Abstract
Posterior fossa ependymoma comprise three distinct molecular variants, termed PF-EPN-A (PFA), PF-EPN-B (PFB), and PF-EPN-SE (subependymoma). Clinically, they are very disparate and PFB tumors are currently being considered for a trial of radiation avoidance. However, to move forward, unraveling the heterogeneity within PFB would be highly desirable. To discern the molecular heterogeneity within PFB, we performed an integrated analysis consisting of DNA methylation profiling, copy-number profiling, gene expression profiling, and clinical correlation across a cohort of 212 primary posterior fossa PFB tumors. Unsupervised spectral clustering and t-SNE analysis of genome-wide methylation data revealed five distinct subtypes of PFB tumors, termed PFB1-5, with distinct demographics, copy-number alterations, and gene expression profiles. All PFB subtypes were distinct from PFA and posterior fossa subependymomas. Of the five subtypes, PFB4 and PFB5 are more discrete, consisting of younger and older patients, respectively, with a strong female-gender enrichment in PFB5 (age: p = 0.011, gender: p = 0.04). Broad copy-number aberrations were common; however, many events such as chromosome 2 loss, 5 gain, and 17 loss were enriched in specific subtypes and 1q gain was enriched in PFB1. Late relapses were common across all five subtypes, but deaths were uncommon and present in only two subtypes (PFB1 and PFB3). Unlike the case in PFA ependymoma, 1q gain was not a robust marker of poor progression-free survival; however, chromosome 13q loss may represent a novel marker for risk stratification across the spectrum of PFB subtypes. Similar to PFA ependymoma, there exists a significant intertumoral heterogeneity within PFB, with distinct molecular subtypes identified. Even when accounting for this heterogeneity, extent of resection remains the strongest predictor of poor outcome. However, this biological heterogeneity must be accounted for in future preclinical modeling and personalized therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genomics has significantly advanced our knowledge of the biology of posterior fossa ependymoma [9,10,11, 14, 20, 23, 24, 26]. Posterior fossa ependymoma comprise three distinct molecular subgroups, defined by gene expression and genome-wide DNA methylation, and they display highly disparate demographics, cytogenetics, and outcomes [6, 11, 12, 14,15,16, 26]. PFA (PF-EPN-A) ependymomas arise in younger patients, primarily infants, and have generally a bland genome and a poor outcome. In contrast, PFB (PF-EPN-B) ependymomas occur in older children and adults, harbor multiple broad cytogenetic aberrations, and generally have a favorable outcome. The third molecular subgroup comprises mostly grade I subependymomas (PF-SE), which occur almost exclusively in adults only and have a good outcome, as well [14]. Currently, the standard of care for all posterior fossa ependymoma in patients under 18 is maximal safe surgical resection followed by conformal radiation to the tumor bed; however, in adults, adjuvant postoperative radiation is variable resulting in a subset of patients who are treated with surgery only [19].
Recently, we demonstrated that there are profound clinical differences and responses to therapy between PFA and PFB [14, 15, 26]. PFA have a poor outcome overall, with an even worse outcome when sub-totally resected. PFB have an excellent outcome overall, where a significant proportion of patients can be cured with a gross total resection alone. Therefore, patients with PFB ependymoma, particularly children are excellent candidates for radiation sparing strategies, where carefully controlled studies are being contemplated of observation alone in the setting of gross total resection. Gain of 1q has been proposed as a marker of poor prognosis in PFA ependymoma; however, its role in PFB is unclear [1, 4, 7, 8, 17, 25]. We have also recently shown that PFA ependymoma is highly heterogeneous entities with two major groups with multiple subtypes [13]. Moreover, PFB has been assumed to be a homogeneous group; however, it is unknown if there exists molecular and/or clinical heterogeneity and additional substructure within PFB ependymoma. As such, unraveling this heterogeneity will be a crucial starting point to the development of personalized therapies within PFB. We assembled the largest cohort to date of clinically annotated PFB ependymomas profiled using genome-wide DNA methylation arrays to assess molecular heterogeneity within the PFB ependymoma subgroup.
