Abstract
We studied the viable but nonculturable (VBNC) state of the human pathogen Escherichia coli O157:H7 during desiccation using genomic, proteomic, and direct methods in a model soil under controlled conditions. During desiccation, the bacterial cells reduced their culturability, protein synthesis, and DNA replication, and changed their morphology. Four days after rewetting of dry soil, the culturability of E. coli recovered, and protein synthesis was restored. A total of 2,324 proteins were differently expressed in the VBNC state compared to the culturable state and identified. Morphological changes during the VBNC state paralleled the downregulation of cytoskeletal proteins and enzymes of the tricarboxylic acid cycle. On the contrary, proteins involved in nutrient transport, membrane, chemotactic, flagellar, virulence, and adhesion were upregulated. Overall, our results indicated that low soil moisture can induce the VBNC state in E. coli O157:H7 but this state can revert to culturability and full metabolic activity upon soil rewetting. These findings can be important aspects in assessing the environmental risks posed by microbial pathogens in soil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
E. coli O157:H7 is a dangerous pathogen, even with a low infective dose (as few as 10 cells) (Tilden Jr. et al. 1996), and can persist in soil from several days to more than 5 months (Islam et al. 2004). Its survival depends on temperature (Semenov et al. 2007), oxygen (Fremaux et al. 2007), pH (Darcan et al. 2009), dissolved organic C (Gitanjali et al. 2018), composition of microbial communities (Semenov et al. 2007), and management practices (Semenov et al. 2009). It is plausible to hypothesize that E. coli O157:H7 may reach man transferred by fruits and vegetables, and this may cause serious risks to human health.
Soil drying-rewetting can affect microbial survival by decreasing the diffusion of resources to microorganisms and causing osmotic stress (Schimel et al. 2007; Sun et al. 2018). Resource availability, such as C substrate availability, is likely the main factor affecting E. coli survival in soil (van Elsas et al. 2010). Under nutrient-limiting conditions, some bacterial cells may become dormant; for example, Gram-positive bacteria can form spores as main survival strategy (Semenov et al. 2007). In hot summers, pathogens, such as Salmonella sp. and fecal coliforms, can become undetectable by culturable techniques due to the decreased soil moisture but these pathogens regrow if soil moisture is increased (Gibbs 1997). When facing unfavorable environments, such as limited nutrients, water availability, or other stress factors, microbes can enter a viable but nonculturable (VBNC) state to survive. The VBNC state was first characterized by in vitro studies of pathogenic bacteria, such as E. coli and Vibrio cholerae (V. cholera) (Xu et al. 1982). E. coli can enter the VBNC state due to starvation, suboptimal temperature, chlorination, osmotic stress, high-pressure CO2, oxidative stress, visible radiation, sunlight, and pH variations (Darcan et al. 2009). The mechanisms responsible for resuscitation from the VBNC state of pathogens, such as E. coli in soil, are poorly known. Both RNA polymerase sigma S (RpoS) and (p)ppGpp modulated by protein RelA play crucial roles in affecting the formation of VBNC cells (Zhao et al. 2017). Magnusson et al. (2005) reported that RpoS expression could promote VBNC-state formation in E. coli O157:H7 under conditions of osmotic and oxidative stress.
Nowadays, techniques based on DNA analysis, mRNA synthesis, de novo sequencing, and microarrays have been used to describe the VBNC state (Coutard et al. 2005; Randa et al. 2004; Rosche et al. 2005), but it is difficult to isolate the short-lived and unstable mRNA (Kim et al. 2014) and only proteomics holds the potential to detect changes in microbial activity because proteins are the final results of gene expression (Nannipieri et al. 2020). In addition, soil proteomics can be used to understand the underlying mechanisms of bacterial transition from the culturable state to VBNC state and vice versa in soil (Giagnoni et al. 2018); however, low protein extraction yields and identification interferences by soil components, such as montmorillonite and humic acids, are technical challenges (Giagnoni et al. 2012, 2013). Soil proteolytic activity and geochemical denaturation also pose challenges in soil proteomics (Renella et al. 2014). In addition, the microbial proteins in soil may influence the identification and quantification of the target protein. Soil is hard to sterilize and the native microbe will regrow even after sterilization via gamma irradiation (Xing et al. 2020), and the proteins of dead cells can persist in soil. Therefore, artificial soil is preferred to the sterilized soil in studies about changes in the proteome due to the VBNC state of bacteria. Indeed, artificial soil was used for this purpose by Giagnoni et al. (2018).
This study was conducted to answer the following questions. (1) Can E. coli O157:H7 enter the VBNC state under dry soil condition? (2) Can E. coli O157:H7 in the VBNC state be resuscitated by wetting dry soil? (3) What are the different morphological and protein expression features of the VBNC state in soil? The starting hypothesis was that the VBNC state and the resuscitation in soil could change bacterial properties including its pathogenicity. To verify this hypothesis, we inoculated E. coli O157:H7 in soils of definite composition until the appearance of the VBNC state under dry conditions. Then we reverted its physiological state to culturability by wetting the dry soil. We studied protein expression and used transmission electron microscopy (TEM), laser scanning confocal microscopy (LSCM) Syto9/PI (Aurass et al. 2011) and TMT-labeled to monitor the VBNC-state E. coli O157:H7 in an artificial soil.
Materials and methods
Bacterial inoculation into artificial soil and its culturability
Artificial soil was prepared by mixing quartz sand (Sigma Aldrich); kaolinite (Clay Minerals Society, USA), with a cation exchange capacity (CEC) of 2 cmol kg−1 and 10 m2 g−1 of surface area; montmorillonite (Clay Minerals Society, USA), with a CEC of 120 cmol kg−1 and 97.4 m2 g−1 of surface area; goethite (Sigma Aldrich); and humic acids (Sigma Aldrich) (Giagnoni et al. 2011). The weight ratio of these components was 78:18:2:1:1.
