Abstract
Objective
To examine whether alcohol intake is causally associated with rheumatoid arthritis (RA).
Methods
We performed a two-sample Mendelian randomization (MR) analysis using the inverse-variance weighted (IVW), weighted median, and MR-Egger regression methods. We used the publicly available summary statistics of alcohol intake frequency from the UK Biobank genome-wide association studies (GWASs; n = 336,965) as the exposure and a GWAS meta-analysis of 5539 autoantibody-positive RA patients and 20,169 controls as the outcome.
Results
We selected 24 single nucleotide polymorphisms (SNPs) associated with alcohol intake frequency at genome-wide significance as instrumental variables (IVs) to improve inference, 16 of which were inversely associated with RA. The IVW method showed no evidence of a causal association between alcohol intake and RA (beta = 0.218, SE = 0.213, p = 0.306). The MR-Egger regression revealed that directional pleiotropy was unlikely to bias the result (intercept = 0.027, p = 0.292). The MR-Egger analysis and the weighted median approach showed no causal association between alcohol intake and RA (beta = −0.778, SE = 0.947, p = 0.420 and beta = −0.286, SE = 0.302, p = 0.344, respectively). Cochran’s Q test did not indicate heterogeneity between IV estimates based on the individual variants, and results from a “leave-one-out” analysis demonstrated that no single SNP was driving the IVW point estimate.
Conclusion
The MR analysis does not support a causal inverse association between alcohol intake and RA occurrence.
Zusammenfassung
Ziel
In der vorliegenden Studie wurde untersucht, ob Alkoholkonsum kausal mit der rheumatoiden Arthritis (RA) zusammenhängt.
Methoden
Es wurde eine Zwei-Stichproben-Mendel-Randomisierungs(MR)-Analyse mit Inverse-Varianz-Gewichtung (IVG), gewichtetem Median und MR-Egger-Regression durchgeführt. Dafür herangezogen wurden die öffentlich zugänglichen statistischen Kennzahlen zur Häufigkeit des Alkoholkonsums aus den genomweiten Assoziationsstudien (GWAS) der UK Biobank (n = 336.965) für die Exposition sowie eine GWAS-Metaanalyse von 5539 Autoantikörper-positiven Patienten mit RA und 20.169 Kontrollpersonen für das Outcome.
Ergebnisse
Insgesamt 24 Einzelnukleotidpolymorphismen (SNP), die mit genomweiter Signifikanz mit der Häufigkeit des Alkoholkonsums assoziiert waren, wurden als Instrumentvariablen (IV) ausgewählt, um bessere Schlussfolgerungen zu ermöglichen. Von diesen waren 16 invers mit RA assoziiert. Die IVG-Methode ergab keinen Hinweis auf einen kausalen Zusammenhang zwischen Alkoholkonsum und RA (beta = 0,218, SE = 0,213, p = 0,306). Die MR-Egger-Regression zeigte, dass eine Verzerrung des Ergebnisses durch eine gerichtete Pleiotropie unwahrscheinlich war (Achsenabschnitt =0,027, p = 0,292). Die MR-Egger-Analyse und der Ansatz mit gewichtetem Median ergaben keinen kausalen Zusammenhang zwischen Alkoholkonsum und RA (beta = −0,778, SE = 0,947, p = 0,420 bzw. beta = −0,286, SE = 0,302, p = 0,344). Der Cochran-Q-Test wies nicht auf eine Heterogenität zwischen IV-Schätzungen auf Grundlage der individuellen Varianten hin, und eine Leave-one-out-Analyse zeigte, dass nicht ein einzelner SNP die IVG-Punktschätzung bestimmte.
Schlussfolgerung
Die MR-Analyse stützt eine kausale inverse Assoziation zwischen Alkoholkonsum und dem Auftreten einer RA nicht.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic synovial joint inflammation that leads to disability and a decreased quality of life. Although the etiology of RA is not fully understood, environmental factors likely play an important role in the development of RA in genetically susceptible individuals [9, 18].