Methods
Patient cohort
Two hundred and twelve posterior fossa ependymomas were obtained through the GENE consortium (Global Ependymoma Network of Excellence), St. Jude’s Children’s Research Hospital, the Collaborative Ependymoma Research Network, the Burdenko Neurosurgical Institute, and the German Cancer Research Center. Both frozen and FFPE samples were collected from diagnosis. Samples were all collected in accordance with the Hospital for Sick Children Research Ethics Board and local institutional research ethics boards. All frozen samples were snap frozen and stored at − 80 °C. Formalin-fixed paraffin-embedded tissue was collected as scrolls or unstained slides. A subtotal resection was defined as more than 5 mm postoperative residual on the postoperative MRI.
Genome-wide DNA methylation profiling
Samples were analyzed on the Illumina Infinium HumanMethylation450 or the HumanMethylationEPIC beadchips at the PM-OICR Translational Genomics Laboratory (Toronto, Ontario) or the German Cancer Research Center (Heidelberg, Germany) according to the manufacturer’s instructions and as previously described [14]. All analysis was conducted in the R Statistical Environment (v3.4.1). Raw data files (.idat) generated by the Illumina iScan array scanner from both frozen and FFPE derived tissue were loaded and preprocessed using the minfi package (v1.22.1). Illumina preprocessing was selected to mimic the normalization performed in Illumina Genome Studio. To account for possible batch effects due to divergent protocols for fresh frozen and FFPE material, a batch adjustment was performed. Batch effects were estimated by fitting a linear model to the log2 transformed intensity values of the methylated and unmethylated channels. After removing the component due to the batch effect, the residuals were backtransformed to intensity scale, and methylation beta values were calculated as described in Illumina’s protocols. Subsequently, the following filtering criteria were applied: removal of probes targeting the X and Y chromosomes, removal of probes containing a single-nucleotide polymorphism (dbSNP132 Common) within five base pairs of and including the targeted CpG site, and probes not mapping uniquely to the human reference genome (hg19) allowing for one mismatch. In total, 431069 probes were kept for analysis. PFB status was determined using the Heidelberg brain tumor classifier (https://www.molecularneuropathology.org/mnp) and unsupervised hierarchical clustering as previously described [2, 14]. Raw and processed methylation data have been deposited into GEO under the accession number GSE117130.
Gene expression profiling
PFB ependymoma samples, for which gene expression profiles generated on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, USA) at the Microarray Department of the University of Amsterdam, The Netherlands, were processed as previously described (GSE64415) [13, 14]. Briefly, data were normalized using the MAS5.0 algorithm, and differentially expressed genes between the five subtypes were generated within the R2: Genomics Analysis and Visualization Platform comparing one (http://r2.amc.nl). TMEV software was used to generate the heatmap displaying the top ten most upregulated genes per subtype [21]. Differentially expressed genes (adj. p < 0.05) when comparing one subtype versus others were used to performed pathway analysis with g:profiler [18].
Unsupervised t-SNE analysis
For unsupervised t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis, we selected the 6749 most variably methylated probes across the data set (s.d. > 0.25). Pairwise sample distances were calculated using 1 minus the weighted Pearson correlation coefficient as the distance measure. Unweighted correlation coefficients provided similar results and as such weighted probes were used to reduce the noise. Pairwise Pearson correlation was calculated using the wtd.cors function of the weights package v.0.85. We used the probe standard deviation subtracted by 0.25 as the weight, giving more variable probes greater influence. The resulting distance matrix was used to perform the t-SNE analysis (Rtsne package v.0.13). The following non-default parameters were used: θ = 0, is_distance = T, pca = F, and max_iter = 2000. Clusters were annotated using the DBSCAN algorithm as implemented in the dbscan package v.1.1.1. The following non-default parameters were used: minPts = 10, eps = 2.4. Subsequently, samples not assigned to any cluster were iteratively merged to their nearest cluster.
