Abstract
Purpose
Global DNA hydroxymethylation is markedly decreased in human cancers, including hepatocellular carcinoma, which is associated with chronic alcohol consumption and aging. Because gene-specific changes in hydroxymethylcytosine may affect gene transcription, giving rise to a carcinogenic environment, we determined genome-wide site-specific changes in hepatic hydroxymethylcytosine that are associated with chronic alcohol consumption and aging.
Methods
Young (4 months) and old (18 months) male C57Bl/6 mice were fed either an ethanol-containing Lieber–DeCarli liquid diet or an isocaloric control diet for 5 weeks. Genomic and gene-specific hydroxymethylcytosine patterns were determined through hydroxymethyl DNA immunoprecipitation array in hepatic DNA.
Results
Hydroxymethylcytosine patterns were more perturbed by alcohol consumption in young mice than in old mice (431 differentially hydroxymethylated regions, DhMRs, in young vs 189 DhMRs in old). A CpG island ~2.5 kb upstream of the glucocorticoid receptor gene, Nr3c1, had increased hydroxymethylation as well as increased mRNA expression (p = 0.015) in young mice fed alcohol relative to the control group. Aging alone also altered hydroxymethylcytosine patterns, with 331 DhMRs, but alcohol attenuated this effect. Aging was associated with a decrease in hydroxymethylcytosine ~1 kb upstream of the leptin receptor gene, Lepr, and decreased transcription of this gene (p = 0.029). Nr3c1 and Lepr are both involved in hepatic lipid homeostasis and hepatosteatosis, which may create a carcinogenic environment.
Conclusions
These results suggest that the location of hydroxymethylcytosine in the genome is site specific and not random, and that changes in hydroxymethylation may play a role in the liver’s response to aging and alcohol.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydroxymethylcytosine has become known as the “sixth base” following methylcytosine as a modified nucleotide found in the mammalian genome [1]. The role of this new epigenetic mark may be twofold. First, hydroxymethylcytosine acts as an intermediate in the removal of methyl groups from cytosine bases, returning the cytosine back to its unmodified form [1, 2]. Additionally, hydroxymethylcytosine may have a role in transcriptional regulation because proteins that bind to the DNA through methylcytosine, the so-called methyl-binding proteins, have less affinity for hydroxymethylcytosine. Because methyl-binding proteins can alter transcription of a gene by binding to the DNA in a gene’s promoter region, hydroxymethylcytosine may also change transcription through this alteration in binding affinity [3, 4]. Hydroxymethylation of DNA in a promoter, CpG island or gene body is associated with an increase in transcription of that gene [5, 6].
The percentage of cytosines that are modified to hydroxymethylcytosine is variable from tissue to tissue, with the most being in the central nervous system [2, 7]. Interestingly, there is a marked decrease in hydroxymethylation in various types of cancer [8]. In the liver, global decreases in hydroxymethylcytosine have been recorded in cases of hepatocellular carcinoma [9–11]. While this research is promising, the genomic locations of which these changes in hydroxymethylcytosine are occurring are currently unknown. Like DNA methylation, gene-specific changes in hydroxymethylation may occur in hepatocellular carcinoma [12].
Chronic alcohol consumption and aging are both risk factors for the development of hepatocellular carcinoma, which is associated with the development of hepatic steatosis and cirrhosis [13, 14]. Additionally, changes in DNA methylation and other epigenetic marks have been identified in alcoholic liver disease, which has been contested as an initial step in the development of hepatocellular carcinoma [15]. It is unknown, however, whether hydroxymethylcytosine may play a role in this stage of liver disease. Both aging and alcohol are known to alter DNA methylation and may also affect DNA hydroxymethylation patterns [16, 17]. We have previously found global fluxes in DNA hydroxymethylation in rodents exposed to a chronic alcohol diet [18]. More specifically, we found that global hepatic DNA hydroxymethylation is significantly decreased in young mice fed an alcohol-containing diet when compared to their young control counterparts, and that aging alone also decreases global DNA hydroxymethylation. This previous study, however, was unable to determine the locations of these changes within the genome, a limitation because alteration in genome-wide hydroxymethylation patterns may alter transcription of specific genes, having the downstream consequence of predisposing the cell to carcinogenesis or other metabolic changes.
