases with induced pluripotent stem cells. Neurotherapeutics 16:979–998. https://doi.org/10.1007/s13311-019-00810-8
Near SL, Whalen LR, Miller JA, Ishii DN (1992) Insulin-like growth factor II stimulates motor nerve regeneration. Proc Natl Acad Sci USA 89:11716–11720. https://doi.org/10.1073/pnas.89.24.11716
O'Riordan CR, Lachapelle AL, Vincent KA, Wadsworth SC (2000) Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV). J Gene Med 2:444–454. https://doi.org/10.1002/1521-2254(200011/12)2:6<444:AID-JGM132>3.0.CO;2-1
Ortega Z, Lucas JJ (2014) Ubiquitin-proteasome system involvement in Huntington's disease. Front Mol Neurosci 7:77. https://doi.org/10.3389/fnmol.2014.00077
Ouchi Y, Banno Y, Shimizu Y, Ando S, Hasegawa H, Adachi K et al (2013) Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J Neurosci 33:9408–9419. https://doi.org/10.1523/JNEUROSCI.2700-12.2013
Pascual-Lucas M, Viana da Silva S, Di Scala M, Garcia-Barroso C, Gonzalez-Aseguinolaza G, Mulle C, et al (2014) Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO Mol Med 6:1246–1262. https://doi.org/10.15252/emmm.201404228
Passini MA, Wolfe JH (2001) Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol 75:12382–12392. https://doi.org/10.1128/JVI.75.24.12382-12392.2001
Pecho-Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, Botta P et al (2014) Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci 17:1064–1072. https://doi.org/10.1038/nn.3761
Plate L, Cooley CB, Chen JJ, Paxman RJ, Gallagher CM, Madoux F et al (2016) Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. Elife. https://doi.org/10.7554/eLife.15550
Porat-Shliom N, Milberg O, Masedunskas A, Weigert R (2013) Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol Life Sci 70:2099–2121. https://doi.org/10.1007/s00018-012-1156-5
Pouladi MA, Xie Y, Skotte NH, Ehrnhoefer DE, Graham RK, Kim JE et al (2010) Full-length huntingtin levels modulate body weight by influencing insulin-like growth factor 1 expression. Hum Mol Genet 19:1528–1538. https://doi.org/10.1093/hmg/ddq026
Pravtcheva DD, Wise TL (2008) Igf2r improves the survival and transmission ratio of Igf2 transgenic mice. Mol Reprod Dev 75:1678–1687. https://doi.org/10.1002/mrd.20909
Proenca CC, Stoehr N, Bernhard M, Seger S, Genoud C, Roscic A et al (2013) Atg4b-dependent autophagic flux alleviates Huntington's disease progression. PLoS ONE 8:e68357. https://doi.org/10.1371/journal.pone.0068357
Ridley AJ (2001) Rho family proteins: coordinating cell responses. Trends Cell Biol 11:471–477. https://doi.org/10.1016/s0962-8924(01)02153-5
Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M et al (2008) Lifeact: a versatile marker to visualize F-actin. Nat Methods 5:605–607. https://doi.org/10.1038/nmeth.1220
Rivera EJ, Goldin A, Fulmer N, Tavares R, Wands JR, de la Monte SM (2005) Insulin and insulin-like growth factor expression and function deteriorate with progression of Alzheimer's disease: link to brain reductions in acetylcholine. J Alzheimers Dis 8:247–268. https://doi.org/10.3233/jad-2005-8304
Ryno LM, Genereux JC, Naito T, Morimoto RI, Powers ET, Shoulders MD et al (2014) Characterizing the altered cellular proteome induced by the stress-independent activation of heat shock factor 1. ACS Chem Biol 9:1273–1283. https://doi.org/10.1021/cb500062n
Safra M, Ben-Hamo S, Kenyon C, Henis-Korenblit S (2013) The ire-1 ER stress-response pathway is required for normal secretory-protein metabolism in C. elegans. J Cell Sci 126:4136–4146. https://doi.org/10.1242/jcs.123000
