Abstract
A pineapple-banana rotation was studied as a model system to investigate the potential emergence of a fungal-mediated disease-suppression in a soil highly infested with the pathogen Fusarium oxysporum causing the banana wilt disease. By using both field and pot experiments, the pineapple-banana rotation system resulted in a significant decrease of the pathogen number and next-stubble banana disease incidence (P < 0.05). This pathogen-suppression phenomenon was linked with detectable shifts in the soil resident fungal taxa tracked in the pineapple season. Most importantly, taxa affiliated with Talaromyces pinophilus and Clonostachys rossmaniae were found to be significantly enriched in the bulk soils due to the pineapple cultivation (P < 0.05). The taxon T. pinophilus was also significantly enriched in the rhizosphere of banana after the rotation (P < 0.05). Later, we used fungal isolation and pot inoculation to validate that both T. pinophilus and C. rossmaniae taxa are able to significantly decrease the pathogen number in the banana rhizosphere soil (P < 0.05), thus confirming their biocontrol effects suppressing the disease. Taken together, this study provides evidence on how crop rotation affects the resident soil microbiome and the development of disease suppressiveness. Besides, this study highlights the importance of understanding the dynamic changes in soil biology mediated by crop rotation and validates the mechanisms underpinning suppression toward promoting practical and directed manipulation of protective microbiomes in agroecosystems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil and plant-associated microbial communities provide key functions that affect plant growth, health, and productivity (Mamet et al. 2019; Zhao et al. 2019). Specific microbial taxa can promote nutrient acquisition in the plant rhizosphere (Mendes et al. 2013), alleviate the pressure imposed by abiotic stressors (Bulgarelli et al. 2015), and induce systemic resistance (Mendes et al. 2014; Meena et al. 2017). However, it is often challenging to identify these complex interacting processes and the mechanisms underpinning them across distinct systems. Even more challenging is the development of effective strategies to properly manipulate microbiomes towards promoting desirable biological functions. To date, research efforts on agricultural sustainability are making a case for the need to better understand how microbes can be used to further support plant health and productivity (Rainmakers and Mazzola 2016; Toju et al. 2018).
Intensive agricultural systems often result in an overall homogenization and depletion of soil microbial diversity, directly impacting the functions they provide. This can often lead to disease outbreaks and the emergence of soil-borne disease (Mendes et al. 2011). The use of agricultural practices, such as conservation agriculture (e.g., no-till) and crop rotation, has been widely adopted to reduce such critical problems that result in soil degradation (Lebeis et al. 2015; Chapelle et al. 2016; Lori et al. 2017). Interestingly, crop rotation systems have been shown to be effective in assisting the maintenance of soil health in sustainable production systems. Previous studies have shown how crop rotation exerts an effect on pathogen-inhibition/disease-suppression by changing soil physicochemical properties, increasing the efficiency of nutrient cycling and nutrient utilization in the system, and inducing dynamic changes in the soil microbiome (Huang et al. 2012; Wang et al. 2015; Hong et al. 2020). However, we still lack a comprehensive understanding of how rotation systems result in the development of suppressive soils acting against soil-borne diseases (Toju et al. 2018; Haskett et al. 2021). Therefore, a better understanding of these dynamic changes and the main players associated with disease suppression is necessary for developing new prospective farming practices.
A set of dynamic interactions between bulk/rhizosphere soil microbiomes and soil-borne diseases have been investigated with direct implications on plant performance (Mendes et al. 2011; Berendsen et al. 2018). The microenvironment of the rhizosphere comprises the millimeter soil layer around plant roots and is characterized by elevated concentrations of organic exudates secreted by plant roots. Thus, it is assumed that plants provide the desired conditions for the growth of specific (often termed as “selected”) microbial taxa (Lebeis et al. 2015; Vieira et al. 2020). Besides, the microbiome of the rhizosphere is known to be mostly defined by the local bulk soil composition (Van der Heijden et al. 2008; Delgado-Baquerizo et al. 2016), and, despite plants can actively select for the dwelling rhizosphere taxa, the active selection has been reported to range from 3 to 17% depending on plant host genotype, physiology, and developmental stage (Fizpatrick et al. 2018; Zheng et al. 2019). Most interestingly, plants living in soil are known to leave a “legacy effect” that significantly modifies the physical and biological environment around the root zone, thus affecting (positively or negatively) the growth and performance of subsequent arriving individuals (Berendsen et al. 2018; Yuan et al. 2018). Hence, identifying how crop rotation systems lead to the promotion of “legacy effects” with potential disease suppression outcomes can result in effective strategies to manipulate beneficial microbiomes in field settings.
The banana Fusarium wilt disease frequently occurs in long-term banana monocropping systems (Butler 2013; Ploetz 2015). In the Hainan province in China, this disease occurs in Cavendish banana systems and it is caused by the pathogen Fusarium oxysporum f. sp. cubense tropical race4 (Foc4) (Lin et al. 2010, 2013). In this study, a soil system highly infested with Foc4 was used to investigate the emergence of soil (microbial-mediated) disease suppression after the introduction of a pineapple-banana rotation system. We set a specific focus on tracking the dynamic changes in the soil microbiome to address the following questions: (i) Does the pineapple-banana rotation result in the emergence of soil suppressiveness or in a lower disease incidence? (ii) What is the effect of the pineapple-banana rotation on the density of the pathogen Foc4 in the soil? (iii) Is the emergence of soil suppressiveness or the lower disease incidence associated with changes in the soil microbiome as a result of the crop rotation? (iv) Are there specific microbial (fungal) taxa associated with the suppression of Foc4?
Materials and methods
Field experimental design
A field experiment was conducted from November 2014 to October 2016 at the Hainan WanZhong Co., Ltd. in Jianfeng town, Ledong County, Hainan Province, China (108°45′E, 18°38′N). The field site has been used to cultivate bananas for the past 8 years. At this site, the incidence of Fusarium wilt disease reached approximately 50% before the start of our experiment. The soil is classified as sandy loam developed from dry red soils, and has a pH of 6.14, with 1.14 g kg−1 of total N, 6.51 g kg−1 of total C, 0.96 g kg−1 of total P, and 0.27 g kg−1 of total K. The field experiment consisted of two treatments: pineapple-banana rotation and banana monoculture. Each treatment had three complete blocks (30 m × 2 m). The field experiment was divided into two periods: pineapple cultivation (FP1) and banana monoculture (FB1), called the rotation period/system; and banana cultivation after the cultivation with pineapple (FP2) and the banana monoculture (FB2), called the next-stubble period. For the rotation period, 24 banana tissue culture seedlings (Musa acuminate AAA Cavendish cv. Brazilian) or 250 pineapple tissue culture seedlings (Golden MD-2) were planted in each block. For the next-stubble period, 24 banana tissue culture seedlings were planted in each block in all treatments. Across the entire experiment, all banana tissue culture seedlings were carefully selected to have similar plant heights and stem diameters before transplanting in the field, to minimize potential confounding effects.
Pot experimental design
To replicate the emergence of the disease-suppression observed in the rotation system in the field experiment, we conducted a confirmatory pot experiment from October 2016 to November 2017 in a well-controlled greenhouse system. The soil was collected from the same field site described above. The entire experiment consisted of 60 polypropylene pots (25 cm × 35 cm, diameter × height), and each pot was filled with 8 kg of well-mixed soil. The soil mixture was prepared by sieving the collected top layer of the soil through a 4-mm sieve to remove plants, roots, and other debris. The treatments established for the pot experiment were the same as those used in the field experiment. Specifically, this experiment was also divided into two periods: pineapple cultivation (PP1) and banana monoculture (PB1), called the rotation period/system; and banana cultivation after the cultivation with pineapple (PP2) and the banana monoculture (PB2), called the next-stubble period. For the rotation period, one pineapple tissue culture seedling (Golden MD-2) or one banana tissue culture seedling (Musa acuminate AAA Cavendish cv. Brazilian) was planted in each pot. For the next-stubble period, one banana tissue culture seedling (Musa acuminate AAA Cavendish cv. Brazilian) was planted in each pot for both treatments.