The artificial soil was sterilized 3 times in an autoclave (20 min at 121 °C, 1 bar pressure) with an intermittent incubation at 20 °C for 2 days between each sterilization step. The artificial soil was then dried in a heater at 50 °C prior to bacterial inoculation. We prepared three independent replicates of each treatment, and the total amount of soil in each microcosm was 10 g. The microcosms consisted of sterile triangular bottles with aseptic sealing film; they were placed into a 37 °C incubator, and the soil pH was 7.0 during the incubation period.
The soil was inoculated with 3 mL containing 108 CFU of Escherichia coli O157:H7 EDL933 (ATCC 43895), giving 107 cells per gram of soil, and subjected to two treatments. In the first treatment, the soil moisture content (w/w) of the microcosms was not adjusted during incubation giving the following values: 30.0% at 0 day (T0); 17.4% at 1 day (T1); 12.3% at 3 days (T3); 7% at 7 days (T7); and 1.6% at 11 days (T11). After, soil moisture was increased to 30% and kept constant until the VBNC cells resuscitated at 15 days (T15). In the second treatment, the soil moisture was kept to 30% and sterile deionized H2O was added if needed; the incubation lasted 11 days.
Counting of total, viable, and culturable cells
The total, viable, and culturable cells were counted after 1 day (T1), 3 days (T3), 7 days (T7), 11 days (T11), and 15 day (T15) of incubation by the heterotrophic plate count (HPC) method (Zhang et al. 2018a).
The viable cell numbers and total cell numbers were enumerated by PMA-quantitative polymerase chain reaction (PMA-qPCR) and quantitative polymerase chain reaction (qPCR), respectively. Soil DNA was extracted using a Power Soil DNA isolation kit (Mo Bio) with minor modifications; the set of PCRs always included negative controls. After DNA extraction from the soil, we applied PMA-qPCR to detect viable E. coli O157:H7 cells (Burkert et al. 2019). The extracted DNA concentrations were determined by using a NanoDrop spectrophotometer (NanoDrop technologies, Wilmington, USA); then samples were stored at − 80 °C before determining the abundance of E. coli O157:H7 by qPCR on a 7500 real-time PCR system (Applied Biosystem, MA, US) according to Burkert et al. (2019). The Z3276 gene was amplified in 10 μL reaction volume containing 5 μL of 2 × Probe Premix (Takara, Dalian, China), 0.2 μM of each primer, 0.35 of μM probe, 0.1 of μL ROX, and 1 μL of DNA template. The qPCR conditions were as follows: activation of TaqMan probe at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 10 s, and annealing at 60 °C for 1 min. Standard curves of the Z3276 gene were generated with serial 10-fold dilutions of plasmids (synthesized by Sangon, Shanghai, China). As qPCR target gene Z3276 is a single copy, the calculation of the cell equivalents was based on the known genome sizes of the Z3276 (Jager et al. 2018). The limit of quantification (LOQ) for all qPCRs ranged from 10 to 100 gene copies per reaction and was implemented as appropriate for each specific run, as described in the supplementary materials.
Morphology assays
The morphology of E. coli O157:H7 cells was analyzed by laser scanning confocal microscopy (LSCM, LSM780, Zeiss), after staining with the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Inc., Eugene, OR), and by transmission electron microscopy (TEM, H-7650, Hitachi). A BacLight solution containing 3.34 mM SYTO 9 (green fluorescence) and 20 mM PI (red fluorescence) in dimethyl sulfoxide (DMSO) was used. SYTO 9 stained all cells green, whereas dead cells were stained red by PI. Briefly, equal volumes of SYTO 9 and PI were mixed thoroughly and 3 μL of mixture was then added to 1 mL of bacterial suspension according to the manufacturer’s instructions. After incubation under darkness at room temperature for 15 min, 5 μL of each suspension was trapped between a slide and a square coverslip, before LSCM observation.
Bacterial protein extraction
Soil samples (T1 and T11) were ground by liquid nitrogen, treated with a lysis buffer (1% Triton X-100, 10 mM dithiothreitol, 1% Protease Inhibitor Cocktail, 50 μM PR-619, 3 μM TSA, 50 mM NAM, and 2 mM EDTA), and then sonicated three times on ice using a high-intensity ultrasonic processor (Scientz). Next, tris-saturated phenol (pH 8.0) of equal volume was added and vortexed for 5 min. The mixture was centrifuged at 4 °C and 5,000g for 10 min, and the upper phenol phase was transferred to a new centrifuge tube. Four volumes of ammonium sulfate–saturated methanol were added to precipitate the proteins at − 20 °C for at least 6 h. After centrifugation at 4 °C for 10 min, the supernatant was discarded. The remaining precipitate was washed with ice-cold methanol one time, and then with ice-cold acetone three times. The protein precipitate was dissolved in 8 M urea and the protein concentrations were determined with a BCA kit (Beyotime) according to the manufacturer’s instructions.
Trypsin digestion
The protein solution was treated with dithiothreitol (to give a concentration of 5 mM), incubated at 56 °C for 30 min, alkylated by adding 11 mM iodoacetamide, and incubated for 15 min at room temperature in the dark. Finally, the urea concentration of the sample was diluted to less than 2 M with distilled water. Trypsin was added at a mass ratio of 1:50 (pancreatin:protein) and the obtained solution was incubated overnight at 37 °C. Trypsin was then added at a mass ratio of 1:100 (pancreatin:protein) and proteins were digested for 4 h (Zhang et al. 2018b).
TMT labeling
The trypsin-digested peptide was desalted by Strata X (Phenomenex) and vacuum-dried. The precipitate was solubilized by 0.5 M TEAB and then peptides were labeled according to the labeling kit (Phenomenex) instructions. Briefly, the labeled reagent was thawed, dissolved in acetonitrile, mixed with peptides, and incubated for 2 h at room temperature. The labeled peptides were mixed, desalted by stage-tip, and dried under a vacuum before the mass spectrometry analysis (1 μ of peptide) (Zhang et al. 2018b).