Saleh N, Moutereau S, Azulay JP, Verny C, Simonin C, Tranchant C et al (2010) High insulinlike growth factor I is associated with cognitive decline in Huntington disease. Neurology 75:57–63. https://doi.org/10.1212/WNL.0b013e3181e62076
Salem L, Saleh N, Desamericq G, Youssov K, Dolbeau G, Cleret L et al (2016) Insulin-like growth factor-1 but not insulin predicts cognitive decline in Huntington's Disease. PLoS ONE 11:e0162890. https://doi.org/10.1371/journal.pone.0162890
Sarkar S, Rubinsztein DC (2008) Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J 275:4263–4270. https://doi.org/10.1111/j.1742-4658.2008.06562.x
Scheper W, Hoozemans JJ (2015) The unfolded protein response in neurodegenerative diseases: a neuropathological perspective. Acta Neuropathol 130:315–331. https://doi.org/10.1007/s00401-015-1462-8
Schmeisser MJ, Baumann B, Johannsen S, Vindedal GF, Jensen V, Hvalby OC et al (2012) IkappaB kinase/nuclear factor kappaB-dependent insulin-like growth factor 2 (Igf2) expression regulates synapse formation and spine maturation via Igf2 receptor signaling. J Neurosci 32:5688–5703. https://doi.org/10.1523/JNEUROSCI.0111-12.2012
Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C et al (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep 3:1279–1292. https://doi.org/10.1016/j.celrep.2013.03.024
Silva D, Dikkes P, Barnes M, Lopez MF (2009) Decreased motoneuron survival in Igf2 null mice after sciatic nerve transection. NeuroReport 20:1414–1418. https://doi.org/10.1097/WNR.0b013e328330b735
Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y et al (2003) Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet 12:1555–1567. https://doi.org/10.1093/hmg/ddg169
Smith HL, Mallucci GR (2016) The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 139:2113–2121. https://doi.org/10.1093/brain/aww101
Soares Martins T, Catita J, Martins Rosa I, Abdces O, Henriques AG (2018) Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 13:e0198820. https://doi.org/10.1371/journal.pone.0198820
Soto C, Pritzkow S (2018) Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci 21:1332–1340. https://doi.org/10.1038/s41593-018-0235-9
Steinmetz AB, Stern SA, Kohtz AS, Descalzi G, Alberini CM (2018) Insulin-like growth factor II targets the mTOR pathway to reverse autism-like phenotypes in Mice. J Neurosci 38:1015–1029. https://doi.org/10.1523/JNEUROSCI.2010-17.2017
Stern SA, Chen DY, Alberini CM (2014) The effect of insulin and insulin-like growth factors on hippocampus- and amygdala-dependent long-term memory formation. Learn Mem 21:556–563. https://doi.org/10.1101/lm.029348.112
Suh HS, Zhao ML, Derico L, Choi N, Lee SC (2013) Insulin-like growth factor 1 and 2 (IGF1, IGF2) expression in human microglia: differential regulation by inflammatory mediators. J Neuroinflammation 10:37. https://doi.org/10.1186/1742-2094-10-37
Taylor RC, Dillin A (2013) XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153:1435–1447. https://doi.org/10.1016/j.cell.2013.05.042
Thiruvalluvan A, P. de Mattos E, Brunsting JF, Bakels R, Serlidaki D, Barazzuol L et al (2020) DNAJB6, a key factor in neuronal sensitivity to amyloidogenesis. Mol Cell (in press)
Torres M, Medinas DB, Matamala JM, Woehlbier U, Cornejo VH, Solda T et al (2015) The protein-disulfide isomerase ERp57 regulates the steady-state levels of the prion protein. J Biol Chem 290:23631–23645. https://doi.org/10.1074/jbc.M114.635565
Trajkovic K, Jeong H, Krainc D (2017) Mutant Huntingtin is secreted via a late endosomal/lysosomal unconventional secretory pathway. J Neurosci 37:9000–9012. https://doi.org/10.1523/JNEUROSCI.0118-17.2017