Using a microarray and bioinformatic analysis, we assessed genome-wide and gene-specific changes in hepatic hydroxymethylcytosine associated with aging and chronic alcohol consumption in hepatic DNA of a mouse model.
Materials and methods
Animal study and diets
This study was reviewed and approved by the Animal Care and Use Committee of the USDA Human Nutrition Research Center on Aging at Tufts University. The manufacture of this array recommends a minimum of three subjects per study group. To our experience, three animals per group are too small of a sample size to compensate for epigenetic variability; therefore, we chose to use four animals per study group. Eight young (4 months old) and eight old (18 months old) male C57Bl/6 mice (Charles River Laboratories, Wilmington MA) were fed a l-amino acid defined Lieber–DeCarli control liquid diet for 3 weeks [19, 20] (Dyets, Bethleham, PA, 210011). Mice were then randomized to continue either the control diet or an isocaloric diet containing ethanol for 5 weeks [21]. Ethanol was included in the diet at a concentration of 3.1 % (v/v), comprising 18 % of total energy, and was introduced over an adaption period of 2 weeks. Maltodextrin was added to the control diet to equal the number of calories delivered from ethanol in the alcohol diet.
Individually housed mice were group pair-fed to decrease variability of energy and nutrient intake within and between dietary groups. The amount of food supplied to each diet group was matched to the mean daily food consumption of the group with the lowest food consumption. Therefore, all mice in each age group were offered the same volume of liquid daily and adjusted when necessary to ensure all mice consumed equal volumes. After 5 weeks of feeding, mice were killed and liver samples were harvested as previously described [22].
Preparation of samples
To determine which locations in the genome contain differentially hydroxymethylated regions in aging or alcohol consumption, a microarray method was conducted. Genomic DNA was extracted via the standard phenol/chloroform/isoamyl alcohol [25:24:1 (v/v/v)] (Invitrogen Life Technologies, Grand Island, NY) method with precipitation by 100 % ethanol and 3 mol/L sodium acetate, pH 5.2. DNA was re-dissolved in Tris–EDTA buffer, the quality was assessed on an Agarose gel, and both 260/280 and 260/230 ratios were measured on a Nanodrop 1000 (Thermo Scientific, Wilmington DE). All samples used in this study had a 260/280 and a 260/230 ratio equal to or greater than 1.8. DNA was then randomly fragmented by sonication into 200- to 1000-bp fragments, which was verified on an Agarose gel. DNA was divided into two aliquots per sample, and one aliquot underwent immunoprecipitation with an antibody (Diagenode, Denville NJ, AF-110-0016) against hydroxymethylated cytosine (hmeDIP), resulting in samples enriched with hydroxymethylcytosine [3]. The immunoprecipitation was verified by qRT-PCR using internal positive and negative controls provided. Non-hmeDIP (input) and hmeDIP DNAs were then amplified using a whole genome amplification kit as described by the manufacturer (Sigma-Aldrich, St. Louis, MO, WGA2).
Hydroxymethyl-enriched samples were fluorescently labeled (Cy5) and competitively hybridized with fluorescently labeled (Cy3) (Roche, Madison WI, 6370250001 & 5583683001) input samples to a high-throughput 3 × 720 k NimbleGen array containing CpG islands and all annotated gene promoters in the mouse genome (Roche, Madison WI, 5924537001). Signals were detected with the NimbleGen microarray scanner MS 200 (Roche, Madison WI).
Microarray quality control and finding differentially hydroxymethylated regions
A modified version of the comprehensive high-throughput array for relative methylation (CHARM) method in R Studio was used to analyze our hmeDIP DNA methylation arrays for all four age and diet contrasts [23]. The original CHARM code was written for samples that underwent methylation-dependent enzyme restriction, yielding samples that are depleted of methylcytosine instead of enriched in methylcytosines. We modified the CHARM R code in R Studio to invert the M ratio to hmeDIP/input and to eliminate the percent methylation calculation to apply CHARM to our hmeDIP-generated DNA methylation arrays. The CHARM package is a freely available open software program from Bioconductor (http://www.bioconductor.org/packages/release/bioc/html/charm.html).
Quality control of DNA hydroxymethylation array
Each slide used in this study contained three microarrays, which increases the possibility that samples will resemble each other if they were on the same slide resulting in slide bias. Principal component analysis (PCA) was used to evaluate the variability contributing to the dataset by each sample following CHARM normalization. Because the three samples on the same slides did not cluster together, we can be confident that there was not an issue of slide bias.