Many environmental factors have been suspected of inducing RA, but their role remains poorly understood. Among these, alcohol intake has been identified as a preventive factor, but the findings regarding its effect on RA development have been inconsistent [15, 22]. Alcohol contains components such as ethanol and antioxidants, which suppress the immune response and decrease the synthesis of proinflammatory cytokines, such as tumor necrosis factor, interleukin-6 (IL-6), and IL-8 [28]. Previous meta-analyses of observational studies have shown that alcohol consumption was inversely associated with the risk of RA [15, 22]. However, observational studies are prone to bias, such as reverse causation and residual confounding, thereby precluding a clear understanding of the effect of alcohol intake on RA [14, 19].
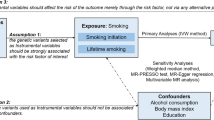
Mendelian randomization (MR) is a technique that uses genetic variants as instrumental variables (IVs) to assess whether an observational association between a risk factor and an outcome is consistent with a causal effect [6]. A two-sample MR estimates causal effects where data on the exposure and outcome have been measured in different samples [17]. This approach is very useful in situations where it is difficult to measure the exposure and outcome in the same set of individuals [17]. No previous study has used the MR approach to test the causal effect of alcohol intake on the risk of RA. Thus, the aim of this study was to examine whether alcohol intake is causally associated with occurrence of RA using a two-sample MR analysis.
Materials and methods
Data sources and selection of genetic variants
We searched the MR Base database (http://www.mrbase.org/), which houses a large collection of summary statistic data from hundreds of genome-wide association studies (GWASs). We used the publicly available summary statistics datasets of GWASs for alcohol intake frequency (increase) from the 500,000 individuals included in the UK Biobank (n = 336,965; https://github.com/Nealelab/UK_Biobank_GWAS) as the exposure. A two-sample MR study of genetic variants associated with alcohol intake was used as the IV to improve inference based on linkage disequilibrium (LD) R2 of 0.001, clumping distance of 10,000 kb, and p-value threshold of 5.00E-08 (genome-wide significance). We obtained summary statistics (beta coefficients and standard errors) for 24 single-nucleotide polymorphisms (SNPs) associated with alcohol intake frequency as the IVs from GWASs from the UK Biobank. We used a GWAS meta-analysis of 5539 autoantibody-positive individuals with RA and 20,169 controls of European descent [25] as the outcome.
Statistical analysis for Mendelian randomization
MR analysis requires genetic variants to be related to, but not potential confounders of an exposure [5]. First, we assessed the independent association of SNPs with alcohol intake frequency. Second, we examined the association between each SNP and risk of RA. Third, we combined these findings to estimate the uncompounded causal association between alcohol intake frequency and risk of RA using MR analysis. We performed two-sample MR, a method used to estimate the causal effect of an exposure (alcohol intake) on outcomes (RA) using summary statistics from different GWASs [11], to assess the causal relationships between alcohol intake frequency and risk of RA, using summary data from alcohol intake frequency and RA GWASs with 24 SNPs as IVs (Table 1).
The IVW method uses a meta-analysis approach to combine the Wald ratio estimates of the causal effect obtained from different SNPs and provide a consistent estimate of the causal effect of the exposure on the outcome when each of the genetic variants satisfies the assumptions of an instrumental variable [21]. Although the inclusion of multiple variants in an MR analysis results in increased statistical power, it has the potential to include pleiotropic genetic variants that are not valid IVs [11]. To explore and adjust for pleiotropy, i. e., the association of genetic variants with more than one variable, the weighted median and MR-Egger regression methods were utilized. MR-Egger regression analysis, which is robust to invalid instruments, tests and accounts for the presence of unbalanced pleiotropy by introducing a parameter for this bias by incorporating summary data estimates of causal effects from multiple individual variants [1]. MR-Egger applies a weighted linear regression of the gene-outcome coefficients on the gene-exposure coefficients [1]. The slope of this regression represents the causal effect estimate, and the intercept can be interpreted as an estimate of the average horizontal pleiotropic effect across the genetic variants [8]. The weighted median estimator provides a consistent estimate of the causal effect, even when up to 50% of the information contributing to the analysis comes from genetic variants that are invalid IVs [2]. The weighted median estimator has the advantage of retaining greater precision in the estimates compared to the MR-Egger analysis [2]. Tests were considered statistically significant at p < 0.05. All MR analyses were performed in the MR Base platform (App version: 1.2.1 e646be (27 June 2018), R version: 3.5.0; ESM 1, Supplementary data: Analysis R code for two-sample MR.; [12]).