Turner BJ, Atkin JD, Farg MA, Zang DW, Rembach A, Lopes EC et al (2005) Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J Neurosci 25:108–117. https://doi.org/10.1523/JNEUROSCI.4253-04.2005
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121. https://doi.org/10.1073/pnas.091062498
Valdes P, Mercado G, Vidal RL, Molina C, Parsons G, Court FA et al (2014) Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc Natl Acad Sci USA 111:6804–6809. https://doi.org/10.1073/pnas.1321845111
Van Raamsdonk JM, Pearson J, Rogers DA, Bissada N, Vogl AW, Hayden MR et al (2005) Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum Mol Genet 14:1379–1392. https://doi.org/10.1093/hmg/ddi147
Van Raamsdonk JM, Pearson J, Slow EJ, Hossain SM, Leavitt BR, Hayden MR (2005) Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington's disease. J Neurosci 25:4169–4180. https://doi.org/10.1523/JNEUROSCI.0590-05.2005
Vidal R, Caballero B, Couve A, Hetz C (2011) Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med 11:1–12. https://doi.org/10.2174/156652411794474419
Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C et al (2012) Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet 21:2245–2262. https://doi.org/10.1093/hmg/dds040
Vonsattel JP, Keller C, Cortes Ramirez EP (2011) Huntington's disease—neuropathology. Handb Clin Neurol 100:83–100. https://doi.org/10.1016/B978-0-444-52014-2.00004-5
Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr (1985) Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol 44:559–577. https://doi.org/10.1097/00005072-198511000-00003
Walsh DM, Selkoe DJ (2020) Amyloid beta-protein and beyond: the path forward in Alzheimer's disease. Curr Opin Neurobiol 61:116–124. https://doi.org/10.1016/j.conb.2020.02.003
Walter HJ, Berry M, Hill DJ, Cwyfan-Hughes S, Holly JM, Logan A (1999) Distinct sites of insulin-like growth factor (IGF)-II expression and localization in lesioned rat brain: possible roles of IGF binding proteins (IGFBPs) in the mediation of IGF-II activity. Endocrinology 140:520–532. https://doi.org/10.1210/endo.140.1.6463
Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086. https://doi.org/10.1126/science.1209038
Wang M, Kaufman RJ (2016) Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529:326–335. https://doi.org/10.1038/nature17041
Wild EJ, Boggio R, Langbehn D, Robertson N, Haider S, Miller JR et al (2015) Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington's disease patients. J Clin Invest 125:1979–1986. https://doi.org/10.1172/JCI80743
Wise TL, Pravatcheva DD (1997) Perinatal lethality in H19 enhancers-Igf2 transgenic mice. Mol Reprod Dev 48:194–207. https://doi.org/10.1002/(SICI)1098-2795(199710)48:2<194:AID-MRD7>3.0.CO;2-N
Wong BKY, Ehrnhoefer DE, Graham RK, Martin DDO, Ladha S, Uribe V et al (2015) Partial rescue of some features of Huntington Disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol Dis 76:24–36. https://doi.org/10.1016/j.nbd.2014.12.030
Wong E, Cuervo AM (2010) Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13:805–811. https://doi.org/10.1038/nn.2575
Zanella ER, Galimi F, Sassi F, Migliardi G, Cottino F, Leto SM et al (2015) IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci Transl Med 7:272ra212. https://doi.org/10.1126/scitranslmed.3010445
Zhang X, Abels ER, Redzic JS, Margulis J, Finkbeiner S, Breakefield XO (2016) Potential transfer of polyglutamine and CAG-repeat RNA in extracellular vesicles in Huntington's disease: background and evaluation in cell culture. Cell Mol Neurobiol 36:459–470. https://doi.org/10.1007/s10571-016-0350-7