Spectral clustering
The same weighted distance matrix used for unsupervised t-SNE analysis was used to perform spectral clustering. Spectral clustering implemented in the SNFtool package (v2.2.1) was run on the patient by patient similarity matrix to obtain groups corresponding to k = 2–8.
Copy number variation (CNV) analysis
Copy-number segmentation was performed from genome-wide methylation arrays using the conumee package (v0.99.4) in the R statistical environment (v3.3.3) as previously described [5, 22]. Broad copy-number events were determined using visual inspection of copy-number plots and significance of the frequency of each broad event was tested using the exact binomial test. Each broad event frequency was compared to the background frequency, which was determined from a robust regression of the observed frequencies with respect to gene content (i.e., number of RefSeq genes) across all chromosomes. This approach was motivated by GISTIC’s broad event analysis [3].
Statistical analysis
Progression-free survival and overall survival were analyzed by the Kaplan–Meier method, and p values reported using the log-rank test. Associations between covariates and risk groups were tested by the Fisher’s exact test. Univariable and multivariable cox proportional hazard regression was used to estimate hazard ratios including 95% confidence intervals. All statistical analyses were performed in the R statistical environment (v3.4.1), using R packages survival (v2.41-3), and ggplot2 (v2.2.1).
Results
DNA methylation profiling identifies five distinct subtypes of PFB ependymoma
Genome-wide methylation profiling was available on 212 primary posterior fossa ependymomas (76 frozen and 136 formalin-fixed paraffin-embedded samples) previously assigned to the PFB subgroup using either the Illumina HumanMethylation450 (n = 180) or HumanMethylationEPIC arrays (n = 32) and the Heidelberg brain tumor classifier (https://www.molecularneuropathology.org/mnp) [2].
To discern the extent of heterogeneity within the PFB subgroup, we applied unsupervised clustering using the most variably methylated probes with a standard deviation over 0.25. A distance matrix was constructed using 1 minus the weighted Pearson correlation, and both t-SNE analysis and spectral clustering were applied to the resulting distance matrix (Fig. 1a, b). Using the t-SNE analysis performing 2000 iterations, five distinct clusters were identified which we term subtypes. Spectral clustering was also applied for k = 2 to k = 8, confirming the optimal clusters at k = 5 (Fig. 1b, Supplemental Fig. 1). The subtypes obtained by t-SNE and the spectral clustering were also largely identical when the t-SNE analysis is compared to spectral clustering at k = 5 (Supplemental Fig. 2A). No major differences in overall methylation levels at probes near the promoter were identified across the five subtypes (Supplemental Fig. 2B). In four instances, paired samples from diagnosis and relapse were available, and we did not observe any switching of subtype at recurrence (1 PFB1, 2 PFB2, and 1 PFB3).
Clustering of PFB reveals five distinct subtypes. a t-SNE plot showing the relative distribution of the five subtypes. b Heatmap obtained from spectral clustering (SpC), whereby yellow represents more similar samples and red represents disparate ones. Color bars at the top indicate the t-SNE cluster at k = 5 and the spectral clustering clusters (SpC) at k = 5; c t-SNE plot showing PFB clustered concomitantly with PF-SE and PFA. d Heatmap representing the expression levels of the ten most differentially expressed genes per subtype. Each column represents one sample, and each lane represents one gene. Gene expression levels are represented by a color scale as indicated
To determine if any of the PFB subtypes were closer to PF-SE subependymoma, particularly the ones with older age, we clustered the 212 PFBs with 34 posterior fossa subependymomas (PF-EPN-SP) and 220 PFAs. The five PFB subtypes clustered together, without any overlap with PF-SE subependymomas or PFAs, suggesting that the heterogeneity within PFB is distinct from the other two groups (Fig. 1c).