LC-MS/MS analysis
Tryptic peptides were solubilized by a solution containing 0.1% formic acid and 2% acetonitrile and separated by the EASY-nLC 1000 ultra-high-performance liquid system with the same solution used to solubilize tryptic peptides. Then tryptic peptides were injected into an NSI ion source for ionization and analyzed using Orbitrap Fusion TM mass spectrometry. The ion source voltage was set to 2.0 kV, and the peptide parent ion and its secondary fragments were analyzed by high-resolution Orbitrap (Zhang et al. 2018b).
Database search
The MS/MS data were processed using the Maxquant search engine (v.1.5.2.8) and matched in the proteome database using the anti-library to rule out false positive rates (FDR) caused by random matches. The minimum peptide length was set to 7 amino acid residues, the maximum number of missing sites was set to 2, the mass tolerance of the primary parent ions was set as 20 mg/L in the first search and 5 mg/L in the main search, and the mass tolerance for the fragment ions was set to 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification and oxidation on Met was specified as variable modifications. The FDR was adjusted to < 1% and the minimum score for peptides was > 40 (Zhang et al. 2018b).
The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. Proteins with fold change ratios of 1.2 and P values of 0.05 were considered to be significantly and differentially expressed. Functional annotations of the proteins were conducted using the Blast 3, 4, 5 GO program against the non-redundant protein database (NR; NCBI). The KEGG database (http://www.genome.jp/kegg/) was used to classify and group the identified proteins. The proteomics and bioinformatics analysis was helped by Jingjie PTM BioLab (Hangzhou) Co. Ltd.
Results
Total, viable, and culturable cell counts and morphological characteristics of culturable and VBNC cells
The number of culturable bacterial cells decreased in both treatments with the incubation time and after 11 days cells were undetectable (< 10 CFU per gram of dry soil) in the dry soil; upon rewetting cells, number increased and accounted for 344 CFU per gram of dry soil after 15 days (Fig. 1). It took 96 h from being undetectable to be counted as 344 CFU per gram of dry soil. If this was due to the regrowth of surviving culturable cells, the generation time of E. coli would be 18.8 h: However, by considering that the generation time was about 2.11 h in a preliminary independent experiment, the growth would have given of 2.6 × 104 CFU per gram of dry soil in only 24 h. Therefore, we suggest that the high generation time (18.8 h) observed in our study is attributable to the resuscitation of VBNC cells, rather than the regrowth of undetected culturable cells. The total and viable cell counts determined by the qPCR and PMA-qPCR methods, respectively, declined in the drying soil being 1.50 × 106 and 1.52 × 105 after 11 days (Fig. 1).
Total, viable, and culturable E. coli cell counts during the incubation of the moist soil (a). Total, viable, and culturable E. coli cell counts during the incubation of drying soil (b). Morphology of E. coli in the wet soil after 11 days (c, d, e). Morphology of E. coli present as VBNC state in the drying soil after 11 days (f, g, h). Morphology of E. coli cells after 15 days, that is after rewetting the dry soil (i, j, k)
The E. coli showed rod-like shapes and dense cytoplasm at 30% soil water content after 11 days (Fig. 1c, d, and e), while the VBNC cells in the air-dried soil were spherical, with thick cell walls, empty cytosol, and intact membranes. And part of the structure of the cell organelles was still visible after 11 days (Fig. 1f, g, and h). The E. coli recovered their culturability after the water content was restored, but were still spherical with some short rod shapes (Fig. 1i, j, and k).
The E. coli cells of soil with constant moisture were mainly the VBNC cells (with green fluorescence), while those of the air-dried soil were mainly dead (with red fluorescence) or close to death (with yellow fluorescence) when both soils were analyzed after 11 days (Fig. 2).
Proteomic analysis
Differentially expressed proteins by comparing the culturable and the VBNC state
The VBNC state of E. coli showed that 1,219 proteins were differentially expressed which makes the data reliable. For the differentially expressed proteins, 802 (66%) were in the cytoplasm, 200 (16%) in the periplasm, 127 (10%) in the cell inner membrane, 54 (5%) in the cell outer membrane, and 36 (3%) extracellular proteins (Fig. 3); 657 proteins were upregulated and 561 downregulated.
GO annotation and GO enrichment of the significant DEPs
All of the identified proteins were subjected to the GO annotation to assess their biological functions (Fig. 4); 1,219 differently expressed proteins in the VBNC state were involved in biological process and molecular functions or were cellular components. At the 3rd level of the GO annotation, it showed that proteins involved in metabolic processes, cell activities, and enzyme activities were the most important in biological process, cellular component, and molecular function, respectively. At the 4th level, proteins involved in macromolecular metabolic processes, intracellular location, and anion binding were the most important in biological process, cellular component, and molecular function, respectively. At the 5th level, proteins related to the biosynthetic processes of organic N compounds, integral components of membrane, and nucleotide binding were the most significant ones by considering the above categories. Interestingly, proteins involved in the functions related to transport, signaling receptors, and flagellum were upregulated, whereas those involved in the DNA repair, carbohydrate metabolism, and regulation of metabolism were downregulated.
To analyze the biological properties and functional alterations of the VBNC state compared to the culturable state, we performed the GO enrichment analysis (Fig. 5). Among the biological processes, the upregulated proteins were mainly those related to cilium or flagellum-dependent cell motility, protein transport, and iron chelate transport while the main downregulated proteins were those related to oxoacid and carboxylic acid metabolisms. As it concerns the cellular location, some upregulated proteins were enriched in periplasmic space, outer membrane, and cell envelope, whereas some downregulated proteins were enriched in the cytoplasm. According to the molecular function analysis, we found that proteins related to transmembrane receptor and signaling activity and transporter activity were primarily enriched as upregulated proteins, while most of the enriched and downregulated proteins were involved in ligase activity, anion binding, and nucleotide binding.