Annotation of DhMRs and bioinformatic analysis
Upon generation of DhMRs for each contrast of interest, regions were annotated to the mouse genome using the freely available genomic regions enrichment of annotations tool (GREAT) [24]. This analysis annotates chromosomal regions back to the UCSC mm9 mouse genome and associates them with the nearest gene [25]. For this study, regions that were 5.0 kb upstream or 1.0 kb downstream of a gene’s transcription start site were mapped to that gene, regardless of other nearby genes. Additionally, curated regulatory domains that have demonstrated experimental evidence of directly regulating a gene that otherwise would fall outside of the above-stated region are included in the mapping process.
Bioinformatic analysis of the gene lists generated in GREAT was conducted using Ingenuity Pathway Analysis, IPA (Ingenuity® Systems, http://www.ingenuity.com). IPA maps each gene to a biological relationship and using published literature classifies these genes into categories of biological function, molecular networks, and canonical pathways. For the network analysis, which finds enriched biological networks in a gene set, a Fisher’s exact test is conducted, which calculates a p value to determine the significance of the enriched network found in the gene list, resulting in a Network score:
For this study, networks were generated using the published literature regarding hepatic mouse data only, and biological networks that contained fewer than five genes were filtered out.
From these networks, the expression of specific genes of interest was determined. Genes of interest were those with a low permutation p value and that were identified to be biologically interesting within the physiological context of aging and alcohol consumption. Furthermore, genes that are transcriptionally regulated by DNA methylation were considered.
Quantification of gene expression
An RNA extraction was conducted using TRIzol® reagent (Invitrogen, Grand Island, NY), and cDNA was synthesized through a standard reverse transcriptase kit as described by the manufacturer (Invitrogen Life Technologies, Grand Island, NY). Quantitative RT-PCR was conducted on the Roche LightCycler® 480 real-time PCR machine (Roche, Madison WI) using TaqMan Fast Advanced Master Mix (Invitrogen, Grand Island, NY) and TaqMan primer assays (Invitrogen Life Technologies, Grand Island, NY). Gene expression was calculated as ΔCt, following normalization to the housekeeping gene GAPDH (ΔCt = Ct GeneX − Ct GAPDH). All expression results are displayed as fold difference (FD = 2−ΔΔCt) relative to the control group for each comparison.
Differences in gene expression were determined using a t test, which was performed in SAS version 9.3 (SAS Institute Inc., Cary NC). The level of significance was p < 0.05 for gene expression analyses.
Results
Genome-wide DNA hydroxymethylation
Four contrasts or group comparisons were performed in this study: Young versus Young + Alcohol, Old versus Old + Alcohol, Young versus Old, and Young + Alcohol versus Old + Alcohol. All probes from the microarray that had a difference in log 2 ratios >0.5 and a t test p values <0.01 were used to generate a heat map. From the clustering of these heat maps, we conclude that samples within the same experimental group had similar hydroxymethylation patterns at specific sites throughout the genome (Fig. 1). Only heat maps from the Young versus Young + Alcohol, and the Young versus Old contrasts are shown. Further analysis identified these groups as having the most differences and the focus of subsequent data interpretations. The heat maps also confer that hydroxymethylation of DNA is occurring at specific sites of DNA, rather than randomly.
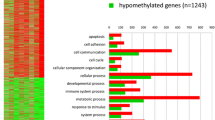
Following per-probe quality control techniques, probes were lumped into differentially hydroxymethylated regions, DhMRs. Genomically neighboring probes are lumped into a DhMR if they both had a p value of <0.01 when calculating the difference in hydroxymethylation of that probe between two groups. The number of DhMRs for each group comparison gives insight into the degree of changes in hydroxymethylation that occurred in the DNA. Overall, comparing hydroxymethylation in the Young group to the Young + Alcohol group gave the highest number of differentially hydroxymethylated regions. This is particularly interesting when compared to the Old versus Old + Alcohol contrast, which had far less DhMRs (431 vs 189), signifying that hydroxymethylation of specific regions in the DNA of young mice is more easily changed by chronic alcohol consumption than in old mice, or that old mice have more variable hydroxymethylation patterns. There are also a large number of regions that had altered hydroxymethylation when comparing the DNA from young mice to old (331 DhMRs). This suggests that aging alone, as compared to aging with alcohol consumption (196 DhMRs), seems to alter hydroxymethylation patterns to a larger degree. Most DhMRs found in this study were near a transcription start site of a gene (Fig. 2), leading us to speculate that hydroxymethylcytosine may affect gene transcription.