Zhao T, Hong Y, Li S, Li XJ (2016) Compartment-dependent degradation of mutant Huntingtin accounts for its preferential accumulation in neuronal processes. J Neurosci 36:8317–8328. https://doi.org/10.1523/JNEUROSCI.0806-16.2016
Ziegler AN, Feng Q, Chidambaram S, Testai JM, Kumari E, Rothbard DE et al (2019) Insulin-like growth factor II: an essential adult stem cell niche constituent in brain and intestine. Stem Cell Reports 12:816–830. https://doi.org/10.1016/j.stemcr.2019.02.011
Zuccato C, Valenza M, Cattaneo E (2010) Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev 90:905–981. https://doi.org/10.1152/physrev.00041.2009
Zuleta A, Vidal RL, Armentano D, Parsons G, Hetz C (2012) AAV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington's disease. Biochem Biophys Res Commun 420:558–563. https://doi.org/10.1016/j.bbrc.2012.03.033
Acknowledgements
We are grateful to Dr. Oliver Bracko for providing IGF2 overexpression constructs. We thank the Harvard Brain Tissue Resource Center for providing HD post-mortem samples. We also thank Drs. Dimitri Krainc and Katarina Trajkovic for feedback on protocols to detect mHtt secretion. We also thank Dr. Cristina Alberini for feedback and providing tools to study IGF2 signaling. We thank Marioly Müller and Dr. Josefina Barrera and her team for their kind help taking the blood samples. We thank Dr. Felipe Oyarzun and Rodrigo Sierpe for the kindly help with the use of the Nanosight NS300. We specially acknowledge Carolina Jerez, Claudia Rivera, Valentina Castillo and Constanza Gonzalez for their technical assistance.
Funding
This work was directly funded by ANID/FONDAP program 15150012 (C.H. and R.L.V.), Millennium Institute P09-015-F (C.H. and R.L.V.), CONICYT-Brazil 441921/2016–7, FONDEF ID16I10223, FONDEF D11E1007 and FONDECYT 1180186 (C.H.). We also thank FONDECYT 3150097 (P.G-H.), FONDECYT 1191003 (R.L.V.), FONDECYT 1150069 (H.G.R.), CONICYT Ph.D. fellowship 21160843 (P.T-E.), CSC and the Postgraduate Student Research and Innovation Project of Jiangsu Province KYLX15_0558 (D.W.), the Hersenstichting and NWO-ALW (S.B.), NIH R01 NS100529 (L.M.E.) and NIH NS092829 (R.L.W.). ISF Legacy Heritage Fund (2394/17) (G.Z.L.)
Author information
Authors and Affiliations
Contributions
PG-H, PT-E, CH and RLV designed the study; PG-H and PT-E designed, performed and analyzed most of the in vitro and in vivo experiments; DW and SB designed, performed and analyzed the pulse-chase experiments; CS and FC analyzed the NTA experiments and assisted with the extracellular vesicle experiments; PC-C provided the information and blood samples from Chilean HD patients; DRH and CG-B designed and performed the experiments of actin remodeling; PT, KAL and BJG performed the microarray analysis using the brains samples from the XBP1cko mice; LP and RLW performed the quantitative proteomics in Neuro2a cells; CS and HGR performed the Ingenuity pathway analysis; SPS produced the AAV2-IGF2 and AAV2-Mock; GZL, NS and MS performed the experiments in HEK293 cells using the full-lenghtHttQ103-myc plasmid; SN and LME designed, performed and analyzed the experiments using MSNs derived from HD patients. AT designed, performed and analyzed the experiments using neurons derived from SCA3 patients.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in this study. AAV-IGF2 for gene therapy and its use in misfolded protein diseases as Huntington’s disease (Chile no. 201603282 and PCT. provisional application no. PCT/CL2017/000040).