KEGG pathway analysis of the significant DEPs
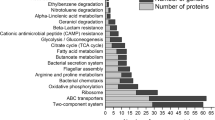
One thousand one hundred five differentially expressed proteins (DEPs) of the VBNC state were enriched according to the KEGG pathways analysis (Fig. 6). Among the upregulated proteins, those related to the ABC transporter, bacterial chemotaxis, flagellar assembly, protein export, and pathogenic E. coli infection pathways were enriched, whereas the enriched and the downregulated proteins were related to the aminoacyl-tRNA biosynthesis, glycine biosynthesis, biofilm formation, glycolysis, citrate cycle (TCA cycle), and DNA replication (Fig. 7 and Fig. S1-S25).
Regulation of differently expressed proteins involved in quorum sensing (a), DNA replication (b), bacterial chemotaxis (c), and flagellar assembly complement and coagulation cascades (d) as showed by the KEGG pathway analysis. The proteins in red were upregulated whereas those in green were downregulated
Network analysis of protein-protein interaction
The network analysis of protein-protein interaction (PPI) identified the major nodes and important linkers among the differentially expressed proteins of the VBNC state. This analysis showed that the main interactions occurred between 150 proteins; those related to 30S and 50S ribosomes were significantly upregulated, while GMP synthase, DNA-directed RNA polymerase, and DNA polymerase were downregulated (Fig. 8).
Discussion
Culturability and morphology of E. coli O157:H7
Cells of C. metallidurans strain CH34 entered the VBNC state by decreasing soil moisture (Giagnoni et al. 2018), probably due to the shortage of available C sources. E. coli O157:H7 needs a high soil moisture content to survive (Nyberg et al. 2014); indeed, cells entered the VBNC state as the artificial soil dried and E. coli O157:H7 regained the culturable state as soil was rewetted. Likely, sharp changes of osmotic pressure and the decrease in nutrient availability led to a decline in the number of culturable cells of E. coli O157:H7. We confirmed what was reported for morphological changes of the chlorine-induced VBNC state of E. coli (Chen et al. 2009) because cells of E. coli O157:H7 in the drying soil assumed globular shapes from long rod shapes, and observation by TEM revealed that the cytoplasm was reduced. Zhao et al. (2016) showed that E. coli O157:H7 could enter the VBNC state under high CO2 pressure and the cell shapes changed from rods to short or to curved rods with a decrease in nuclear and cytoplasmic density. In addition, these cells showed loose nuclear material, a gap between the intima and the adventitia, and the decreased ribosome number. Other researches also reported that poor nutrition (Chen et al. 2009), low temperature (Chen et al. 2009), or extreme pH (Giotis et al. 2007) decreased cell size with transitions from bacilli to cocci in the VBNC state (Colwell 2000; Krebs and Taylor 2011). These changes enabled cells to have the largest surface area for nutrient uptake while maintaining the least amount of cell mass (Krebs and Taylor 2011). When conditions changed to favorable ones, the cells returned to their normal shapes (Chiu et al. 2008).
Our study mainly focused on protein expression. The fact that a total of 2,324 proteins were identified indicates that artificial soil was a good model for studying the proteomic features of E. coli in the soil environment. Proteins related to the cell morphology, such as the MreB protein, a cytoskeletal protein present in all non-spheroidal bacteria and involved in cell polarity, chromosome separation, sporulation, and cell-shape control (Chiu et al. 2008), were downregulated in the VBNC cells in this research. The starvation state of V. parahaemolyticus decreased the MreB expression and this changed the morphology of cells from rod shapes to small spheres (Chiu et al. 2008). In our study, the peptidoglycan polymerase MrdB protein was upregulated 1.45-fold in the VBNC E. coli compared to the culturable cells. This protein is involved in the cell wall elongation and the peptidoglycan biosynthesis pathway which is important for the cell wall synthesis. Besides, the DacB protein involved in the cell morphological changes (Hung et al. 2013) was also upregulated, and this may be a potential reason for the formation of small spheres in VBNC cells in our study. Likely, the cell walls of E. coli under the VBNC state thickened to resist the hostile environment.
Membrane proteins
Distinguishing between viable (including culturable and VBNC) and dead cells by measuring the integrity of the cell membranes is a widely accepted approach, because only dead cells lose integrity of the cell membranes (Vives-Rego et al. 2000). Membranes of VBNC cells were stained by SYTO9 providing that they were intact.
The outer membrane proteins of gram-negative bacteria, like E. coli O157:H7, play important roles in many cellular and physiological processes (Qian et al. 2008) and for adapting cells to variations in environmental conditions (Xu et al. 2005), and their expression changes when bacteria are in the VBNC state (Zhang et al. 2019a; Zhao et al. 2016). We observed that several membrane proteins, mainly involved in transport functions, were differentially expressed in the VBNC cells compared to culturable cells. This was the behavior of three representative outer membrane proteins, CusC, OmpW, and OmpR. CusC was involved in the excretion of monovalent copper ions or monovalent silver ions from the cytoplasm to outside (Kim et al. 2011a). The efflux of copper ions may help microorganisms to withstand damage caused by any reactive oxygen produced, for example by phagocytic cells (Munson et al. 2000). Therefore, upregulation of CusC may be beneficial for the survival of the VBNC cells under oxidative stress. OmpF is one of the most abundant proteins in the outer membrane of E. coli, exists as a stable trimer, forms an electronegative channel on the outer membrane, and is responsible for the diffusion of positively charged molecules with a molecular weight lower than 600 Da to pass through cell membrane. OmpF is also directly involved in the absorption of large toxic proteins such as colicin (Housden et al. 2010; Nikaido 2003), and it is mainly expressed under nutrient deficiencies and hypotonic pressures at room temperatures, thereby contributing to the efficient absorption of nutrients by E. coli in nutrient-deficient media (Yoshida et al. 2006). Likely, the upregulated expression of OmpF (2.04-fold) might have maintained the integrity of the extracellular membrane and allowed the cells to acquire nutrients in large quantities for a rapid recovery when conditions were appropriate (Zhao et al. 2016). The E. coli can increase OmpW protein expression under H2O2 oxidative stress to survive; it is important to underline that the ompw mutant strain was more resuscitable under VBNC state than the wild strain (Asakura et al. 2008).