Determination of Fusarium wilt disease incidence and soil and rhizosphere sampling
After 1 month, the incidence of banana Fusarium wilt disease started to be visual, and the final disease incidence was determined once the incidence became stable. Typical disease symptoms were adopted to determine the banana Fusarium wilt disease (Shen et al. 2019). Disease incidence was quantified as the percentage of infected plants relative to the total number of plants.
Bulk soils were randomly collected at a depth of 10–30 cm at the end of each season. Briefly, three banana plants (at least 3 m apart in the field experiment) were selected in each sample collection. We collected three cores under the trunk base of each banana plant in the field and during the pot experiment. Two composite samples were collected from each block in the banana monoculture and in the pineapple rotation. Similar sampling was performed for both the field and pot experiments, and a total of six mixed samples in each season were obtained from each treatment. The fresh roots of three healthy plants (i.e., without disease symptoms) at the same site as the bulk soil sampling were collected and mixed to obtain a rhizosphere sample. All bulk soil and rhizosphere samples were transferred to plastic packaging bags, kept on ice, and transported to the laboratory (< 24 h). The bulk soils were sieved through a 2-mm mesh and thoroughly homogenized. To collect the rhizosphere, the soil loosely adhered to the roots was shaken off and discarded. Then, the roots were transferred into sterile Erlenmeyer flasks containing a saline solution and shook for 30 min at 170 r/min. The obtained solution was centrifuged at 18,514 × g (12,000 r/min) for 10 min and the precipitated material was collected as the rhizosphere sample (Fu et al. 2016).
After processing, each soil sample was divided into two parts: one part was stored at − 80 °C for subsequent DNA extraction, and the other part was stored at 4 °C to be used for short-term experiments. For the field experiment, the following different soil samples were collected: bulk and rhizosphere soils from the banana monoculture of the rotation period (FB1S and FB1R, respectively), bulk and rhizosphere soils from pineapple of the rotation period (FP1S and FP1R, respectively), rhizosphere soils from the banana monoculture of the next-stubble period (FB2R), and rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (FP2R). The detail of treatments of the pot experiment is described as follows: bulk and rhizosphere soils from the banana monoculture of the rotation period (PB1S and PB1R, respectively), bulk and rhizosphere soils from pineapple of the rotation period (PP1S and PP1R, respectively), rhizosphere soils from the banana monoculture of the next-stubble period (PB2R), and rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (PP2R). The determination of soil physicochemical properties was performed as previously reported by Bao et al. (1986).
DNA extraction and Illumina MiSeq sequencing
Total soil DNA was extracted using the PowerSoil DNA Isolation Kit (MoBio Laboratories Inc., USA). Each DNA sample was extracted from each soil sample, and a total of six DNA replicates in each season were obtained per treatment. The fungal sequencing libraries were constructed based on the manufacturer’s protocols and following the previous literature (Caporaso et al. 2010; Kozich et al. 2013). The primer set ITS1F and ITS2R was used to amplify the fungal ITS1 region using the Thermo Scientific®Phusion High-Fidelity PCR Master Mix (New England Biolabs, UK) (Nilsson et al. 2018). PCR amplifications were carried out as previously described by Shen et al. (2019). The concentration and final library quality were measured using an Agilent 2100 Bioanalyzer Instrument (Agilent Technologies Co. Ltd, USA) and the KAPA Library Quantification Kit (Kapa Biosystems, USA), respectively. All constructed libraries were sequenced on an Illumina MiSeq 2000 platform at the Personal Biotechnology Company (Shanghai, China).
Bioinformatic analyses
Raw sequences were demultiplexed and adaptor and primer sequences were trimmed. The forward and reverse sequences of each soil sample were merged after removing low-quality sequences (Caporaso et al. 2011; Bokulich et al. 2013). All obtained sequences were processed using DADA2 in QIIME2 to generate the final ASV table (Hall and Beiko 2018). ASV representative sequences were selected from the obtained table and classified using the UNITE and Warcup fungal ITS databases (Deshpande et al. 2014; Nilsson et al. 2018). All raw sequence data were uploaded to the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA724805.
The number of taxonomic groups obtained per sample was standardized in R using the package GUniFrac. Principal coordinate analysis (PCoA) based on Bray–Curtis distances and analysis of similarities (ANOSIM) were conducted with the R package vegan (Oksanen et al. 2012). The differentially abundant microbial taxa were detected using linear discriminant analysis and DESeq2 (Segata et al. 2011; Love et al. 2014). Linear discriminant analysis of the effect size was performed using the online interface Galaxy (http://huttenhower.sph.harvard.edu/lefse/), with an alpha value < 0.05 and LDA score > 3 (Bulgarelli et al. 2012; Amato et al. 2019), and DESeq2 was conducted with the R package DESeq2.
Quantitative PCR analysis
The absolute abundance of Fusarium oxysporum f. sp. cubense tropical race4 (Foc4) was determined using quantitative polymerase chain reaction (qPCR) based on the taxa-specific primers FocSc-1/FocSc-2 (Huang et al. 2015). Each standard curve was established with tenfold serial dilutions of the target gene inserted into a plasmid. The qPCR reactions contained 2 μl (~ 20 ng) of template DNA from the soil samples, 10 μl SYBR® Green premix Ex Taq™ (2 ×), 6 pmol of each primer (0.4 μl of each primer), 0.4 μl ROX Reference Dye II, and 7.2 μl of sterile nuclease-free water. All results were expressed as log10 values (target gene copy numbers g−1 soil). These results were further used to conduct statistical analyses.
Isolation of suppressive fungal taxa, identification, and Foc4 inhibitory assays
Taxa-specific fungi with potential inhibitory effect on Foc4 were isolated and identified as follows: Clonostachys rosea, Clonostachys rossmaniae, and Talaromyces pinophilus. A total of 20 g of well-mixed soil collected from each treatment within the field and pot experiments in the rotation period was added to 180 ml of sterile water and shaken for 30 min. The soil suspensions were serially diluted from 10−2 to 10−4 and plated on Petri dishes containing PDA medium (Hopebio Company, Qingdao, China) supplemented with 25 mg ml−1 of chloramphenicol, and RBA (Rose Bengal Agar, Hopebio Company, Qingdao, China) solid medium (Wang et al. 2013). Each dilution was plated using three replications. Fungal colonies were counted and numbered continuously after incubation at 28 °C. Once the number of fungal colonies stabilized, a total of 60 fungal colonies were randomly chosen (based on the number) from three plates in each treatment and purified three times. Their respective DNAs were extracted using the EZNA fungal DNA kit (Omega Bio-tek, Doraville, GA, USA), and targeted PCR amplifications were performed using the primer set ITS1 and ITS4. These primers amplify the ITS1-5.8S-ITS2 and a partial fragment of the 18S and 28S. These amplicons were sequenced at TSINGKE Biological Technology Company (Beijing, China) (Singh et al. 2018). Based on the above results, the pathogen-inhibiting effects of three C. rossmaniae, three T. pinophilus, and three C. rosea isolates on Foc4 colony growth were tested using dual-culture experiments.