Data and materials availability
MTA may be required for access to AAV-IGF2 and some vectors. All data are available upon request from C.H.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 (AVI 1209 kb) Supplementary video 1. Neuro2a treated with mock medium
Supplementary file2 (AVI 1561 kb) Supplementary video 2. Neuro2a treated with IGF2-enriched medium
401_2020_2183_MOESM3_ESM.tif
Supplementary file3 (TIF 1831 kb) Supplementary fig. 1 XBP1 deficiency does not alter Igf1 mRNA levels. a Igf2 mRNA levels were measured by real-time PCR in cDNA generated from striatal tissue of 9-month-old XBP1cKO-YAC128 mice. Data represent the average and SEM of the analysis of three animals per group. b Igf1 mRNA levels were measured by real-time PCR in cDNA generated from striatal tissue of 9-month-old XBP1cKO-YAC128 mice. Data represent the average and SEM of five animals. Statistically significant differences detected by one-tailed unpaired t-test (**: p < 0.01)
401_2020_2183_MOESM4_ESM.tif
Supplementary file4 (TIF 2666 kb) Supplementary fig. 2 IGF2 expression reduces polyQ79 levels. a PolyQ79-EGFP monomers from Fig. 2B were analyzed in whole cell extracts by western blot analysis using an anti-GFP antibody (left panel). PolyQ79-EGFP monomer levels after 24 and 48 h of expression were quantified and normalized to Hsp90 levels (middle and right panel, respectively) b Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock). 24 h later, cells were collected in Trizol and Gfp mRNA levels were measured by semi-quantitative PCR. Values represent the mean and SEM of at least three independent experiments. c Quantification of filter trap experiment from Fig. 2c (time point 48 h)
401_2020_2183_MOESM5_ESM.tif
Supplementary file5 (TIF 7168 kb) Supplementary fig. 3 IGF2 does not inhibit global protein synthesis or degradation. a HEK293T cells were co-transfected with GFP-mHttQ43 expression vector with IGF2 plasmid (right, upper panel) or empty vector (left, upper panel) and then pulse labeled with 35S for indicated time points. Autoradiography (AR) represents total proteins that were synthesized in the time of harvesting cells. All data were normalized to time point 0 h (lower panel). b HEK293T cells were co-transfected with GFP-mHttQ43 expression vector with IGF2 plasmid (right, upper panel) or empty vector (Mock) (left, upper panel) and then pulse labeled with 35S for indicated time points to follow the decay of the labeled protein while chasing with unlabeled precursor. Autoradiography (AR) represents total extracts in each time (upper panel). All data were normalized to time point 1 h (lower panel). c HEK293T cells were transfected with GFP-mHttQ43 and IGF2 expression vectors. Pulse was performed 24 h after transfection. Cells were treated with 30 μM chloroquine (CQ) or 1 µM bortezomib (Bort) at the beginning of the chasing for additional 21 h (upper panel). Ubiquitin, p62 and LC3 were measured as control of the autophagy inhibition
401_2020_2183_MOESM6_ESM.tif
Supplementary file6 (TIF 16029 kb) Supplementary fig. 4 Proteasome, autophagy or conventional secretion are not involved in the reduction in polyQ79 levels after IGF2 expression (control experiments). a Neuro2a cells were co-transfected with expression vectors for polyQ79-EGFP and IGF2 or empty vector (Mock). After 8 h, cells were treated with 1 μM MG132 for additional 16 h. PolyQ79-EGFP inclusions were visualized by fluorescence microscopy (upper panel) and polyQ79-EGFP aggregation levels measured by filter trap (lower panel). Image cropped from the same membrane and film. b Filter trap assay was performed to detect polyQ79-EGFP aggregates using the same cells extracts of Fig. 4e using an anti-GFP antibody (upper panel) and quantified (lower panel). Image cropped from the same membrane and film. c Neuro2a cells were transfected with IGF2 for 8 h and then treated with 1 μM lactacystin (Lact) or 1 μM MG132 for additional 16 h. Ubiquitin accumulation was measured by western blot to confirm the activity of the inhibitors. Hsp90 expression was measured as loading control. d Neuro2a cells where co-transfected with IGF2 plasmid or empty vector (Mock) in the presence or absence of polyQ79-EGFP expression vector for 8 h, and then treated with 30 μM chloroquine (CQ) for additional 16 h. p62 levels were monitored by western blot using anti-p62 antibody. Hsp90 