Proteins involved in the DNA replication and recombination
Under the VBNC state, DnaA, DnaE, DnaG, DnaN, and DnaX proteins, which are involved in DNA polymerase activity (Cohen-Fix and Livneh 1994), were all downregulated. The DnaA protein is the starting protein for chromosome replication in E. coli and plays an important role in the initiation and regulation of chromosome replication. The dnaE gene mutations cause defects in DNA replication and elongation, decreasing the rate of DNA synthesis and inhibiting cell division (Strauss et al. 2004). The DnaG protein is an RNA polymerase and catalyzes the synthesis of short RNA molecules acting as primers for DNA polymerase during DNA replication. The DNA polymerase III tau subunit encoded by the dnaX gene is part of a beta sliding clamp loading complex that hydrolyzes ATP to load beta clamp onto the primed DNA. The downregulation of the above proteins reduced the rate of DNA synthesis and inhibited cell division, and this seems to explain why the VBNC cells could not divide.
The SeqA that is encoded by the seqA gene affects DNA replication in the VBNC cells (Zhao et al. 2016). Likely, its upregulation inhibited the initiation of DNA replication and delayed the separation of nuclei and cell division, leading to the formation of VBNC state. In addition, the presence of loose nuclei in VBNC cells might also depend on the upregulated expression of SeqA.
Proteins involved in carbohydrate transport and metabolism
The proteins involved in the tricarboxylic acid cycle (TCA cycle), glyoxylate cycle, glycolysis, and disaccharide metabolism were downregulated in the VBNC state, indicating that the carbohydrate metabolism was inhibited.
The carbohydrate phosphotransferase system is the major carbohydrate uptake system that transports and phosphorylates a variety of carbohydrates by consuming phosphoenolpyruvate in bacteria (Jamal et al. 2013). The expressed proteins of the VBNC state, which were related to the phosphotransferase system such as enzyme I (E1) of phosphotransferase system (encoded by PtsI, PtsP, and PtsA), were downregulated, thus decreasing carbohydrate metabolism. The phosphotransferase system is involved not only in the absorption of carbohydrates but also in the control of bacterial organic C dynamic. A decrease in the activity of the phosphotransferase system under the VBNC state increased the amount of PEP; besides, it has been reported that PEP content was increased in persister and starved-state E. coli (Radzikowski et al. 2016). In addition, the growth rate of E. coli was reduced after mutation of the ptsI gene (Nilsson et al. 2003). Therefore, the downregulation of EI expression may be one of the causes for the inability of VBNC cells to be culturable.
In E. coli, glucose-6-phosphate is mainly involved in the glycolytic pathway and pentose phosphate pathway, both of which are regulated by glucose-6-phosphate dehydrogenase (G6PDH) encoded by the zwf gene (Sandoval et al. 2011). The overexpression of G6PDH increased the C flux through the pentose phosphate pathway (Kim et al. 2011b; Nicolas et al. 2007). In our experiment, the G6PDH was downregulated under the VBNC state, because the glycolysis pathway was downregulated and the overall metabolism was slowed down under unfavorable conditions.
The network analysis of the protein interaction in this study showed that the pyruvate kinase, encoded by pyka and pykf and catalyzing the rate-limiting reaction in the glycolysis (Xie et al. 2016), was a high-integration-degree kinase suggesting that pyruvate and the glycolysis pathway was probably important in the formation of the VBNC state. In addition, pyruvate kinase M2 (PKM2) plays an important role in maintaining redox homeostasis in cells and this role has not received enough attention so far (Liang et al. 2016). Monahan and Harry (2016) reported that the pyruvate kinase enzyme (PKY), which produces pyruvate in the final reaction of glycolysis, is involved in the Z-ring formation of B. subtilis cells. The addition of exogenous pyruvate restored normal division in the absence of the pyruvate kinase activity, thus suggesting that pyruvate was a key metabolite in the coordination of bacterial growth. The pyruvate accumulation was involved in the elimination of ROS, whose over-accumulation caused the VBNC state (Zhang et al. 2019b). In our study, the two pyruvate kinases and the eight proteins involved in pyruvate synthesis were all downregulated, suggesting that this overall downregulation might have also caused the VBNC-state formation.
Proteins of the transport systems
ABC transporters responsible for the transport of extracellular substances into cells represent the main nutrient uptake by E. coli. In addition, they can export toxic molecules out of cells (Davidson et al. 2008). In our study, the ABC transporters were all upregulated in the VBNC state. The TonB protein was upregulated by 2.38-fold, and the TonB system–like proteins were significantly upregulated and enriched in the VBNC state. The TonB system is generally involved in the transport of many important substances, such as iron, heme, vitamin B12, carbohydrates, and various transition metal elements in Gram-negative bacteria (Schauer et al. 2008). The main 5 MCPs proteins of E. coli, namely, Tsr, Tar, Trg, Tap, and Aer, were all upregulated as well as most of the chemotaxis proteins under the VBNC state in our study.