Effects of root exudates on the growth of Foc4 and potentially suppressive taxa
Plant root exudates were obtained using the washing method (Fang et al. 2016). First, the banana and pineapple seedlings were planted in a 1-L polypropylene pot filled with silica sand. After each plant sprouted a sufficient volume of fresh roots, we used sterile deionized water to wash the silica sand every 7 days. To eliminate impurities and microbes in the extracted solution, the obtained solutions containing plant root exudates were filtered with a 0.22-μm filter membrane. The obtained filtered liquids were freeze-dried (Li et al. 2016; Monda et al. 2017) and 1 g of dry root exudate of each crop was dissolved in 9 ml of sterile deionized water. After that, 10 ml of solution was added to 90 ml of potato dextrose agar solid medium (1%). Three treatments were established as follows: addition of banana exudates (BE), addition of pineapple exudates (PE), and addition of the same volume of sterile deionized water (CKE). A 3-mm diameter fresh hyphal mat of Foc4 and the isolated potentially suppressive taxa described above were inoculated at the center of Petri dishes in each treatment/replicate. Each strain was tested using three replicates. Finally, plates containing the Foc4 and the potentially suppressive taxa were incubated at 28 °C, and the diameters of the colonies were measured after three days.
Carbon metabolic profiles of Foc4 and potentially suppressive taxa
Biolog FF MicroPlate™ (Biolog Company, America) was used to measure the carbon metabolic profiles of Foc4, C. rosea, T. pinophilus, C. rossmaniae, and three other strains (identified as Talaromyces sp. that showed no antagonistic effects against Foc4; these were assumed as controls). All strains were cultured on PDA medium to obtain spores, and the spores of each strain were added to FF inoculation fluid (FF-IF) with sterile swabs to obtain spore suspensions. One hundred microliters of the spore suspension from each strain (adjusted to 75% ± 2% with a turbidity meter in FF inoculation fluid, FF-IF) was added to each well of a Biolog FF MicroPlate™, and each strain had three replicates. All Biolog FF MicroPlates were placed in an aerobic Omnilog incubator plate reader at 20 ℃ (Wang et al. 2018), and all colorimetric values of the inoculated plates at 590 nm were determined at 24 h, 36 h, 48 h, 60 h, 72 h, 96 h, 120 h, 144 h, and 168 h. The final colorimetric value of each well was subtracted from the value of the blank well in each plate. The results were considered positive only when the colorimetric values between the first observation and the last observation of all replicates had significant differences. The percentage of shared C source types between each key strain and the pathogen Foc4 was obtained by the formula (A ∩ B)/B × 100%, where A is the available C source of an antagonistic microbe and B is the available C source of Foc4.
Validation of the Foc4 suppression by isolated antagonistic taxa
To test the suppressive potential of the isolated fungal taxa, we used a highly infested soil from the field to conduct a confirmatory pot experiment with the inoculation of three T. pinophilus (TS-28, TS-49, and TS-53), three C. rossmaniae (CE-55, CE-67, and CE-71), three C. rosea (CA50, CA64, and CA65), and three additional Talaromyces spp. (T. aculeatus, T. angelious, T. verruculosus) known to have no apparent antagonistic effects on the pathogen Foc4. The treatments were as follows: inoculation with T. pinophilus: TS28, TS49, and TS53; inoculation with C. rossmaniae: CE55, CE67, and CE71; inoculation with C. rosea: CA50, CA64, and CA65; and inoculation with the three Talaromyces spp.: T. aculeatus, T. angelious, T. verruculosus; inoculation with T. pinophilus and C. rossmaniae: TS28 + CE67, TS49 + CE55, TS53 + CE71; inoculation with T. pinophilus and C. rosea: TS28 + CA65, TS49 + CA50, TS53 + CA64; inoculation with T. pinophilus, C. rossmaniae, and C. rosea: TS28 + CE67 + CA65, TS49 + CE55 + CA50, TS53 + CE71 + CA64. A treatment consisting of inoculation with Escherichia coli was used as one control (EC), and adding the same volume of sterile deionized water was treated as another (CKW). The Potato Dextrose Broth (PDB, Hopebio Company, Qingdao, China) was used to grow these fungal inoculants in a shaker for seven days (28 ℃ at 170 r/min). The final spore concentration of each inoculum reached ca. of 108 CFU ml−1. These inoculants were prepared by successive centrifugation and re-suspension of the spores using sterile deionized water. Then, the spores or cells of key microbes were added to the soil at a final concentration of 105 CFU g−1 dry soil. Each treatment contained six pots (11 cm × 15.5 cm, diameter × height), and each pot contained 1.5 kg of soil and one transplanted banana seedling. After 45 days, six well-mixed soil samples were collected from each treatment, and the abundance of Foc4 in the banana rhizosphere soil was determined using qPCR, as described above.
Statistical analyses
Non-normally distributed data were square-rooted or log10 transformed before statistical analyses. The linear models based on stepwise regression selection (the function of step () in R) simultaneously used the Akaike information criteria (AIC) and the Bayesian information criterion (BIC) to find the best explanatory variables (Zhao et al. 2019). A structural equation model (path analysis) was used to relate the direct and indirect factors impacting pathogen density and disease incidence, and this analysis was conducted in R using the package sem (Mamet et al. 2019). Boxplots were generated using the R package ggpolt2, and correlograms were generated in R using the packages reshape2, ggpolt2, and psych. The respective P-values were adjusted for false discovery rate (FDR) (Wickham 2012; Revelle 2014; Wickham and Chang 2015). The histograms and curve charts were plotted in R using the packages ggpolt2 and Rmisc (Hope 2013; Wickham and Chang 2015). Tukey’s HSD tests (P < 0.05) were performed using the SPSS 22.0 software (IBM, USA). The phylogenetic trees were constructed “de novo” with MEGA7 using the sequences derived from potentially suppressive taxa and the twenty best-matched sequences obtained from the NCBI database.
Results
Pathogen density and disease incidence in the field experiment
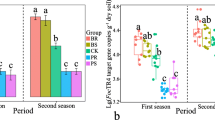
At the next-stubble period (banana cultivation in all treatments), the banana disease incidence of the FP2 treatment was significantly lower than that of the FB2 treatment in different sites (P < 0.05) (Fig. 1a). And the banana disease incidence in the northern Hainan Province (Lingao County) also showed a similar trend (Fig. S1a). Pineapple cultivation in the highly infested soil effectively decreased the pathogen copy number in the treatments FP1S and FP1R, compared with that of the FB1S and FB1R treatments, respectively (P < 0.05). The pathogen density in FP2R was also significantly lower than that of FB2R (P < 0.05) (Fig. 1b). In addition, the pathogen numbers in FP1S and FP1R were the lowest across all bulk and rhizosphere soils, respectively. Last, we found a significant correlation between the pathogen number in the rhizosphere soil and the disease incidence in the next-stubble period (r = 0.89, P < 0.001) (Fig. S1b).
Bar chart displaying the banana disease incidence in the field experiment (a). Foc4 gene copies per gram of dry weight soil in the field experiment (b). Bar chart displaying the banana disease incidence in the pot experiment (c). Foc4 gene copies per gram of dry weight soil in the pot experiment (d). FP2: banana cultivation after the cultivation with pineapple, and FB2: banana monoculture in the next-stubble period (field experiment). FB1S and FB1R: bulk and rhizosphere soils from the banana monoculture of the rotation period, respectively; FP1S and FP1R: bulk and rhizosphere soils from pineapple of the rotation period, respectively; FB2R: rhizosphere soils from the banana monoculture of the next-stubble period; FP2R: rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (field experiment). PP2: banana cultivation after the cultivation with pineapple, and PB2: banana monoculture in the next-stubble period (pot experiment). PB1S and PB1R: bulk and rhizosphere soils from the banana monoculture of the rotation period, respectively; PP1S and PP1R: bulk and rhizosphere soils from pineapple of the rotation period, respectively; PB2R: rhizosphere soils from the banana monoculture of the next-stubble period; PP2R: rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (pot experiment). Bar charts display standard errors, and lowercase letters indicate statistically significant differences (p < 0.05) based on Tukey's HSD test
Pot validation experiment
The pot experiment corroborated the results obtained in the field, that is, the cultivation of pineapple in the highly infested soil significantly decreased the disease incidence (P < 0.05) (Fig. 1c), and the pathogen copy number in the PP1S and PP1R treatments, compared with the values observed in the PB1S and PB1R treatments, respectively (P < 0.05) (Fig. 1d). The pathogen copies in PP2R were still significantly lower than that of PB2R (P < 0.05) (Fig. 1d), showing a significant correlation with the disease incidence (r = 0.93, P < 0.001) (Fig. S1c). Besides, we found the disease incidence in the banana monoculture system to increase with the planting season (Fig. S2).