Differential gene expression across PFB subtypes
Array-based gene expression analyses of the five PFB subtypes (n = 25) reveal the top upregulated genes per subtypes (Fig. 1d), Differential expression analysis identifies in total 693 genes as significantly differentially expressed (adj. p < 0.05) between one versus the other four subtypes. Similar to the methylation-based clustering, PFB4 and PFB5 had significantly more differentially expressed genes compared to PFB1, 2, and 3 which exhibited fewer differences. Subsets of these genes, which included transcription factors, developmental patterning pathways, and potential drug targets, showed upregulated expression within the five subtypes. Gene-ranked pathway enrichment analysis of subtype-specific genes indicated several general and specific developmental processes and developmental patterning pathways identified as deregulated processes, specifically within PFB4 and PFB5 (Supplemental Fig. 3) [18].
Demographic differences across PFB subtypes
Annotation of demographics of the five subtypes reveals significant age differences between the five subtypes, specifically, an enrichment of younger patients in PFB4 (median 15 years, IQR 10.1–23.5 years) and enrichment of older patients in PFB5 (median 40.6, IQR 28.5–50 years, Fig. 2a p = 0.011). PFB1, PFB2, and PFB3 had similar median age at diagnosis (PFB1 25.9 years, IQR 13.4–42 years; PFB2: median 26 years, IQR 17–41 years; PFB3: median 29.1 years, IQR 13.2–41 years, Fig. 2a). Moreover, there are significant gender differences, wherein PFB2 and PFB4 have a high male bias, whereas PFB3 and PFB5 have a high female patient bias (Fig. 2b, p = 0.04).
PFB subtype-specific chromosomal aberrations
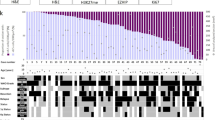
We generated genome-wide DNA copy-number profiles, using the combined intensity of the methylated and unmethylated probes as previously described [5]. Many samples displayed aneuploidy without any recurrent amplifications or deletions, consistent with the previous descriptions of PFB ependymoma [26]. Moreover, the overall patterns of broad copy-number changes were similar to the previous reports, including enrichment of 1q gain (12%), monosomy 6 (61.3%), monosomy 10 (38.7%), monosomy 17 (33.5%), trisomy 5 (31.1%), trisomy 8 (23.5%), trisomy 18 (51.9%), and 22q loss (48.1%) [14]. However, when we repeated this analysis across the PFB subtypes, we observed several subtype-specific aberrations like monosomy 2 in PFB2 (PFB1 17/75, PFB2 18/36, PFB3 10/54, PFB4 4/30, PFB5 1/17, p = 0.0014), monosomy 3 in PFB1 (PFB1 38/75, PFB2 5/36, PFB3 11/54, PFB4 4/30, PFB5 2/17, p = 3.8 × 10−5), monosomy 6 in PFB1,2,3 (PFB1 51/75, PFB2 26/36, PFB3 41/54, PFB4 8/30, PFB5 4/17, p = 1.2 × 10−6), monosomy 8 in PFB3 (PFB1 1/75, PFB2 5/36, PFB3 25/54, PFB4 2/30, PFB5 1/17, p = 1.3 × 10−11), gain of chromosome 11 in PFB4 (PFB1 17/75, PFB2 13/36, PFB3 14/54, PFB4 21/30, PFB5 0/17, p = 4.4 × 10−6), and enrichment of 1q gain (PFB1 19/75, PFB2 5/36, PFB3 1/54, PFB4 0/30, PFB5 1/17, p = 1.6 × 10−4) and monosomy 17 (PFB1 44/75, PFB2 10/36, PFB3 9/54, PFB4 5/30, PFB5 3/17, p = 1.6 × 10−6) within PFB1 (Fig. 3a, b). Copy-number solutions for k = 3 and 4 suggested that PFB1, 2, and 3 were more similar groups overall, but, at k = 5, certain broad events were further enriched, like monosomy 2 in PFB2, monosomy 8 in PFB3, monosomy 3 in PFB1, and gain of 1q in PFB1 (Supplemental Fig. 3). T-SNE analysis and NMF clustering of broad copy-number aberrations could not recapitulate any of the observed subtypes (Supplemental Fig. 4).