In dry soil environments, the migration of organic and inorganic substrates and extracellular enzymes is restricted, thereby affecting microbial access to nutrients. As the soil water potential and water film thickness decreased, microbial activity and passive mobility also decreased. Despite that the intracellular metabolism slowed down in the VBNC state, the expression of TonB and the ABC transporters was upregulated, likely because the cells could be immediately active to uptake nutrients when they resuscitate from the VBNC state.
Proteins involved in the flagellar movement
The flagellum is a key feature affecting the morphology, chemotaxis, and survival of bacteria (Chilcott and Hughes 2000). Several reports have shown that flagella directly mediates bacterial adhesion (Haiko and Westerlund 2013).
In our research, 19 proteins involved in the flagellar synthesis, rotary motor, hook (universal joint), and fibril (spiral propeller) were all upregulated in the VBNC state. Bacteria steer the direction of rotation of the flagellar motor to avoid adverse environments, and the change in the direction of rotation of the flagellar motor depends on the chemotaxis signal (Fukuoka et al. 2007; Kojima and Blair 2004). Among the stator proteins located on the cell membrane and responsible for the flagellar movement, the FliN, FliG, and FliM proteins together formed a “rotating complex” involved in flagella assembly, rotation, and clockwise-counterclockwise conversion (Morimoto et al. 2010). In this study, FliN and FliG were upregulated, which was likely to increase the sensitivity of the VBNC cells to chemotactic signals. It also suggests that those cells, once resuscitating from the VBNC state, were ready to start searching for nutrients especially under nutrient limitations. Indeed, the genes related to “flagella motility” were all downregulated when E. coli responded to five different perturbations (Jozefczuk et al. 2010). Zhong et al. (2018) also reported that the proteins associated with the flagellum were significantly downregulated in the VBNC state of Vibrio parahaemolyticus when the state was induced by low temperature and oligotrophic conditions. The different results were probably due to the fact that those studies have been conducted in pure culture with uniform nutrient distribution and without concentration. On the contrary, soils are solid and nutrients are not evenly distributed. Such concentration differences may lead to chemotaxis of microorganisms toward nutrients using flagella. A similar study showed that E. coli had the ability to chemotactically move toward glucose even when it entered dormancy state (Yamasaki et al. 2020).
Moreover, the flagellar cascade inhibitor protein DksA was downregulated in our study. Lemke et al. (2009) reported that DksA/ppGpp of E. coli inhibited the expression of the flagellar cascade during the stationary phase and following starvation. Likely, DksA/ppGpp coordinated flagella and ribosome assembly to save energy, thus confirming what was observed for the flagellar protein data in this study.
Proteins involved in pathogenicity
E. coli O157:H7 is known as one of the most dangerous foodborne pathogenic bacteria due to its ability to cause diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (Harris 1990). All of these diseases arise from the A/E damage on the surface of intestinal epithelial cells via the production of Shiga toxin, hemolysin, and adhesion (Donnenberg et al. 1993; Elliott et al. 1998).
The proteins related to pathogen properties of E. coli O157:H7 examined in our study can be broadly classified into three categories. The first category participates in the synthesis and function of the type III secretion system (T3SS). We observed a 1.6-fold downregulation of type III structural proteins such as sepQ. The tir gene encoding the type III secreted protein was upregulated in the VBNC state. This multifunctional protein is required for efficient pedestal formation in host epithelial cells during infection (Battle et al. 2014). Tir, Ler (type III secretion system LEE master regulator), and Espd (LEE-encoded effector which is involved in pili synthesis) were all upregulated in the VBNC state in our study.
The second category concerned pili chaperone proteins expressed by the espD gene and the fimC gene (Low et al. 2006); these proteins were upregulated. They accelerate the folding and stabilization of the pili subunit; in addition, they are involved in the transport of pili subunits through the periplasmic space to the pili assembly station of the outer membrane (Larsson et al. 2005). The upregulation of those two proteins suggested the enhanced cell adhesion ability in the VBNC state, which likely depended on the uneven distribution of humic acids in the artificial soil; E. coli cells might have adhered to the surface of humic acids’ particles because humic acids are a nutrient source among the surface-reactive particles of the artificial soil; this adhesion may be one of the causes of E. coli survival in the dry soil thus maintaining the risk of potential pathogenicity.
The third category is pathogenicity-related virulence factors. In the present study, we have analyzed maltoporin encoded by the lamB gene (Werts et al. 1992), which was downregulated 1.29-fold. This category also includes hydroxymethyl oxoglutarate aldolase encoded by the eda gene (Chattopadhyay et al. 1991), which was upregulated 1.69-fold, and the outer membrane protein OmpA and virulence protein, encoded by the stx1A, stx2A, and stxB genes (Kim et al. 2010), which were upregulated.
Yaron and Matthews (2002) observed that virulence genes stx1 and stx2 were expressed in the VBNC state of E. coli O157:H7. Lothigius et al. (2010) found that although enterotoxins were not produced when E. coli entered the VBNC state, the expression of virulence genes eltB and estA encoding the LT and STh enterotoxins was maintained. In addition, VBNC cells still present potential danger for causing fatal diseases because of their rapid resuscitation and activation of the virulent proteins under suitable conditions (Du et al. 2007). Patrone et al. (2013) observed that C. jejuni in the VBNC state still maintained the ability to adhere to intestinal cells. These studies demonstrated that the virulence genes in VBNC cells could still be expressed.