Heterogeneity and sensitivity test
We assessed heterogeneities between SNPs using Cochran’s Q‑statistics [10] and I2 statistic [3, 13]. We also performed a “leave-one-out” analysis to investigate the possibility that the causal association was driven by a single SNP.
Results
Studies included in the meta-analysis
Instrumental variables for Mendelian randomization
We selected 24 independent SNPs from alcohol intake GWASs as the IVs. All of them are associated with alcohol intake frequency at genome-wide significance (Table 1; Fig. 1). Sixteen of the 24 SNPs were inversely associated with RA, and the association with rs2159935 was statistically significant (Table 1; ESM 2, Supplementary data: Original and harmonized datasets). In all, 0.5% of variance in the exposure (value of R2 statistic) was explained by the genetic variants serving as IVs.
Mendelian randomization results
The IVW method found no evidence to support a causal association between alcohol intake and RA (beta = 0.218, SE = 0.213, p = 0.306; Table 2; Figs. 1 and 2). The intercept represents the average pleiotropic effect across the genetic variants, i. e., the average direct effect of a variant with the outcome. If the intercept is not zero (the MR-Egger test), there is evidence of directional pleiotropy. The MR-Egger regression revealed that directional pleiotropy was unlikely to be biasing the result (intercept = 0.027, p = 0.292). The MR-Egger analysis found no causal association between alcohol intake and RA (beta = −0.778, SE = 0.947, p = 0.420; Table 2; Fig. 2). The weighted median approach did not provide evidence of a causal association between alcohol intake and RA either (beta = −0.286, SE = 0.302, p = 0.344; Table 2; Fig. 2). IVW was positive, while the more robust estimates were negative. However, they did not still differ significantly between each other based on the confidence intervals (Table 2).
Scatter plots of genetic associations with alcohol intake against the genetic associations with rheumatoid arthritis (RA). The slopes of each line represent the causal association for each method. The blue line represents the inverse-variance weighted (IVW) estimate, the green line represents the weighted median estimate, and the dark blue line represents the MR-Egger estimate. SNP single nucleotide polymorphism
Heterogeneity and sensitivity test
Cochran’s Q test and the funnel test indicated no evidence of heterogeneity between IV estimates based on the individual variants (Table 2). Heterogeneity is the variability in the causal estimates obtained for each SNP (i. e. how consistent is the causal estimate across all SNPs). Low heterogeneity suggests increased reliability of MR estimates. Our results of I2 values showed low heterogeneity, indicating increased reliability of MR estimates (Table 2). Results from the “leave-one-out” analysis demonstrated that no single SNP was driving the IVW point estimate. The MR estimates generated using IVW, weighted median, and MR-Egger regression analyses were consistent. Therefore, the MR analysis results do not support a causal association between alcohol intake and RA.
Discussion
We used three different estimation methods for the MR analyses: the inverse variance weighting method, the weighted median method, and MR-Egger regression. The MR estimates using all three methods were consistent; they do not support a causal inverse association between alcohol intake and occurrence of RA.