Arm-level cytogenetic events across PFB subtypes. a Frequency and significance of arm-level gains and losses across the five PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test). b Frequency and significance of whole chromosome gain and losses across the five PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test)
Extent of surgical resection is the strongest predictor of progression-free survival across PFB ependymoma
Survival across the five PFB groups was determined using a Kaplan–Meier survival analysis, and no significant difference in overall or progression-free survival was observed between the five subtypes (Supplemental Fig. 5A, B). Progression-free survival was available on 137 cases, and progression events were evenly distributed across the five subtypes. Late relapses were common in this cohort, whereby 13 of the 36 relapses occurred after 5 years and five relapses even after 10 years, highlighting the importance of long-term follow-up in this group. Late relapses were evenly distributed across the five subtypes. Sixteen deaths were observed in the cohort, nine events in PFB1, and five events in PFB3, with one death in each of PFB2 and PFB4, and no deaths observed in PFB5.
The rates of incomplete resection and upfront adjuvant radiation were not significantly different across the five subtypes (Supplemental Fig. 6C, D). Consistent with our previous observations, in this cohort overall, a subtotal resection portended to a worse progression-free survival in a univariable analysis (STR: HR 3.01 95% CI 1.51–5.98 p = 1.7 × 10−3). Interestingly in this cohort, no upfront radiation and 1q gain were not significant predictors of poor outcome in a univariable analysis, although both have high hazard ratios, suggesting that they may have a trend to a worse outcome (no upfront radiation: HR 1.76 95% CI 0.83–3.75 p = 0.15; 1q gain: 1.84 95% CI 0.76–4.48 p = 0.18) (Supplemental Table 2).
To discern if other copy-number abnormalities confer any prognostic relevance, an exploratory univariable analysis was undertaken, whereby survival was assessed across all copy-number aberrations with at least a 10% incidence. Surprisingly, loss of 13q was significant in a univariable analysis (Fig. 4, log-rank p = 0.01); a univariable analysis of 13q loss compared to both balanced 13q and 13q gain confirmed a significant effect size (HR 2.66, 95% CI 1.262–5.591, p = 0.01). A multivariable analysis was then undertaken incorporating 13q and extent of resection, and the prognostic value of 13q loss remains significant (HR 2.73, 95% CI 1.18–5.32 p = 0.017). To further determine the relationship between 13q loss and extent of resection can potentially predict progression-free survival, a Kaplan–Meier analysis was performed comparing 13q balanced/gain versus 13q loss stratified by extent of resection, which suggests that subtotal resected 13q loss tumors may represent a novel high-risk group of PFB ependymoma. Loss of 13q was not significant in predicting overall survival (HR 0.92, 95% CI 0.20–4.20, p = 0.92).
To determine if extent of surgical resection and chromosome 13q loss remained prognostic when correcting for PFB subtypes, age, gender, 1q gain, and upfront radiation, a multivariable analysis of progression-free survival was performed (Table 1). Both an incomplete resection and 13q loss remained significant, when correcting for co-variants. Interestingly, upfront radiation and 1q gain were marginally significant in the multivariable model; however, a significant interaction factor could not be discerned between a subtotal resection, upfront radiation, and 1q gain. Only an incomplete resection was a risk factor for worse overall survival (Table 1). Overall outcome was excellent across the whole cohort with only 16 deaths across 142 patients, and none of the variables above were predictive of death, although longer follow-up is likely required.
Discussion
Across the largest cohort of PFB ependymoma assembled to date, we show that PFB is comprised of five distinct molecular subtypes. These five subtypes display distinct demographics and copy-number profiles, suggesting that there exists a significant molecular heterogeneity within PFB ependymoma. We also show that none of the five subtypes of PFB ependymoma overlap with either subependymoma or any of the PFA ependymoma subtypes, further strengthening the existence of three major molecular subgroups of posterior fossa ependymoma.
PFB ependymoma has been considered for de-escalation of therapy due to their excellent overall survival [12]. Previously, we have shown that 50% of gross totally resected PFB ependymoma can be cured with surgery alone, suggesting that this group could potentially be spared the long-term side effects of radiation and/or chemotherapy [15]. Our current data suggest that even when correcting for heterogeneity within PFB ependymoma, this group can still benefit from de-escalation of therapy. However, the observation that late relapses are common in PFB highlights that long-term follow-up is required for any eventual trial of de-escalation of therapy. Overall survival in the cohort was excellent, suggesting that successful salvage is possible; however, the observation that deaths were enriched in PFB1 and 3 requires further evaluation in future cohorts.