expression was monitored as loading control (upper panel). p62 levels were quantified and normalized to Hsp90 (lower panel). e Neuro2a cells were co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock) for 8 h and then treated with 30 μM CQ for additional 16 h. p62 and LC3 levels were measured by western blot as control for autophagy inhibitors in experiment shown in Fig. 4h. Hsp90 levels were determined as loading control. f HEK293T cells were co-transfected with polyQ79-EGFP and IGF2 expression vectors or empty vector (Mock). After 8 h, cells were treated with 30 μM chloroquine (CQ) for additional 16 h. PolyQ79-EGFP aggregation was analyzed in whole cells extracts by western blot using anti-GFP antibody. Hsp90 expression was monitored as loading control (left panel). PolyQ79-EGFP HMW species levels were quantified and normalized to Hsp90 levels (right panel). g Filter trap assay was performed using the same cells extracts analyzed in (f), and polyQ79-EGFP HMW species were detected using anti-GFP antibody (upper panel) and quantified (lower panel). h HEK293T cells were transiently transfected with IGF2 expressing vector (IGF2) or empty plasmid (Mock) for 8 h and then treated with 30 μM CQ for additional 16 h. p62 and LC3 levels were measured by western blot as control for autophagy inhibitors in experiment shown in Fig. 4f and Fig. 4g. Hsp90 levels were determined as loading control. i, j Neuro2a cells were co-transfected with polyQ79-EGFP and IGF2 or empty vector (Mock) for 8 h and then treated with 200 nM bafilomycin A1 (Baf) or 250 mM 3-methyladenine (3-MA) for additional 16 h. PolyQ79-EGFP HMW species were analyzed in cell lysates using western blot with an anti-GFP antibody. Hsp90 or GAPDH expressions were monitored as loading control. In all quantifications, average and SEM of at least three independent experiments are shown. Statistically significant differences detected by two-tailed unpaired t-test (**: p < 0.01; *: p < 0.05). k HEK293 cells were co-transfected with FL-Htt103Q and IGF2 plasmid or empty vector (Mock). 24 h later, culture media was collected and loaded for dot blot detection of FL-Htt103Q using anti-Htt antibody. l Neuro2a cells were co-transfected with a polyQ79-EGFP expression vector and IGF2 plasmid or empty vector (Mock). 48 h later, cell death was measured by FACS after propidium iodide staining. m NanoSight profile of exosomes purified form IGF2 expressing cells. Isolated exosomes from Neuro2a cells co-transfected with polyQ79-EGFP and IGF2 plasmid or empty vector (Mock) were analyzed by NanoSight nanotracking analysis and plotted by size to confirm proper purification method. n Western blot analysis of enriched fractions of microvesicles (MV) and exosomes using the markers Alix and CD63
401_2020_2183_MOESM7_ESM.tif
Supplementary file7 (TIF 3537 kb) Supplementary fig. 5 Gene therapy to deliver IGF2 into the brain. a Neuro2a cells were co-transfected with expression vectors for mHtt588Q95-RFP and IGF2 plasmid or empty vector (Mock) using an AAV back bound. After 24 h, mHttQ95-RFP inclusions were visualized by fluorescence microscopy (left panel). Protein aggregation was analyzed in whole cells extracts by western blot (right panel). Hsp90 was measured as loading control. b Igf2-HA expression was confirmed by conventional PCR in cDNA obtained from dissected striatum of animals co-injected with AAV mHtt588Q95-RFP and AAV-IGF2 or AAV-Mock showed in Fig. 8a. c Igf2-HA expression was confirmed by conventional PCR in cDNA obtained from dissected striatum of YAC128 animals injected with AAV-IGF2 or AAV-Mock intra-cerebroventricular at neonatal stage P1-P2 shown in Fig. 8c. d Disease progression was monitored once every two weeks, using the rotarod test. YAC128 and littermate control mice were injected with AAVs as indicated in Fig. 8c. Motor performance was monitored once every two weeks from 2-month to 8-month old. The analysis shows the average of the group at each time point
Rights and permissions
About this article
Cite this article
García-Huerta, P., Troncoso-Escudero, P., Wu, D. et al. Insulin-like growth factor 2 (IGF2) protects against Huntington’s disease through the extracellular disposal of protein aggregates. Acta Neuropathol 140, 737–764 (2020). https://doi.org/10.1007/s00401-020-02183-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-020-02183-1