The contrasts of Young to Young + Alcohol, and Young to Old yielded the most DhMRs with greater differences in hydroxymethylcytosine compared to the DhMRs found in the Old versus Old + Alcohol and the Young + Alcohol versus Old + Alcohol contrasts. For these reasons, the Young versus Young + Alcohol and the Young versus Old contrasts are the focus of subsequent analysis presented here.
Gene-specific changes in DNA hydroxymethylation
After the determination of DhMRs, each region was mapped back to the mouse genome. Some regions were between multiple genes; therefore, we included all of these genes in our downstream network analysis. Gene-specific changes in hydroxymethylation for each contrast were analyzed using a network analysis in IPA (Ingenuity® Systems; http://www.ingenuity.com). From these networks, we can determine biologically relevant gene systems that may be affected by either alcohol consumption or aging.
Gene-specific changes in hydroxymethylcytosine associated with alcohol consumption in young mice
Conducting a network analysis in IPA in the Young and the Young + Alcohol groups resulted in two networks with the biological functions of: (1) lipid metabolism, molecular transport, small molecule biochemistry; and (2) cell death and survival, liver necrosis/cell death, cellular development (Table 1). Within these networks, alcohol seemed to increase DNA hydroxymethylation of these genes, with a few exceptions. This may be due to the increased oxidative environment caused by exposure to alcohol, or it may be a programmed biological response.
The gene nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (Nr3c1; permutation p = 0.055) is a central node in network 1 (Fig. 3). The glucocorticoid receptor is involved in cellular differentiation and acts as a transcription factor that can bind to glucocorticoid response elements to activate transcription [26]. Both of these genes are well conserved among mammalian species and have been previously demonstrated as being epigenetically regulated [27, 28].
Network of genes involved in lipid metabolism that contained a DhMR in the Young versus Young + Alcohol contrast. Red demonstrates genes that were more hydroxymethylated in the Young group, and green demonstrates genes that were more hydroxymethylated in the Young + Alcohol group. White demonstrates a gene within the biological network that did not contain a DhMR. Solid line represents a direct relationship between genes; dashed line represents an indirect relationship between genes
The region of the glucocorticoid receptor gene, in which our DhMR falls, is at a CpG island roughly 2.5 kb upstream of the gene’s transcription start site and has increased hydroxymethylation in the Young + Alcohol group relative to the Young. This gene also had significantly increased mRNA expression in the Young + Alcohol group relative to the Young (p = 0.015; Fig. 4). The increased hydroxymethylation of the CpG islands in the Young + Alcohol group may be altering the binding of proteins in that region, thereby increasing transcription.
Gene-specific changes in hydroxymethylcytosine associated with aging
Overall, three biological networks encompassing two biological functions were enriched in our dataset (two of these networks of genes are involved in lipid metabolism, but are separated for easier viewing) (Table 2). This suggests that aging may have an effect on DNA hydroxymethylation of genes involved in lipid metabolism in hepatic DNA even without the consumption of alcohol.
Leptin is an important peptide hormone involved in feeding patterns and energy expenditure. Decreases in either leptin, or the leptin receptor, are associated with obesity [29–31]. A region of DNA ~1 kb upstream of the leptin receptor gene (Lepr) was found to have decreased hydroxymethylation in old mice relative to young (permutation p = 0.044; Fig. 5). Old mice also had a significant decrease in the expression of the leptin receptor relative to the young mice (p = 0.029; Fig. 6).
Network of genes involved in protein synthesis and hepatic system development that contained a DhMR in the Young versus Old contrast. Red demonstrates genes that were more hydroxymethylated in the Young group, and green demonstrates genes that were more hydroxymethylated in the Old group. White demonstrates a gene within the biological network that did not contain a DhMR. Solid line represents a direct relationship between genes, and dashed line represents an indirect relationship between genes
Discussion
Through the analysis of genome-wide and gene-specific hydroxymethylcytosine patterns, we can better understand the degree and directionality of changes occurring in the hepatic hydroxymethylome that are associated with aging and chronic alcohol consumption. Chronic alcohol consumption and aging are both associated with increased risk of hepatocellular carcinoma. The genomic and gene-specific alterations in hydroxymethylcytosine patterns presented here may shed light on the early stages of carcinogenesis as well as metabolic shifts that occur with the aging the process and the dysregulation caused by alcohol.