Proteins regulating VBNC state
RpoS protein is a key regulator in the formation of the VBNC state (Zhao et al. 2017). The network analysis of the protein interaction showed that the DNA-directed RNA polymerase subunit alpha, encoded by rpoA, rpoB, and rpoC genes, had the highest degree of interaction and it was downregulated, suggesting the decreased transcription which is important in the formation of VBNC state. The SpoT and RelA proteins catalyze the formation of ppGpp, which played a key role in the formation of E. coli VBNC state (Ayrapetyan et al. 2015). Both proteins were downregulated in our research. In addition, the proteins regulated by the relA gene interacted with 33 proteins in the PPI network analysis, indicating that the expression of the relA gene may be important in the VBNC-state formation. The polyphosphate kinase is involved in various cell functions, including cell survival, stress responses, host colonization, and virulence (Brown and Kornberg 2008; Gangaiah et al. 2010). Gangaiah et al. (2009) reported that polyphosphate kinase 1 (PPK1) could promote the entry of Campylobacter jejuni into the VBNC state by increasing the accumulation of Poly-P. In our research, polyphosphate kinase, expressed by the ppk gene, was downregulated. The PPK is involved in various intracellular reactions, whose role in the formation of the VBNC state is still unknown (Pinto et al. 2015).
Pu et al. (2019) reported that the ATP content of E. coli was gradually reduced under nutrient-limiting conditions. In addition, many proteins involved in cellular functions formed aggresomes, when the cells entered the VBNC state. The ATP content of cells was replenished upon restoring optimal nutrient conditions and with the resuscitation from the VBNC state. The DnaK-ClpB bichaperone system can facilitate the disaggregation. In our study, the proteins encoded by the clpB and dnaK genes were downregulated in the VBNC state suggesting that some aggresomes may have formed during the VBNC state. Three possible mechanisms regarding the formation of VBNC E. coli in this study are as follows:
-
1.
Dry conditions lead not only to a decrease in water content but also to the decrease in nutrient availability in the soil. When cells were stressed to produce a stringent response (SR), RelA and SpoT as alarm signal (p)ppGpp (alarmone nucleotides) synthase proteins were downregulated. This process further leads to the downregulation of downstream RpoS, which causes most of the E. coli to die whereas the survivors entered the VBNC state.
-
2.
Water and nutrient stress result in the ROS accumulation in cells and antioxidant enzymes were upregulated to remove ROS. When ROS accumulation reached a certain level, some E. coli will enter the VBNC state to survive in adverse environment.
-
3.
Both water and nutrient stress decrease the ATP content of E. coli cells. Moreover, DnaK and ClpB proteins were downregulated and protein aggregates were condensed, thus causing them to enter the VBNC state.
Conclusions
This study demonstrated that E. coli O157:H7 could enter the VBNC state after soil drying and could be resuscitated by rewetting. Furthermore, when compared with cultivable cells, cells in VBNC state were spherical, with thicker cell walls and emptier cytosol. Besides, the proteomic profiles of the VBNC cells showed that the proteins involved in the DNA replication and recombination, carbohydrate transportation, and metabolism were downregulated, which implies the low metabolic activity and unculturable state of VBNC E. coli O157:H7. However, the proteins related to the flagellar movement, pathogenicity, and nutrient uptake were all upregulated in the VBNC cells, indicating that VBNC E. coli could still cause fatal diseases. In addition, because of their rapid resuscitation when suitable conditions were again established in soil, the VBNC E. coli O157:H7 can still pose potential environmental risk.
Despite that this study was conducted in an artificial soil, it has the potential to understand the mechanisms underlying the VBNC state of E. coli in soil where the soil proteomic approach is still a technical challenge due to several factors including the huge amounts of expressed proteins, which cannot all be extracted from soil and detected. The search of expressed proteins in soil might be limited to those playing an important role in soil quality as those involved in the expression of pathogenicity. According to Schloter et al. (2018), molecular indicators of soil quality are required; these novel indicators should represent soil status in an accurate manner and indicate the range of values being acceptable for defining soil quality. Future research should select proteins representing microbial pathogenicity to be included in novel framework for soil quality assessment.
References
Asakura H, Kawamoto K, Haishima Y, Igimi S, Yamamoto S, Makino SI (2008) Differential expression of the outer membrane protein W (OmpW) stress response in enterohemorrhagic Escherichia coli O157:H7 corresponds to the viable but non-culturable state. Res Microbiol 159:709–717. https://doi.org/10.1016/j.resmic.2008.08.005
Aurass P, Prager R, Flieger A (2011) EHEC/EAEC O104:H4 strain linked with the 2011 German outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environ Microbiol 13:3139–3148. https://doi.org/10.1111/j.1462-2920.2011.02604.x
Ayrapetyan M, Williams TC, Baxter R, Oliver JD (2015) Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect Immun 83:4194–4203. https://doi.org/10.1128/iai.00404-15
Battle SE, Brady MJ, Vanaja SK, Leong JM, Hecht GA (2014) Actin pedestal formation by enterohemorrhagic Escherichia coli enhances bacterial host cell attachment and concomitant type III translocation. Infect Immun 82:3713–3722. https://doi.org/10.1128/IAI.01523-13
Brown MRW, Kornberg A (2008) The long and short of it - polyphosphate, PPK and bacterial survival. Trends Biochem Sci 33:284–290. https://doi.org/10.1016/j.tibs.2008.04.005