Beta-diversity of fungal communities and identification of differentially abundant ASVs
For the field experiment, based on the principal coordinate analysis (PCoA) and the analysis of similarities (ANOSIM), the fungal community composition in the FP1R and FP1S treatments was significantly different from those of FB1R and FB1S treatments, respectively (P < 0.001). In addition, the banana rhizosphere fungal community in the FP2R treatment was also significantly different from that of the FB2R (P < 0.05) (Fig. 2a). In accordance with the field experiment, the pot validation experiment exhibited similar results. The cultivation with pineapple in the highly infested soil significantly altered the fungal community composition in the bulk soil during the rotation period and further influenced the next-stubble banana rhizosphere microbiome (Fig. 2c).
Principal coordinates analysis (PCoA) of soil fungal communities based on Bray–Curtis distances for the field experiment (a). Differential abundance analyses based on LDA and DESeq2 for the field experiment (b). PCoA of soil fungal communities based on Bray–Curtis distances for the pot experiment (c). Differential abundance analyses based on LDA and DESeq2 for the pot experiment (d). FB1S and FB1R: bulk and rhizosphere soils from the banana monoculture of the rotation period, respectively; FP1S and FP1R: bulk and rhizosphere soils from pineapple of the rotation period, respectively; FB2R: rhizosphere soils from the banana monoculture of the next-stubble period; FP2R: rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (field experiment). PB1S and PB1R: bulk and rhizosphere soils from the banana monoculture of the rotation period, respectively; PP1S and PP1R: bulk and rhizosphere soils from pineapple of the rotation period, respectively; PB2R: rhizosphere soils from the banana monoculture of the next-stubble period; PP2R: rhizosphere soils from banana after the cultivation with pineapple of the next-stubble period (pot experiment). ANOSIM: analysis of similarities
Compared with the banana monoculture, the rotation with pineapple in the highly infested soil resulted in the higher relative abundance of specific fungal taxa in the rhizosphere (FP1R and PP1R) and bulk (FP1S and PP1S) soils (both in the field and pot experiments). We also found some taxa to occur at higher relative abundances in the next-stubble period in the banana rhizosphere after the cultivation with pineapple (FP2R and PP2R). The banana monoculture also displayed some ASVs at higher relative abundances in the rhizosphere soil after the rotation period (FB1R and PB1R), bulk soil (FB1S and PB1S), and next-stubble rhizosphere soil (FB2R and PB2R) (Fig. 2b and 2d). A detailed view on these results can be seen in Fig. S3 and S4, respectively.
Despite several physicochemical properties (i.e., pH, EC, TOC, NH4+, NO3−, AK, and AP) in the bulk soils of the rotation system had no significant association with differences in the soil microbiome, Foc4 showed a positive association with microbial community composition under banana monoculture in the rotation (P < 0.01) (Fig. S5a and S5c). In the next-stubble period, Foc4 and banana disease incidence (DI) both were found to be positively related to microbial community composition under the banana monoculture (P < 0.01) (Fig. S5b and S5d).
Identification of potential taxa involved in disease suppression
Correlation analyses between Foc4 copy numbers and the frequency of differentially abundant taxa were conducted to identify potential pathogen-suppressive taxa. In the field experiment, ASV7, ASV18, ASV90, ASV92, and ASV147 in the bulk soil of pineapple showed a significantly negative correlation with Foc4 copy numbers (P < 0.01). ASV184 in the bulk soil of the banana monoculture showed a significantly positive correlation with Foc4 copy numbers (Fig. 3a). In the next-stubble period, ASV18 in the rhizosphere soil of the banana rotation system showed a significant negative correlation with Foc4 copy numbers. Additionally, ASV13 had a significantly positive correlation with Foc4 copy numbers in the rhizosphere soil, and these taxa were found to be at significantly higher relative abundances in the banana monoculture (Fig. 3b). In the validation experiment, the correlations of ASV18 and ASV92 in the rotation bulk soil and next-stubble soil were found to be consistent with those of the field experiment (Fig. 3c and 3d).
Differential abundance and correlation analyses of fungal ASVs that were significantly related to Foc4 copy numbers in the bulk soil of the rotation period in the field experiment (a). Differential abundance and correlation analyses of fungal ASVs that were significantly related to Foc4 copy numbers in rhizosphere soils of the next-stubble period in the field experiment (b). Differential abundance and correlation analyses of fungal ASVs that were significantly related to Foc4 copy numbers in the bulk soil of the rotation period in the pot experiment (c). Differential abundance and correlation analyses of fungal ASVs that were significantly related to Foc4 copy numbers in rhizosphere soil of the next-stubble period in the pot experiment (d). Correlation analyses were based on Spearman, and the obtained P-values were corrected for false discovery rate (FDR). *P < 0.05, **P < 0.01, and ***P < 0.001. Bulk soils from pineapple (FP1S) and the banana monoculture (FB1S) of the rotation period (field experiment). Rhizosphere soils from banana after the cultivation with pineapple (FP2R) and the banana monoculture (FB2R) of the next-stubble period (field experiment). Bulk soils from pineapple (PP1S) and the banana monoculture (PB1S) of the rotation period (pot experiment). Rhizosphere soils from banana after the cultivation with pineapple (PP2R) and the banana monoculture (PB2R) of the next-stubble period (pot experiment)
These ASVs of potentially suppressive taxa were further included in linear models to evaluate their potential relation with the overall pathogen suppression. First, the residuals of these models were in accordance with the normal distribution (Shapiro–Wilk test, P > 0.05), and most differences in pathogen suppression were explained in the models (R2 > 0.8, P < 0.001). Each variance inflation factor (VIF) value of the final taxa was lower than 10, collectively indicating the validity and strength of the linear models (Tables 1 and 2). For the field experiment, ASV18 (r < 0, P < 0.001) and ASV92 (r < 0, P < 0.01) were the top two ASVs most significantly correlated with the pathogen suppression in the bulk soil of the rotation period, and ASV18 (r < 0, P < 0.001) was the strongly related with pathogen suppression in the next-stubble period. Besides, ASV184 negatively correlated with the pathogen density increase in the rotation (r < 0, P < 0.1) and ASV13 positively correlated with the pathogen density increase in next-stubble period (r < 0, P < 0.1) (Table 1). In the validation experiment, ASV18 (r < 0, P < 0.001) and ASV92 (r < 0, P < 0.001) showed a similar negative relationship with the pathogen. The differences were that ASV139 (r < 0, P < 0.01) had a significant positive association with pathogen suppression in the rotation bulk soil and in the next-stubble rhizosphere soil. The classification information of these key taxa can be seen in the supplementary table (Table S1). By using a combined annotation based on the UNITE and Warcup databases and “de novo” phylogenetic trees based on NCBI (National Center of Biotechnology Information), ASV18, ASV92, and ASV139 were taxonomically affiliated with Talaromyces pinophilus, Clonostachys rossmaniae, and Clonostachys rosea (Table S1 and Fig. S6a, S6b, S6c), respectively.