By comparing the number of regions that are differentially hydroxymethylated between experimental groups, we conclude that alcohol has a larger effect on hydroxymethylation in young mice relative to old. The larger effect on young mice is similar to our previous DNA methylation study [21] and suggests that the response to alcohol in young mice is stronger than in old mice. This may create an environment in young mice that is either protective against or prone to the alcoholic liver injury, but the clinical significance needs to be clarified in future studies. Aging alone is associated with large genome-wide alterations in hydroxymethylcytosine. Furthermore, consumption of alcohol may mask the effects of aging on the hydroxymethylome. These changes largely occur within 5 kb up or downstream of the transcription start site of genes, which suggests that those hydroxymethylcytosine changes (DhMRs) are genomically near the transcription starting sites, which are heavily regulated regions yielding functionally relevant changes in gene expression.
To our knowledge, this is the first report of gene-specific changes in hydroxymethylation of hepatic DNA that are associated with aging and chronic alcohol consumption. Our finding that chronic alcohol consumption in young mice increases hydroxymethylation at a CpG island in the promoter of the glucocorticoid receptor gene, Nr3c1, and is associated with increased levels of glucocorticoid receptor mRNA is novel. Glucocorticoid receptor signaling is metabolically important in the liver and may play a role in hepatic lipogenesis and steatosis, both of which are increased during chronic alcohol consumption [32, 33]. Glucocorticoids are involved in array of physiological pathways, from glucose homeostasis to apoptosis to the stress response [26, 34]. The activity of the glucocorticoid receptor is tissue specific. For example, increased lymphocyte glucocorticoid receptor expression has been associated with anxiety and post-traumatic stress disorder [35], while decreased glucocorticoid receptor expression in the brain is associated with helplessness and depression [36, 37]. In other tissues, the methylation of the glucocorticoid receptor gene, Nr3c1, seems to be sensitive to external environmental exposures such as stress and the diet [38, 39] and can be passed onto future generations [40, 41]. Glucocorticoid receptor-mediated apoptosis is a mechanism by which some cancer treatments suppress growth of cells in blood cancers [42]. Importantly, the glucocorticoid signaling pathway has previously been implicated in the development of hepatic steatosis and hepatic lipogenesis [33, 43], both of which are associated with alcohol intake [32]. Therefore, the changes in hydroxymethylcytosine and mRNA expression we report here may be an important step in the liver’s response to chronic alcohol consumption. Moreover, because we only see changes in Nr3c1 hydroxymethylation and expression when alcohol is consumed by the young mice, it is possible that the older mice have an impaired cellular response to chronic alcohol consumption.
We report that aging is associated with a decrease in hydroxymethylcytosine at a region located ~1 kb upstream of the transcription start site of the leptin receptor gene, Lepr, and is also associated with lowered levels of Lepr mRNA. Previous studies have reported a decline in leptin hormone associated with aging, although this is interrupted by increased fat mass [44, 45]. An age-associated change in Lepr transcription has not been previously reported. In addition to its association with obesity, rodents with a deficiency in leptin or the leptin receptor have a tendency to develop hepatic steatosis [46, 47]. Conversely, leptin is a mediator of hepatic fibrosis, and patients with steatohepatitis have elevated serum leptin levels [48, 49]. These findings indicate that the leptin signaling pathway is important in hepatic lipid homeostasis and fibrosis and may be important in carcinogenesis. Through a series of studies, we found that alcohol and aging, cancer risk factors, can alter DNA hydroxymethylation. Our earlier studies found that alcohol and aging can alter global DNA hydroxymethylation in mice [18]. We have also found global hydroxymethylation changes in alcohol-associated hepatocellular carcinoma in humans [11]. This study aimed to reveal how altered genome-wide and region-specific patterns of DNA hydroxymethylation may be an initial step in the development of alcohol-associated liver cancer. Taken together, the results presented here provide novel insight into the epigenetic ramifications of aging and chronic alcohol consumption. These findings demonstrate that hydroxymethylcytosine is site specific and may regulate transcription of genes. These early investigations identify associations that could be further followed up with functional assays and longer-term hepatic cancer studies. Future research may validate these findings and determine whether changes in protein–DNA binding are occurring at the genomic locations of these DhMRs.