The findings of previous meta-analyses have reported a protective effect of alcohol on RA. An inverse relationship between alcohol intake and RA risk is biologically plausible. Alcohol inhibited the onset of collagen-induced inflammatory arthritis through downregulating leukocyte migration and reducing nuclear factor-κB (NF-κB) activation [16]. Alcohol was also shown to have anti-inflammatory effects in humans through reducing NF-kB-driven inflammatory mediator production by monocytes, which is a key cellular pathway in RA [20].
A meta-analysis demonstrated that low to moderate alcohol consumption may reduce the risk of developing RA in a dose-dependent manner, indicating the existence of a J-shaped nonlinear trend between alcohol consumption and risk of RA [15]. However, individuals with low to moderate alcohol intake have healthier lifestyles compared with complete abstainers, and RA could therefore result from confounding lifestyle factors. Another meta-analysis found that alcohol intake is inversely associated with anti-citrullinated protein antibody (ACPA)-positive RA [22]. However, this result was obtained only from retrospective case–control studies, which are subject to both selection and recall bias; thus, there is a high possibility the finding was affected by bias.
MR minimizes the possibility of bias inherent to observational studies due to residual confounding or reverse causality [23]. However, MR studies are susceptible to bias from pleiotropy (association of genetic variants with more than one variable) [27]. Although the inclusion of multiple variants in MR analysis typically leads to increased statistical power, it also results in the potential inclusion of pleiotropic genetic variants that are not valid IVs [24]. To eliminate pleiotropy, we employed a weighted median estimator, which provides valid estimates even if 50% of the SNPs are not valid instruments [2], and we used MR-Egger regression to provide a test for unbalanced pleiotropy and a causal estimate of the influence of the exposure on the outcome in its presence [1]. The MR-Egger approach showed no evidence of unbalanced pleiotropy, as indicated by the intercept p-value. The MR-Egger analysis results in a loss of precision and power, but our weighted median estimator results were similar to the IVW estimator, providing additional confidence in these associations. Our data do not support a causal association between alcohol intake and RA risk. Previously reported associations between alcohol intake and occurrence of RA may be the result of bias or confounding factors inherent to observational studies, such as misclassification of alcohol consumption, reverse causation, a small number of studies of small sizes, recall bias, and selection bias.
The present study has several limitations. First, our analysis included a relatively small number of SNPs as IVs and might have had limited power to detect an association. The statistical power can be increased, and a more precise causal estimate can be obtained by combining multiple genetic variants together [7]. Second, most genetic variants only have a modest effect on a given exposure, because genetic variants might only explain a very small proportion of variance in a particular exposure [26]. Explaining 0.5% of variance in the exposure is a small value, as also the number of 24 SNPs we used as IVs is quite small. We estimated study power to test whether our study was adequately powered to detect clinically relevant changes in RA risk [4]. The power of the MR analysis had a limited power (0.6). This means that very large numbers of cases are required to detect a causal relationship for the outcome of interest. Third, the RA studies were based on participants of European ancestry. As causality may depend on ethnicity and selection bias, further MR studies involving other populations are needed to account for this. Fourth, only autoantibody-positive individuals with RA were included in this MR analysis. Thus, the causal association between alcohol intake and RA risk should also be explored in autoantibody-negative RA patients. Nevertheless, this meta-analysis has its strengths. Although alcohol has been studied as a potential protective factor for RA, an MR has never been performed. This is the first such study on the causal relationship between alcohol intake and RA.
In conclusion, the MR analysis results do not support a causal association between alcohol intake and RA risk. Epidemiological evidence for an inverse association of alcohol intake with a lower risk of RA does not appear to be causal. However, well-designed epidemiological studies and MR studies using more variants that account for a larger proportion of alcohol consumption are warranted to confirm or rule out causality.