Structural equation modeling
The two models obtained for the field and pot experiments were statistically supported (P > 0.46, RMSEA < 0.08, GFI > 0.90), and explained the largest differences in Fusarium wilt disease in both experiments (R2 = 0.87 and R2 = 0.91, respectively) (Fig. 4a and 4b). For the field experiment, the microbiome of the rhizosphere in the rotation period significantly influenced that of the relative bulk soil (ρ = 0.72, P < 0.05). The pathogen number in the bulk soils of the rotation system was significantly correlated with pathogen density in the next-stubble rhizosphere soil (ρ = 0.69, P < 0.05), subsequently resulting in an increase of Fusarium wilt disease (ρ = 0.56, P < 0.05). The cultivation of pineapple in the highly infested soil significantly increased the abundances of T. pinophilus and C. rossmaniae in the rotation bulk soil (ρ = 0.82, P < 0.05; ρ = 0.55, P < 0.05, respectively), which notably reduced the pathogen density (ρ = − 0.48, P < 0.05; ρ = − 0.41, P < 0.05, respectively). T. pinophilus abundance in the next-stubble rhizosphere soil showed a significant pathogen-inhibiting effect (ρ = − 0.32, p < 0.05) and disease-suppressive potential (ρ = − 0.40, P < 0.05) (Fig. 4a). For the pot experiment, the cultivation of pineapple in the highly infested soil showed a similar pathogen-inhibiting and disease-suppressive dynamics based on statistical associations. The differences were that the pineapple cultivation significantly increased the abundance of C. rosea, which seems to inhibit the growth of the pathogen in the bulk soil of the rotation period. This taxon was also found to be present at higher relative abundance in the next-stubble rhizosphere soil (Fig. 4b).
Structural equation model (SEM) linking the microbiome with the abundances of T. pinophilus, C. rossmaniae, and Foc4 (copy numbers), and their relations with the disease incidence in the field experiment (a). Structural equation model (SEM) linking the microbiome with the abundances of T. pinophilus, C. rossmaniae, C. rosea, and Foc4 (copy numbers), and their relations with the disease incidence in the pot experiment (b). R2, P-values, χ2, RMSEA (root mean square error of approximation), GFI (goodness-of-fit index), and Df (degree of freedom) denoting the fit of the models
Isolation of fungal suppressive strains and growth assays on root exudates growth
The isolation of potentially suppressive fungal taxa resulted in the identification of T. pinophilus, C. rossmaniae, and C. rosea (Fig. S6d, S6e and S6f), with proportions of 11.67%, 5%, and 5%, respectively. All these taxa have higher relative abundances in the rotation compared with the banana monoculture (Fig. 5a). Colony diameters of Foc4, T. pinophilus, C. rossmaniae, and C. rosea in PDA medium supplemented with root exudates showed that the Foc4 colony diameter of the PE treatment was significantly larger compared with that of the CKE treatment (P < 0.05), but relatively smaller than the observed in BE. We found the PE treatment to have a positive effect on the growth of T. pinophilus, C. rossmaniae, and C. rosea compared with the BE and CKE treatments (Fig. 5b). Detailed data on the growth of Foc4, T. pinophilus, C. rossmaniae, and C. rosea are provided in Fig. S7.
Proportions of T. pinophilus, C. rossmaniae, and C. rosea isolates obtained in each treatment (n of individual taxa/60 isolates per treatment) (a). Growth diameters of T. pinophilus, C. rossmaniae, C. rosea, and Foc4 on PDA medium supplemented with different plant root exudates. B1: the rotation period with banana cultivation, P1: the rotation period with pineapple cultivation (b). BE: the addition of banana root exudates, PE: the addition of pineapple root exudates. CK: control with sterile water. Bar charts display standard errors and lowercase letters indicate statistically significant differences (P < 0.05) based on Tukey’s HSD test
Inhibition experiment and metabolic profiles of T. pinophilus, C. rossmaniae, and C. rosea
Based on the dual-culture experiment, T. pinophilus and C. rossmaniae, but not C. rosea, had direct antagonistic effects on the growth of Foc4 (Fig. S8a, S8b and S8c), while C. rosea showed a stronger growth capacity relative to that of Foc4 after 10 days (Fig. S8d). The metabolic profiles of these taxa showed that all of them reached stable growth conditions after 96 h. Specifically, C. rosea isolates had a greater overlap in terms of C utilization with Foc4 than that showed by Talaromyces spp. and C. rossmaniae isolates (Fig. S9a and S9b). Worth mentioning, there was a larger overlap in C utilization with Foc4 from 24 to 72 h than after 96 h of incubation (Fig. S10a, S10b, S10c and S10d). Additionally, C. rosea also competed for more C source with Foc4 between 36 and 72 h than other taxa, and the differences in C source types at 48 h and 60 h were greater than those observed at any other measured time points (Table S2).
Inoculation experiment with potential suppressive taxa
Pot experiments were conducted to test the pathogen suppression potential of T. pinophilus (TS), C. rossmaniae (CE), and C. rosea (CA). Compared to the controls (i.e., CKW—no inoculation, EC-F—E. coli inoculation, and other Talaromyces spp. with no antagonistic effects), the inoculation with T. pinophilus, C. rosea, and C. rossmaniae significantly inhibited the disease incidence (based on the Foc4 copy numbers in the soil). There were no significant differences across these treatments displaying suppression (P > 0.05) (Fig. 6). Moreover, the inoculation with two or three strains significantly decreased the pathogen density in the banana rhizosphere compared to controls, and the combination of T. pinophilus and C. rossmaniae showed the highest pathogen-inhibiting effects across all sets of inoculation tested (P < 0.05) (Fig. S11).
Absolute quantification of Foc4 copy numbers in the inoculated soils in the pot experiment. EC: control inoculation with Escherichia coli. CK: control inoculation with sterile water. We tested the inoculation with single isolates of T. pinophilus (TS28, TS49, and TS53), C. rossmaniae (CE55, CE67, and CE71), and C. rosea (CA50, CA64, and CA65). Fungal isolates with no antagonistic effects on the pathogen were also tested, as follows: T-no: Talaromyces aculeatus, T. angelious, and T. verruculosus. Lowercase letters indicate statistically significant differences (P < 0.05) based on Tukey’s HSD test. See Fig. S11 for additional detail
Discussion
The cultivation of pineapple in the highly pathogen-infested soil significantly decreased the pathogen density and disease incidence; this result was in line with our previous reported findings (Wang et al. 2015). We further showed in a pot experiment performed under well-controlled greenhouse conditions that these results were replicable in terms of disease-suppressive effects and the enrichment of specific suppressive fungal taxa in the soil. As previously hypothesized, the banana monoculture system resulted in a continuous increase in the pathogen density and subsequent disease incidence (Fu et al. 2016; Shen et al. 2019), whereas crop rotation led toward the development of a suppressive soil (Bernard et al. 2014; Larkin and Lynch 2018). Interestingly, we found both banana and pineapple root exudates to significantly enhance Foc4 growth, albeit the effect of banana exudates was greater. These results are thought-provoking since most of the suppressive effects of crop rotation driven by root exudates were found to be imposed by inhibitory effects on pathogen growth (Fang et al. 2016; Li et al. 2020). Our results, however, showed that these positive effects on pathogen growth mediated by root exudates might also affect the growth of specific suppressive taxa. In doing so, such taxa can directly outcompete the pathogen resulting in a pathogen-suppression outcome that reduces the pathogen density and disease incidence.
The cultivation of pineapple in the highly infested soil significantly changed the composition of the soil fungal communities in the rotation and next-stubble period. These results also nicely align with our previous findings (Wang et al. 2015). Dynamic changes in the relative abundances of specific fungal taxa in agroecosystems are known to occur as a result of crop rotation and due to differences in the history of practices applied across sites, that is, leading to potential “legacy effects” (Hartman et al. 2018; Somenahally et al. 2018). In addition, the RDA results also revealed that Foc4 density and banana disease incidence were closely connected with shifts in fungal taxa relative abundances and community dynamics, rather than directly associated with potential changes in soil physicochemical properties. This phenomenon also corroborates with findings reported in other systems (Teste et al. 2017; Hu et al. 2018). Collectively, these results confirm that the cultivation with pineapple as a rotation crop directly affected the soil microbial community composition, which further exerted an effect on the next-stubble rhizosphere microbiomes.