Several studies over the past 15 years have suggested 1q gain to be a marker of poor prognosis across all ependymomas as a whole [7, 8]. However, when 1q is evaluated in a subgroup-specific manner, it is only prognostic for progression-free and overall survival in PFA, with no prognostic relevance in the RELA subgroup of supratentorial ependymoma [14]. Indeed, currently, 1q-gained PFA are being prioritized urgently for new and novel upfront approaches. Our previous work suggested that 1q gain was prognostic for progression-free survival but not overall survival within PFB [14]. Our current results in a much larger cohort suggest that there may be only limited utility to 1q gain as a prognostic marker specifically for selection of patients who would benefit from radiation sparing strategies, and this requires further evaluation in prospective cohorts. However, the current results are consistent with our previous observations that the overall survival is not significantly different in PFB patients harboring 1q-gained tumors. The relatively high hazard ratio for 1q gain suggests that its role as a prognostic marker may warrant further evaluation. However, for the next generation of clinical trials within posterior fossa ependymoma, 1q should be used for stratification only within PFA, and highlights the importance of molecularly informed studies.
We show, for the first time, that identification of chromosome 13q loss may represent a more reliable prognostic marker across PFB ependymoma than 1q gain. Indeed, when stratifying chromosome 13q loss by extent of resection, gross totally resected PFB ependymoma with 13q loss appears to still have a poor outcome. The absence of other large cohorts of PFB ependymoma precludes its independent validation; however, it does warrant further study particularly in the context of ongoing and upcoming prospective cooperative group studies.
Most importantly, however, the identification of multiple subtypes of PFB, analogous to recent work across PFA ependymoma, shows clearly that there exists a significant biological heterogeneity within this group [13]. The current cohort is significantly limited by a lack of sufficient gene expression data, but the available data support the heterogeneity within PFB as determined by methylation profiling. Prospective collection of frozen tissue particularly in adults with posterior fossa ependymoma will be crucial to determine which specific biological processes underlie the observed subtypes. As such, our identification of five subtypes provides strong support for the prospective collection of tissue in this group for further next-generation integrated analysis. Moreover, the observation that late relapses and deaths are not infrequent in this group highlights a need to identify and validate actionable pathways. Indeed, our identification of multiple subtypes of PFB has profound implications for future targeted therapies, as any “PFB” specific therapies may be restricted to specific subtypes. This is especially important in the context of the observed subtype-specific copy-number aberrations which suggest that PFB tumors do not emerge from a single driving event. As of today, zero recurrent mutations have been found in PFB ependymoma and a clear driver for these tumors is still lacking [10]. By focusing on specific PFB subtypes and by making use of additional molecular data like large gene expression profiling and more comprehensive next-generation-sequencing enriching for each subtype, future studies may be able to elucidate the underlying events that ultimately lead to the development of these tumors [10].
Further delineating the biology and prognostic markers of PFB ependymoma will require extensive collaboration across adult and pediatric centres, as currently adult ependymoma is an often-neglected entity, with a paucity of frozen tissue available. However, future basic science and translational science efforts on PFB should account for the heterogeneity within PFB ependymoma in the development of more targeted therapeutics.