Abbreviations
- DhMR:
-
Differentially hydroxymethylated region
- CpG:
-
Cytosine guanine
- IPA:
-
Ingenuity pathway analysis
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- hmeDIP:
-
Hydroxymethyl DNA immunoprecipitation
- CHARM:
-
Comprehensive high-throughput array for relative methylation
- PCA:
-
Principal component analysis
- GREAT:
-
Genomic regions enrichment of annotations tool
References
Münzel M, Globisch D, Carell T (2011) 5-Hydroxymethylcytosine, the sixth base of the genome. Angew Chem Int Ed 50(29):6460–6468
Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5(12):e15367
Jin SG, Kadam S, Pfeifer GP (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38(11):e125
Valinluck V, Tsai H-H, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32(14):4100–4108
Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473(7347):398–402
Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25(7):679–684
Li W, Liu M (2011) Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011:870726
Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP (2011) 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71(24):7360–7365
Chen ML, Shen F, Huang W, Qi JH, Wang Y, Feng YQ, Liu SM, Yuan BF (2013) Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole time-of-flight mass spectrometry. Clin Chem 59(5):824–832
Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, Yang Z, Wu L, Xia F, Bie P (2013) Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS ONE 8(5):e62828
Udali S, Guarini P, Moruzzi S, Ruzzenente A, Tammen SA, Guglielmi A, Conci S, Pattini P, Olivieri O, Corrocher R, Choi SW, Friso S (2015) Global DNA methylation and hydroxymethylation differ in hepatocellular carcinoma and cholangiocarcinoma and relate to survival rate. Hepatology 62(2):496–504
Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A (2008) Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 47(3):908–918
Morgan TR, Mandayam S, Jamal MM (2004) Alcohol and hepatocellular carcinoma. Gastroenterology 127(5):S87–S96
El–Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132(7):2557–2576
Mandrekar P (2011) Epigenetic regulation in alcoholic liver disease. World J Gastroenterol 17(20):2456–2464
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5(8):e1000602
Davis CD, Uthus EO (2004) DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med 229(10):988–995
Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Sauer J, Friso S, Choi SW (2014) Aging and alcohol interact to alter hepatic DNA hydroxymethylation. Alcohol Clin Exp Res 38(8):2178–2185
DeCarli LM, Lieber CS (1967) Fatty liver in the rat after prolonged intake of ethanol with a nutritionally adequate new liquid diet. J Nutr 91(3 Suppl):331–336
Lieber CS, DeCarli LM, Sorrell MF (1989) Experimental methods of ethanol administration. Hepatology 10(4):501–510
Sauer J, Jang H, Zimmerly EM, Kim KC, Liu Z, Chanson A, Smith DE, Mason JB, Friso S, Choi SW (2010) Ageing, chronic alcohol consumption and folate are determinants of genomic DNA methylation, p16 promoter methylation and the expression of p16 in the mouse colon. Br J Nutr 104(1):24–30
Choi SW, Friso S, Dolnikowski GG, Bagley PJ, Edmondson AN, Smith DE, Mason JB (2003) Biochemical and molecular aberrations in the rat colon due to folate depletion are age-specific. J Nutr 133(4):1206–1212
Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP (2008) Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18(5):780–790
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28(5):495–501
Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A (2011) The UCSC genome browser database: update 2011. Nucleic Acids Res 39(suppl 1):D876–D882
Yudt MR, Cidlowski JA (2002) The glucocorticoid receptor: coding a diversity of proteins and responses through a single gene. Mol Endocrinol 16(8):1719–1726
Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, Tymowska-Lalanne Z, Plagge A, Powles-Glover N, Kelsey G (2006) Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38(3):350–355
Suderman M, McGowan PO, Sasaki A, Huang TC, Hallett MT, Meaney MJ, Turecki G, Szyf M (2012) Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci 109(Supplement 2):17266–17272
Clément K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte J-M (1998) A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 392(6674):398–401
Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM (1995) Weight-reducing effects of the plasma protein encoded by the obese gene. Science 269(5223):543–546
Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, Collins F (1995) Effects of the obese gene product on body weight regulation in ob/ob mice. Science 269(5223):540–543
Lieber CS (2004) Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol 34(1):9–19
Mueller KM, Kornfeld JW, Friedbichler K, Blaas L, Egger G, Esterbauer H, Hasselblatt P, Schlederer M, Haindl S, Wagner KU (2011) Impairment of hepatic growth hormone and glucocorticoid receptor signaling causes steatosis and hepatocellular carcinoma in mice. Hepatology 54(4):1398–1409
Mueller KM, Themanns M, Friedbichler K, Kornfeld J-W, Esterbauer H, Tuckermann JP, Moriggl R (2012) Hepatic growth hormone and glucocorticoid receptor signaling in body growth, steatosis and metabolic liver cancer development. Mol Cell Endocrinol 361(1–2):1–11
Shaffer D, Giller EL Jr (1991) Lymphocyte glucocorticoid receptor number in posttraumatic stress disorder. Am J Psychiatry 148(4):499–504
Ridder S, Chourbaji S, Hellweg R, Urani A, Zacher C, Schmid W, Zink M, Hörtnagl H, Flor H, Henn FA (2005) Mice with genetically altered glucocorticoid receptor expression show altered sensitivity for stress-induced depressive reactions. J Neurosci 25(26):6243–6250
McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ (2009) Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12(3):342–348
de Rooij SR, Costello PM, Veenendaal MV, Lillycrop KA, Gluckman PD, Hanson MA, Painter RC, Roseboom TJ (2011) Associations between DNA methylation of a glucocorticoid receptor promoter and acute stress responses in a large healthy adult population are largely explained by lifestyle and educational differences. Psychoneuroendocrinology 37(6):782–788
Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC (2007) Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 97(6):1064–1073
Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM (2008) Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 3(2):97–106
Radtke K, Ruf M, Gunter H, Dohrmann K, Schauer M, Meyer A, Elbert T (2011) Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatry 1(7):e21
Schlossmacher G, Stevens A, White A (2011) Glucocorticoid receptor-mediated apoptosis: mechanisms of resistance in cancer cells. J Endocrinol 211(1):17–25
Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA (2004) Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology 40(6):1322–1332
Gabriely I, Ma XH, Yang XM, Rossetti L, Barzilai N (2002) Leptin resistance during aging is independent of fat mass. Diabetes 51(4):1016–1021
Wang ZW, Pan WT, Lee Y, Kakuma T, Zhou YT, Unger RH (2001) The role of leptin resistance in the lipid abnormalities of aging. FASEB J 15(1):108–114
Diehl AM (2005) Lessons from animal models of NASH. Hepatol Res 33(2):138–144
Sahai A, Malladi P, Pan X, Paul R, Melin-Aldana H, Green RM, Whitington PF (2004) Obese and diabetic db/db mice develop marked liver fibrosis in a model of nonalcoholic steatohepatitis: role of short-form leptin receptors and osteopontin. Am J Physiol-Gastrointest Liver Physiol 287(5):G1035–G1043
Uygun A, Kadayifci A, Yesilova Z, Erdil A, Yaman H, Saka M, Deveci MS, Bagci S, Gulsen M, Karaeren N (2000) Serum leptin levels in patients with nonalcoholic steatohepatitis. Am J Gastroenterol 95(12):3584–3589
Leclercq IA, Farrell GC, Schriemer R, Robertson GR (2002) Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J Hepatol 37(2):206–213
Acknowledgments
This work is based upon work supported by the U.S. Department of Agriculture–Agricultural Research Service (ARS) under Agreement No. 58-1950-0-014. Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Dept of Agriculture. This work is also supported in part by the Korean Food Research Institute (S.W.C.). All authors participated in the design of the research project. S.A.T. and L.K.P. adapted the microarray methods and computational analysis for use in this study. S.A.T. and S.W.C. conducted the research and drafted manuscript. G.G.D., L.M.A., and S.F. designed the research, interpreted the results of experiment, and revised manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors do not have any conflicts of interest.
Rights and permissions
About this article
Cite this article
Tammen, S.A., Park, L.K., Dolnikowski, G.G. et al. Hepatic DNA hydroxymethylation is site-specifically altered by chronic alcohol consumption and aging. Eur J Nutr 56, 535–544 (2017). https://doi.org/10.1007/s00394-015-1098-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-015-1098-4