The integration of differential abundance analysis of fungal taxa with linear models resulted in the indication of species potentially associated with the disease suppression, namely T. pinophilus and C. rossmaniae. These taxa significantly explained the differences in pathogen inhibition between the rotation and monoculture system, in both the field and pot experiments. According to the literature, plants often recruit beneficial microbes via signaling molecules released in the root exudates (Zhalnina et al. 2018; Lin et al. 2019). In this study, we found pineapple root exudates to significantly promote the growth of T. pinophilus and C. rossmaniae isolates. Despite these experiments were performed in silico, it demonstrates a potential mechanism on how plants can drive the establishment of disease suppressive microbiomes. Importantly, T. pinophilus were also significantly enriched in the next-stubble banana rhizosphere soil after the cultivation with pineapple, thus suggesting that taxa enriched during the rotation system can assemble in the banana rhizosphere in the next season resulting in a protective effect. This result is supported by previous studies showing that protective microbiomes can be enriched in soils with temporal beneficial effects on plants in subsequent cycles (Berendsen et al. 2018; Yuan et al. 2020). Moreover, in the pot experiment, C. rosea, which was not observed in the field experiment, also showed the same suppressive capacity as the above taxa; this result might be related to differences in environmental factors between the field and pot experiments (Garcia-Pichel et al. 2013; Frindte et al. 2019). Importantly, the results of SEM analyses also corroborated the above findings. The pineapple-banana rotation system resulted in changes in the fungal community by promoting the increase in the abundance of specific pathogen-suppressive taxa from the resident soil microbiome. This was similarly found to exert disease suppression in the rotation and next-stubble rhizosphere soil. Previous studies indicate that specific agricultural practices, including crop rotation, have become an effective way to promote soil health and enhance crop growth (Yin et al. 2010; Lupwayi et al. 2017; Banerjee et al. 2018). Our results advance such an argument by showing the mechanism by which a specific rotation system promotes the emergence of a disease-suppressive soil status.
By addressing the density of these fungal suppressive taxa using standard culture-dependent methods, we surprisingly found the results with respect to the increase in the abundance of these taxa to be consistent with the sequencing results. The pathogen-suppressive ability of these isolates was further tested in a well-controlled pot experiment. Worth mentioning, the taxon T. pinophilus was previously shown to inhibit other fungal pathogens, such as Pythium and Rhizoctonia, by secreting antifungal compounds (Kazerooni et al. 2019). However, to the best of our knowledge, this is the first report on the suppressive potential of this species on Fusarium wilt disease. Here, we observed that only taxa phylogenetically close to ASV18 displayed a direct suppressive effect on Foc4. Giving that several non-suppressive isolates of Talaromyces sp. are known to be unable to secrete antifungal compounds, it is tempting to speculate that the suppression observed in our suppressive taxa might be—to some extent—mediated by antifungal substances that affect negatively the pathogen density in the soil. Besides, as these inoculated taxa seem to antagonize the pathogen, it is plausible that some level of competition might also exist with other resident taxa within the microbiome. For instance, this can lead to indirect changes that also might account for the soil suppressive status. This is a topic of research that nicely links with principles of invasion ecology and has been more fundamentally discussed elsewhere in terms of the causes and consequences of direct microbial inoculations in soils and other systems (de-Bashan et al. 2020; Mawarda et al. 2020). The fungal isolates affiliated to C. rosea, which appeared as a potential suppressive taxon only in the pot experiment, are important biocontrol fungi that control different plant diseases, including Fusarium wilt disease, via multiple mechanisms (Roberti et al. 2008; Tian et al. 2014; Mukesh et al. 2020). Here, we did not find a direct inhibitory effect of C. rosea on the pathogen Foc4; however, this taxon was able to outcompete and overgrow the colony of the pathogen Foc4 in the dual-culture experiment. This likely indicates that by utilizing similar resources to grow, C. rosea might be able to ecologically displace Foc4 in the soil, thus indirectly resulting in the pathogen and disease control (Ghoul and Mitri 2016; Rivett et al. 2016). This assumption was further confirmed by the Biolog data, showing that C. rosea shares the highest similarity in terms of metabolic profile of C source utilization with the pathogen Foc4, when compared to other suppressive strains, T. pinophilus, C. rossmaniae. Worth mentioning, C. rosea is able to compete with Foc4 for the key C sources, such as γ-hydroxybutyric acid and succinic acid, which might relate to spore germination and mycelium growth and the pathogenicity of Fusarium sp. (Kamilova et al. 2006; Wang et al. 2016). Moreover, T. pinophilus and C. rosea are known growth-promoting taxa that ameliorate plant biotic and abiotic stresses via the production of secondary metabolites that enhance crop performance. As such, it is plausible that these taxa might also exert an influence on plant disease suppression (Sutton et al. 2008; Zhao et al. 2021). Together, these pieces of evidence lead to the fact that suppression of soil-borne diseases mediated by the resident microbiome might be attributed to several factors and mechanisms rather than by a single one operating in isolation (de-Bashan et al. 2020).
Last, it is worth noting that the combined inoculation of T. pinophilus, C. rossmaniae, and C. rosea did not show the greater pathogen-suppressing capacity in our inoculation experiment (Fig. S11). Rather, the combination of two taxa, T. pinophilus and C. rossmaniae, showed the best result. According to the literature, the use of microbial consortia is expected to have higher efficiency due to multiple ecological functions that result from greater biological diversity, thus enhancing plant growth or mediating the suppression of soil-borne diseases (Sarma et al. 2015; Hu et al. 2017). To add to that, a more holistic perspective would integrate the physicochemical conditions and biological properties of the soil with the nature and density of the inoculum, all of which dynamically affect the microbiome status and its potential activity against pathogens (Bashan et al. 2020). As such, advancing our understanding of the mechanisms underpinning a given phenomenon at the levels of ecological interactions and organism physiology is critical towards developing effective strategies to manage beneficial microbiomes in agroecosystems.
Conclusions
In summary, this study focused on manipulating the resident fungal microbiome in a soil highly infested with the pathogen Fusarium oxysporum via crop rotation toward the development of a disease suppressive status. The rotation system significantly changed the soil microbiome and reduced both the pathogen density and the disease incidence via the enrichment of specific disease-suppressive fungal taxa. In particular, we found this suppressive effect to be mediated by the enrichment of taxa that can directly (T. pinophilus and C. rossmaniae via the secretion of antifungal substances) and indirectly (C. rosea via competitive exclusion) antagonize the pathogen (Fig. 7). Later, we also showed that these effects were not cumulative and that an optimum design of a protective consortium must take into consideration the ecology of the taxa within it and their interaction with the resident soil community. Therefore, exploring the ecological mechanisms mediating the assembly of a protective microbiome, as well as better understanding the biological interactions among taxa involved in the pathogen suppression, is fundamental to advance our ability to prospectively manipulate microbiomes in field settings.