References
Araki A, Chocholous M, Gojo J, Dorfer C, Czech T, Heinzl H et al (2016) Chromosome 1q gain and tenascin-C expression are candidate markers to define different risk groups in pediatric posterior fossa ependymoma. Acta Neuropathol Commun 4:88. https://doi.org/10.1186/s40478-016-0349-9
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31(737–754):e736. https://doi.org/10.1016/j.ccell.2017.05.005
Gojo J, Lotsch D, Spiegl-Kreinecker S, Pajtler KW, Neumayer K, Korbel P et al (2017) Telomerase activation in posterior fossa group A ependymomas is associated with dismal prognosis and chromosome 1q gain. Neuro Oncol 19:1183–1194. https://doi.org/10.1093/neuonc/nox027
Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R et al (2013) Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913–916. https://doi.org/10.1007/s00401-013-1126-5
Khatua S, Ramaswamy V, Bouffet E (2017) Current therapy and the evolving molecular landscape of paediatric ependymoma. Eur J Cancer 70:34–41. https://doi.org/10.1016/j.ejca.2016.10.013
Kilday JP, Mitra B, Domerg C, Ward J, Andreiuolo F, Osteso-Ibanez T et al (2012) Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children’s Cancer Leukaemia Group (CCLG), Societe Francaise d’Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP). Clin Cancer Res 18:2001–2011. https://doi.org/10.1158/1078-0432.ccr-11-2489
Korshunov A, Witt H, Hielscher T, Benner A, Remke M, Ryzhova M et al (2010) Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol 28:3182–3190. https://doi.org/10.1200/jco.2009.27.3359
Mack SC, Agnihotri S, Bertrand KC, Wang X, Shih DJ, Witt H et al (2015) Spinal myxopapillary ependymomas demonstrate a warburg phenotype. Clin Cancer Res 21:3750–3758. https://doi.org/10.1158/1078-0432.ccr-14-2650
Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang X et al (2018) Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553:101–105. https://doi.org/10.1038/nature25169
Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM et al (2014) Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506:445–450. https://doi.org/10.1038/nature13108
Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A et al (2017) The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol 133:5–12. https://doi.org/10.1007/s00401-016-1643-0
Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B et al (2018) Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. https://doi.org/10.1007/s00401-018-1877-0
Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F et al (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW et al (2016) Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol 34:2468–2477. https://doi.org/10.1200/jco.2015.65.7825
Ramaswamy V, Taylor MD (2016) Treatment implications of posterior fossa ependymoma subgroups. Chin J Cancer 35:93. https://doi.org/10.1186/s40880-016-0155-6
Rand V, Prebble E, Ridley L, Howard M, Wei W, Brundler MA, Fee BE, Riggins GJ, Coyle B, Grundy RG (2008) Investigation of chromosome 1q reveals differential expression of members of the S100 family in clinical subgroups of intracranial paediatric ependymoma. Br J Cancer 99:1136–1143. https://doi.org/10.1038/sj.bjc.6604651
Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J (2016) g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44:W83–W89. https://doi.org/10.1093/nar/gkw199
Ruda R, Reifenberger G, Frappaz D, Pfister SM, Laprie A, Santarius T et al (2018) EANO guidelines for the diagnosis and treatment of ependymal tumors. Neuro Oncol 20:445–456. https://doi.org/10.1093/neuonc/nox166
Ryall S, Guzman M, Elbabaa SK, Luu B, Mack SC, Zapotocky M, Taylor MD, Hawkins C, Ramaswamy V (2017) H3 K27M mutations are extremely rare in posterior fossa group A ependymoma. Childs Nerv Syst 33:1047–1051. https://doi.org/10.1007/s00381-017-3481-3
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. https://doi.org/10.1016/j.ccr.2012.08.024
Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P et al (2005) Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 8:323–335. https://doi.org/10.1016/j.ccr.2005.09.001
Thompson YY, Ramaswamy V, Diamandis P, Daniels C, Taylor MD (2015) Posterior fossa ependymoma: current insights. Childs Nerv Syst 31:1699–1706. https://doi.org/10.1007/s00381-015-2823-2
Ward S, Harding B, Wilkins P, Harkness W, Hayward R, Darling JL, Thomas DG, Warr T (2001) Gain of 1q and loss of 22 are the most common changes detected by comparative genomic hybridisation in paediatric ependymoma. Genes Chromosomes Cancer 32:59–66. https://doi.org/10.1002/gcc.1167
Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R et al (2011) Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20:143–157. https://doi.org/10.1016/j.ccr.2011.07.007
Acknowledgements
VR is supported by operating funds from the Canadian Institutes of Health Research, American Brain Tumor Association, the Garron Family Cancer Center, Meagan’s Walk, b.r.a.i.n.child, the Brain Tumor Foundation of Canada and a Collaborative Ependymoma Research Network (CERN) basic science fellowship. FMGC is supported by the Stephen Buttrum Brain Tumor Research Fellowship, granted by Brain Tumor Foundation of Canada. MZ is supported by Garron Family Cancer Center Fellowship, Meagan’s Walk Neuro-Oncology Fellowship and Restracomp from Research Training Center at The Hospital for Sick Children. MDT is supported by funds from the Garron Family Chair in Childhood Cancer Research at The Hospital for Sick Children and The University of Toronto, and operating funds from the National Institutes of Health (R01CA159859 and R01CA148699), and the Pediatric Brain Tumor Foundation. EB is supported by funds from the Garron Family Chair in Childhood Cancer Research at The Hospital for Sick Children and The University of Toronto. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 to Memorial Sloan Kettering Cancer Center. KWP is supported by a CERN Research Fellowship.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Florence M. G. Cavalli, Jens-Martin Hübner, and Tanvi Sharma are co-first authors.