References
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44(2):512–525
Bowden J, Davey Smith G, Haycock PC et al (2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40(4):304–314
Bowden J, Del Greco F, Minelli C et al (2016) Assessing the suitability of summary data for Mendelian randomization analyses using MR-Egger regression: the role of the I^ 2 statistic. Int J Epidemiol 45(6):1961–1974
Brion M-JA, Shakhbazov K, Visscher PM (2012) Calculating statistical power in Mendelian randomization studies. International journal of epidemiology 42(5):1497–1501
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37(7):658–665
Burgess S, Daniel RM, Butterworth AS et al (2014) Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol 44(2):484–495
Burgess S, Dudbridge F, Thompson SG (2016) Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 35(11):1880–1906
Burgess S, Thompson SG (2017) Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 32(5):377–389
Edwards C, Cooper C (2006) Early environmental factors and rheumatoid arthritis. Clinical & Experimental. Immunology 143(1):1–5
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315(7121):1533–1537
Hartwig FP, Davies NM, Hemani G et al (2016) Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol 45(6):1717–1726
Hemani G, Zheng J (2016) Wade K et al MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. Acta Neurochir (Wien) 16:78972 (Dec)
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Cochran WG, Rubin DB (1973) Controlling bias in observational studies: A review. Sankhyā: Indian J Stat Series A:417–446
Jin Z, Xiang C, Cai Q et al (2014) Alcohol consumption as a preventive factor for developing rheumatoid arthritis: a dose-response meta-analysis of prospective studies. Ann Rheum Dis 73(11):1962–1967
Jonsson M, Verdrengh M, Brisslert M et al (2007) Ethanol prevents development of destructive arthritis. Proc Natl Acad Sci 104(1):258–263
Lawlor DA (2016) Commentary: Two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol 45(3):908
Lee YH, Bae S‑C, Choi SJ et al (2012) Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep 39(12):10627–10635
Lee YH, Bae SC, Song GG (2013) Hepatitis B virus (HBV) reactivation in rheumatic patients with hepatitis core antigen (HBV occultcarriers) undergoing antitumor necrosis factor therapy. Clin Exp Rheumatol 31(1):118–121
Mandrekar P, Jeliazkova V, Catalano D et al (2007) Acute alcohol exposure exerts anti-inflammatory effects by inhibiting IκB kinase activity and p65 phosphorylation in human monocytes. J Immunol 178(12):7686–7693
Pierce BL, Burgess S (2013) Efficient design for Mendelian randomization studies: subsample and 2‑sample instrumental variable estimators. Am J Epidemiol 178(7):1177–1184
Scott IC, Tan R, Stahl D et al (2013) The protective effect of alcohol on developing rheumatoid arthritis: a systematic review and meta-analysis. Rheumatology 52(5):856–867
Smith GD, Ebrahim S (2003) ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease?. Int J Epidemiolog 32(1):1–22
Smith GD, Ebrahim S (2004) Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33(1):30–42
Stahl EA, Raychaudhuri S, Remmers EF et al (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42(6):508
Swerdlow DI, Kuchenbaecker KB, Shah S et al (2016) Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol 45(5):1600–1616
Thompson JR, Minelli C, Bowden J et al (2017) Mendelian randomization incorporating uncertainty about pleiotropy. Stat Med 36(29):4627–4645
Waldschmidt TJ, Cook RT, Kovacs EJ (2008) Alcohol and inflammation and immune responses: summary of the 2006 Alcohol and Immunology Research Interest Group (AIRIG) meeting. Alcohol 42(2):137–142
Acknowledgements
This study was supported in part by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health and Welfare, Republic of Korea (HI15C2958).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
S.-C. Bae and Y. H. Lee declare that they have no competing interests.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Redaktion
U. Müller-Ladner, Bad Nauheim
U. Lange, Bad Nauheim
Caption Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Bae, S., Lee, Y.H. Alcohol intake and risk of rheumatoid arthritis: a Mendelian randomization study. Z Rheumatol 78, 791–796 (2019). https://doi.org/10.1007/s00393-018-0537-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00393-018-0537-z
Keywords
- Alcohol intake
- Rheumatoid arthritis
- Mendelian randomization
- Genetic predisposition to disease
- Genome-wide association study