References
Amato KR, Sanders JG, Song SJ, Nute M, Metcalf JL, Thompson LR, Morton JT, Amir A, Mckenzie VJ, Humphrey G, Gogul G, Gaffney J, Baden AL, Britton GAO, Cuozzo FP, Fiore AD, Dominy NJ, Goldberg AG, Kowalewski MM, Lewis RJ, Link A, Sauther ML, Tecot S, White BA, Nelson KE, Stumpf RM, Knight R, Leigh SR (2019) Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J 13:576–587
Banerjee S, Schlaeppi K, Van der Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576
Bao SD, Su HY, An ZS et al (1986) Soil agrochemical analysis, 2nd edn. China agriculture Press, Beijing
Bashan Y, Prabhu SR, de-Bashan LE, Kloepper JW, (2020) Disclosure of exact protocols of fermentation, identity of microorganisms within consortia, formation of advanced consortia with microbe-based products. Biol Fertil Soils 56:443–445
Berendsen RL, Vismans G, Yu K, Song Y, De Jonge R, Burgman WP, Burmølle M, Herschend J, Bakker PAHM, Pieterse CMJ (2018) Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J 12:1496–1507
Bernard E, Larkin RP, Tavantzis S, Alyokhin EMS, A, Gross SD, (2014) Rapeseed rotation, compost and biocontrol amendments reduce soilborne diseases and increase tuber yield in organic and conventional potato production systems. Plant Soil 374:611–627
Bokulich N, Subramanian S, Faith J, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, CMHardy A, Schulze-Lefert P, (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17:392–403
Bulgarelli D, Rott M, Schlaeppi K, Van der Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95
Butler D (2013) Fungus threatens top banana. Nature 504:195–196
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chapelle E, Mendes R, Bakker PAH, Rainmakers JM (2016) Fungal invasion of the rhizosphere microbiome. ISME J 10:265–268
de-Bashan LE, Nannipieri P, Antoun H, Lindermann RG (2020) Application of beneficial microorganisms and their effects on soil, plants, and the environment: the scientific legacy of Professor Yoav Bashan. Biol Fertil Soils 56:439–442
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK (2016) Microbial diversity drives multi-functionality in terrestrial ecosystems. Nat Commun 7:10541
Deshpande V, Wang Q, Greenfield P, Charleston M, Porras-Alfaro A, Kuske CR, Cole JR, Midgley DJ, Tran-Dinh N (2014) Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 108:1–5
Fang Y, Zhang L, Jiao Y, Liao J, Luo L, Ji S, Li J, Dai K, Zhu S, Yang M (2016) Tobacco Rotated with Rapeseed for Soil-Borne Phytophthora Pathogen Biocontrol: Mediated by Rapeseed Root Exudates. Front Microbiol 7:894
Fizpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ (2018) Assembly of ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci USA 115:E1157–E1165
Frindte K, Pape R, Werner K, Löffler J, Knief C (2019) Temperature and soil moisture control microbial community composition in an arctic–alpine ecosystem along elevational and micro-topographic gradients. ISME J 13:2031–2043
Fu L, Ruan Y, Tao C, Li R, Shen Q (2016) Continuous application of bioorganic fertilizer induced resilient culturable bacteria community associated with banana Fusarium wilt suppression. Sci Rep-UK 6:27731
Garcia-Pichel F, Loza V, Marusenko Y, Mateo PM (2013) Potrafka R. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science 340:1574–1577
Ghoul M, Mitri S (2016) The Ecology and evolution of microbial competition. Trends Microbiol 24:833–845
Hall M, Beiko RG (2018) 16S rRNA gene analysis with QIIME2. Microbiome Analysis: Methods in Molecular Biology 1849:113–129
Hartman K, Van der Heijden MGA, Wittwer RA, Banerjee S, Walser JC, Schlaeppi K (2018) Cropping practices manipulate abundance, patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14
Haskett TL, Tkacz A, Poole PS (2021) Engineering rhizobacteria for sustainable agriculture. ISME J 15:949–964
Hong S, Jv H, Lu M, Wang B, Zhao Y, Ruan Y (2020) Significant decline in banana Fusarium wilt disease is associated with soil microbiome reconstruction under chilli pepper-banana rotation. Eur J Soil Biol. 97:103154
Hope RM (2013) Package 'Rmisc': Ryan Miscellaneous. Version 1.5. https://cran.rproject.org/web/packages/Rmisc/index.html.
Hu J, Wei Z, Weidner S, Friman VP, Xu YC, Shen QR, Jousset A (2017) Probiotic Pseudomonas communities enhance plant growth and nutrient assimilation via diversity-mediated ecosystem functioning. Soil Biol Biochem 113:122–129
Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, Van der Heijden MGA, Schlaeppi K, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:2738
Huang XQ, Wen T, Zhang JB, Meng L, Zhu TB, Liu LL (2015) Control of soilborne pathogen Fusarium oxysporum by biological soil disinfestation with incorporation of various organic matters. Eur J Plant Pathol 143:223–235
Huang YH, Wang RC, Li CH, Zuo CW, Wei YR, Zhang L, Yi GJ (2012) Control of Fusarium wilt in banana with Chinese leek. Eur J Plant Pathol 134:87–95
Kamilova F, Kravchenko LV, Shaposhnikov AI, Makarova N, Lugtenberg B (2006) Effects of the tomato pathogen Fusarium oxysporum f. sp. Radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol Plant Microbe in 19:1121–1126
Kazerooni EA, Rethinasamy V, Al-Sadi AM (2019) Talaromyces pinophilus inhibits Pythium and Rhizoctonia-induced damping-off of cucumber. J Plant Pathol 101:377–383
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq Illumina sequencing platform. Appl Environ Microb 79:5112–5120
Larkin RP, Lynch RP (2018) Use and Effects of Different brassica and other rotation crops on soil borne diseases and yield of potato. Horticulturae 4:37
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatiti S, Del Rio TG, Jones CD, Tringe SG, Dangl JL (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864
Li C, Tian Q, Rahman MK, Wu F (2020) Effect of anti-fungal compound phytosphingosine in wheat root exudates on the rhizosphere soil microbial community of watermelon. Plant Soil 456:223–240
Li CY, Mostert G, Zuo CW, Beukes I, Yang QS, Sheng O, Kuang RB, Wei YR, Hu CH, Rose L, Karangwa P, Yang J, Deng GM, Liu SW, Gao J, Viljoen A, Yi GJ (2013) Diversity and distribution of the banana wilt pathogen Fusarium oxysporum f. Sp. Cubense in China. Fungal Genom Biol: 1–16. https://doi.org/10.4172/2165-8056.1000111
Li H, Qiu Y, Wang X, Liu W, Chen G, Ma Y, Xing B (2016) Suspension stability and aggregation of multi-walled carbon nanotubes as affected by dissolved organic matters extracted from agricultural. Environ Pollut 210:323–329
Lin F, Gao J, Zeng T, Zeng H (2010) Isolation and identification of banana vasicular wilt in Hainan Province and determination of biological characteristics of strains Focr1 and Focr4. Genom Appl Biol 29:314–321
Lin Y, Ye G, Kuzyakov Y, Liu D, Fan J, Ding W (2019) Long-term manure application increases soil organic matter and aggregation, and alters microbial community structure and keystone taxa. Soil Biol Biochem 134:187–196
Lori M, Symnaczik S, Mäder P, Deyn DG, Gattinger A (2017) Organic farming enhances soil microbial abundance and activity-a metaanalysis and meta-Regression. PLoS ONE 12:1–25
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol 15:550
Lupwayi NZ, Larney FJ, Blackshaw RE, Kanashiro DA, Pearson DC (2017) Phospholipid fatty acid biomarkers show positive soil microbial community responses to conservation soil management of irrigated crop rotations. Soil till Res 168:1–10
Mamet SD, Redlick E, Brabant M, Lamb EG, Helgason BL, Stanley K, Siciliano SD (2019) Structural equation modeling of a winnowed soil microbiome identifies how invasive plants re-structure microbial networks. ISME J 13:1988–1996
Mawarda PC, Roux XL, van Elsas JD, Salles JF (2020) Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol Biochem. 148:107874
Meena KK, Sorty AM, Bitla UM, Choudhary K, Gupta P, Pareek A, Singh DP, Prabha R, Sahu PK, Gupta VK, Singh HB, Krishanani KK, Minhas PS (2017) Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci 8:172