Marcel Kool and Vijay Ramaswamy are co-senior authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2018_1888_MOESM1_ESM.eps
Supplemental Fig. 1A–F: Spectral clustering of PFB at k = 2–7. Heatmap of the similarity matrix obtained by methylation data on the 212 primary PFB samples. Yellow represents more similar and red represents dissimilar. Color bar at the top represents the final PFB1–5 cluster assignation of the samples (EPS 38612 kb)
401_2018_1888_MOESM2_ESM.eps
Supplemental Fig. 2: Genomic comparison of PFB subtypes. A) Comparison of subtype assignments between spectral clustering and t-SNE analysis. Relationships (indicated by the grey bars between columns) between the groups of samples obtained by t-SNE analysis (3 to 5 clusters) and the groups obtained by spectral clustering (SpC, 2–5 clusters). B) Density plot of overall promoter methylation levels across the five PFB subtypes (EPS 2173 kb)
401_2018_1888_MOESM3_ESM.eps
Supplemental Fig. 3: List of differentially regulated pathways across PFB subtypes. Enrichment of specific pathways within the PFB subtypes using significantly differentially expressed genes (adjusted p < 0.05). No pathway were identified for PFB2 and 3, since those two subtypes had a low number of significantly expressed genes (EPS 1839 kb)
401_2018_1888_MOESM4_ESM.eps
Supplemental Fig. 4: Unsupervised clustering of broad copy-number events across PFB subtypes. A) t-SNE analysis of broad copy-number profiles across 212 PFB ependymomas with corresponding methylation t-SNE subtype assigned. B) Unsupervised consensus clustering of broad copy-number events using negative matrix factorization (NMF) from k = 2 to k = 6 and a plot of the cophenetic coefficient (EPS 31557 kb)
401_2018_1888_MOESM5_ESM.eps
Supplemental Fig. 5: Arm-level cytogenetic events at k = 4 PFB subtypes from t-SNE analysis. A) Frequency and significance of arm-level gains and losses across the 4 PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test). B) Frequency and significance of whole chromosome gain and losses across the four PFB subtypes. Darker bars show significant arm-level events (q value ≤ 0.1, Chi-squared test) (EPS 1472 kb)
401_2018_1888_MOESM6_ESM.eps
Supplemental Fig. 6: Clinical characteristics of PFB subtypes. A) Progression-free and B) overall survival across PFB subtypes. P values determined using the log-rank test. C) Pie chart of the administration of upfront radiation across PFB subtypes and D) Pie chart of the incidence of gross total resection across PFB subtypes. P values determined using the Fisher’s exact test. Overall survival stratified by E) 1q gain and F) 13q status. p values determined using the log-rank test (EPS 2421 kb)
Rights and permissions
About this article
Cite this article
Cavalli, F.M.G., Hübner, JM., Sharma, T. et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol 136, 227–237 (2018). https://doi.org/10.1007/s00401-018-1888-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1888-x