Mendes LW, Kuramae EE, Navarrete AA, Van Veen JA, Tsai SM (2014) Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8:1577–1587
Mendes R, Garbeva P, Rainmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663
Mendes R, Kruijt M, De Bruijn I, Dekkers E, Van der Voort M, Schneider JHM, Piceno YM, Desantis TZ, Andersen GL, Bakker PAHM, Rainmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Monda H, Cozzolino V, Vinci G, Spaccini R, Piccolo A (2017) Molecular characteristics of water-extractable organic matter from different composted biomasses and their effects on seed germination and early growth of maize. Sci Total Environ 590–591:40–49
Mukesh D, Heriberto V, Martin B, Funck JD, Magnus K (2020) Lysm proteins regulate fungal development and contribute to hyphal protection and biocontrol traits in Clonostachys rosea. Front Microbiol 11:679
Nilsson RH, Anslan S, Bahram M, Wurzbacher P Baldrian, Tedersoo L (2019) Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 17:95–109
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K (2018) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens, MHH, Wagner H (2012) Vegan: community ecology package. R Package ed Version n 2.5–7. https://cran.r-project.org, https://github.com/vegandevs/vegan
Ploetz RC (2015) Fusarium wilt of banana. Phytopathology 105:1512–1521
Rainmakers JM, Mazzola M (2016) Soil immune responses. Science 352:1392–1393
Revelle W (2014) Package psych: Procedures for personality and psychological research. R package version 1.4.3. http://cran.r-project.org/web/packages/psych/psych.pdf
Rivett DW, Scheuerl T, Culbert CT, Mombrikot SB, Johnstone E, Barraclough TG, Bell T (2016) Resource-dependent attenuation of species interactions during bacterial succession. ISME J 10:2259–2268
Roberti R, Veronesi A, Cesari A, Cascone A, Berardino ID, Bertini L, Caruso C (2008) Induction of PR proteins and resistance by the biocontrol agent Clonostachys rosea in wheat plants infected with Fusarium culmorum. Plant Sci 175:339–347
Sarma BK, Yadav SK, Singh S, Singh HB (2015) Microbial consortium-mediated plant defense against phytopathogens: readdressing for enhancing efficacy. Soil Biol Biochem 8:25–33
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Shen Z, Xue C, Penton CR, Thomashow LS, Zhang N, Wang B, Ruan Y, Li R, Shen Q (2019) Suppression of banana Panama disease induced by soil microbiome reconstruction through an integrated agricultural strategy. Soil Biol Biochem 128:164–174
Singh A, Lasek-Nesselquist E, Chaturvedi V, Chaturvedi S (2018) Trichoderma polysporum selectively inhibits white-nose syndrome fungal pathogen Pseudogymnoascus destructans amidst soil microbes. Microbiome 6:139
Somenahally A, dupont JI, Brady J, Mclawrence J, Northup B, Gowda P, (2018) Microbial communities in soil profile are more responsive to legacy effects of wheat-cover crop rotations than tillage systems. Soil Biol Biochem 123:126–135
Sutton JC, Liu W, Ma J, Brown WG, Stewart JF, Walker GD (2008) Evaluation of the fungal endophyte Clonostachys rosea as an inoculant to enhance growth, fitness and productivity of crop plants. Acta Hortic 782:279–286
Teste FP, Kardol P, Turner BL, Wardle DA, Zemunik G, Renton M, Laliberté E (2017) Plant–soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science 355:73–176
Tian T, Li S, Sun M (2014) Synergistic effect of dazomet soil fumigation and Clonostachys rosea against cucumber Fusarium wilt. Phytopathology 104:1314–1321
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET (2018) Core microbiomes for sustainable agroecosystems. Nat Plants 4:247–257
Van der Heijden MGA, Bardgett RD, Van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Vieira S, Sikorski J, Dietz S, Herz K, Schrump M, Bruelheide H, Scheel D, Friedrich MW, Overmann J (2020) Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J 14:463–475
Wang B, Li R, Ruan Y, Ou Y, Zhao Y, Shen Q (2015) Pineapple-banana rotation reduced the amount of Fusarium oxysporum more than maize-banana rotation mainly through modulating fungal communities. Soil Biol Biochem 86:77–86
Wang B, Yuan J, Zhang J, Shen Z, Zhang M, Li R, Ruan Y, Shen Q (2013) Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol Fertil Soils 49:435–446
Wang H, Wang J, Chen Q, Wang M, Hsiang T, Shang S, Yu Z (2016) Metabolic effects of azoxystrobin and kresoxim-methyl against Fusarium kyushuense examined using the Biolog FF microplate. Pestic Biochem Physi 130:52–58
Wang K, Yin X, Mao H, Chu C, Tian Y (2018) Changes in structure and function of fungal community in cow manure composting. Bioresource Technol 255:123–130
Wickham H (2012) reshape2: Flexibly reshape data: a reboot of the reshape package. R package version 1. http://cran.ms.unimelb.edu.au/
Wickham H, Chang W (2015) ggplot2: an implementation of the grammar of graphics. R package version 1. http://ggplot2.org, https://github.com/hadley/ggplot2
Yin C, Jones KL, Peterson DE, Garrett KA, Hulbert SH, Paulitz TC (2010) Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol Biochem 42:2111–2118
Yuan J, Wen T, Zhang H, Penton ZM, CR, Thomashow LS, Shen Q, (2020) Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J 14:2936–2950
Yuan J, Zhao J, Wen T, Zhao M, Li R, Goossens P, Huang Q, Bai Y, Vivanco JM, Kowalchuk GA, Berendsen RL, Shen Q (2018) Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6:156
Zhalnina K, Louie KB, Hao Z, Mansoori N, Da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, Firestone MK, Northen TR, Brodie EL (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480
Zhao W, Shi X, Xian P, Feng Z, Yang J, Yang X (2021) A new fusicoccane diterpene and a new polyene from the plant endophytic fungus Talaromyces pinophilus and their antimicrobial activities. Nat Prod Res 35:124–130
Zhao Z, He J, Geisen S, Han L, Wang J, Shen J, Wei W, Fang Y, Li P, Zhang L (2019) Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 7:33
Zheng T, Liang C, Xie H, Zhao J, Yan E, Zhou X, Bao X (2019) Rhizosphere effects on soil microbial community structure and enzyme activity in a successional subtropical forest. FEMS Microbiol Ecol 5:iz043
Acknowledgements
We sincerely thank all those who have assisted us with any part of this paper and Hainan WanZhong Co., Ltd. for providing access to the experimental equipment and greenhouse facilities.
Funding
This work was supported by the National Natural Science Foundation of China (42090065 and 31972509), the Fundamental Research Funds for the Central Universities (KYXK202009 and KYXK202008), the Hainan Provincial Natural Science Foundation of China (320RC483), the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD), the 111 project (B12009), and the Innovative Research Team Development Plan of the Ministry of Education of China (IRT_17R56).
Author information
Authors and Affiliations
Contributions
The experiment was designed by Rong Li. All experiments were performed by Xianfu Yuan. Data collection, analysis, and writing were performed by Xianfu Yuan, Rong Li, and Francisco Dini-Andreote. Beibei Wang, Wu Xiong, Zongzhuan Shen, Yunze Ruan, Qirong Shen, Francisco Dini-Andreote, and Shan Hong contributed to the intellectual input and assistance to this study and manuscript preparation.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors have approved the manuscript in its entirety and agreed for its publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, X., Wang, B., Hong, S. et al. Promoting soil microbial-mediated suppressiveness against Fusarium wilt disease by the enrichment of specific fungal taxa via crop rotation. Biol Fertil Soils 57, 1137–1153 (2021). https://doi.org/10.1007/s00374-021-01594-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-021-01594-w