Burkert A, Douglas TA, Waldrop MP, Mackelprang R (2019) Changes in the active, dead, and dormant microbial community structure across a Pleistocene permafrost chronosequence. Appl Environ Microbiol 85:7. https://doi.org/10.1128/aem.02646-18
Chattopadhyay MK, Ghosh AK, Sengupta S (1991) Control of methionine biosynthesis in Escherichia-coli k12 - a closer study with analog-resistant mutants. J Microbiol 137:685–691. https://doi.org/10.1099/00221287-137-3-685
Chen SY, Jane WN, Chen YS, Wong HC (2009) Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int J Food Microbiol 129:157–165. https://doi.org/10.1016/j.ijfoodmicro.2008.11.009
Chilcott GS, Hughes KT (2000) Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. https://doi.org/10.1128/MMBR.64.4.694-708.2000
Chiu SW, Chen SY, Hc W (2008) Localization and expression of mreB in Vibrio parahaemolyticus under different stresses. Appl Environ Microbiol 74:7016–7022. https://doi.org/10.1128/aem.01020-08
Cohen-Fix O, Livneh Z (1994) In vitro UV mutagenesis associated with nucleotide excision-repair gaps in Escherichia coli. J Biol Chem 269:4953–4958. https://www.jbc.org/content/269/7/4953.short
Colwell RR (2000) Viable but nonculturable bacteria: a survival strategy. J Infect Chemother 6:121–125. https://doi.org/10.1007/pl00012151
Coutard F, Pommepuy M, Loaec S, Hervio-Heath D (2005) mRNA detection by reverse transcription-PCR for monitoring viability and potential virulence in a pathogenic strain of Vibrio parahaemolyticus in viable but nonculturable state. J Appl Microbiol 98:951–961. https://doi.org/10.1111/j.1365-2672.2005.02534.x
Darcan C, Ozkanca R, Idil O, Flint KP (2009) Viable but non-culturable state (VBNC) of Escherichia coli related to EnvZ under the effect of pH, starvation and osmotic stress in sea water. Polish J Microbiol 58:307–317. http://www.pjm.microbiology.pl/archive/vol5842009307.pdf
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364, table of contents. https://doi.org/10.1128/MMBR.00031-07
Donnenberg MS, Yu J, Kaper JB (1993) A 2nd chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia-coli to epithelial-cells. J Bacteriol 175:4670–4680. https://doi.org/10.1128/jb.175.15.4670-4680.1993
Du M, Chen J, Zhang X, Li A, Li Y, Wang Y (2007) Retention of virulence in a viable but nonculturable Edwardsiella tarda isolate. Appl Environ Microbiol 73:1349–1354. https://doi.org/10.1128/AEM.02243-06
Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB (1998) The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 28:1–4. https://doi.org/10.1046/j.1365-2958.1998.00783.x
Fremaux B, Muller MLD, Combaret CP, Gleizal A, Rozand CV (2007) Growth and survival of non-O157:H7 Shiga-toxin-producing Escherichia coli in cow manure. J Appl Microbiol 102:89–99. https://doi.org/10.1111/j.1365-2672.2006.03059.x
Fukuoka H, Sowa Y, Kojima S, Ishijima A, Homma M (2007) Visualization of functional rotor proteins of the bacterial flagellar motor in the cell membrane. J Mol Biol 367:692–701. https://doi.org/10.1016/j.jmb.2007.01.015
Gangaiah D, Kassem II, Liu Z, Rajashekara G (2009) Importance of polyphosphate kinase 1 for Campylobacter jejuni viable-but-nonculturable cell formation, natural transformation, and antimicrobial resistance. Appl Environ Microbiol 75:7838–7849. https://doi.org/10.1128/AEM.01603-09
Gangaiah D, Liu Z, Arcos J, Kassem II, Sanad Y, Torrelles JB, Rajashekara G (2010) Polyphosphate kinase 2: a novel determinant of stress responses and pathogenesis in Campylobacter jejuni. PLoS One 5:e12142. https://doi.org/10.1371/journal.pone.0012142
Giagnoni L, Magherini F, Landi L, Taghavi S, Bini AML, Nannipieri P, lelie DV, Renella G (2011) Extraction of microbial proteome from soil: potential and limitations assessed through a model study. Eur J Soil Sci 62:74–81. https://doi.org/10.1111/j.1365-2389.2010.01322.x
Giagnoni L, Magherini F, Landi L, Taghavi S, van der Lelie D, Puglia M, Bianchi L, Bini L, Nannipieri P, Renella G, Modesti A (2012) Soil solid phases effects on the proteomic analysis of Cupriavidus metallidurans CH34. Biol Fertil Soils 48:425–433. https://doi.org/10.1007/s00374-011-0641-6
Giagnoni L, Migliaccio A, Nannipieri P, Renella G (2013) High montmorillonite content may affect soil microbial proteomic analysis. Appl Soil Ecol 72:203–206. https://doi.org/10.1016/j.apsoil.2013.07.010
Giagnoni L, Arenella M, Galardi E, Nannipieri P, Renella G (2018) Bacterial culturability and the viable but non-culturable (VBNC) state studied by a proteomic approach using an artificial soil. Soil Biol Biochem 118:51–58. https://doi.org/10.1016/j.soilbio.2017.12.004
Gibbs RA (1997) Regrowth of faecal coliforms and Salmonellae in stored biosolids and soil amended with biosolids. Water Sci Technol 35:269–275. https://doi.org/10.1016/S0273-1223(97)00271-0
Giotis ES, McDowell DA, Blair IS, Wilkinson BJ (2007) Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl Environ Microbiol 73:997–1001. https://doi.org/10.1128/AEM.00865-06
Gitanjali NK, Christie AA, Sébastien V, Brözel VS (2018) Growth and extended survival of Escherichia coli O157:H7 in soil organic matter. Front Microbiol 9:762. https://doi.org/10.3389/fmicb.2018.00762
Haiko J, Westerlund WB (2013) The role of the bacterial flagellum in adhesion and virulence. Biology 2:1242–1267. https://doi.org/10.3390/biology2041242
Harris AA (1990) Hemorrhagic colitis and Escherichia coli O157:H7 - identifying a messenger while pursuing the message. Mayo Clin Proc 65:884–888. https://doi.org/10.1016/s0025-6196(12)62579-8
Housden NG, Wojdyla JA, Korczynska J, Grishkovskaya I, Kirkpatrick N, Brzozowski AM, Kleanthous C (2010) Directed epitope delivery across the Escherichia coli outer membrane through the porin OmpF. P Natl Acad Sci Usa 107:21412–21417. https://doi.org/10.1073/pnas.1010780107
Hung WC, Jane WN, Wong HC (2013) Association of a D-alanyl-D-alanine carboxypeptidase gene with the formation of aberrantly shaped cells during the induction of viable but nonculturable Vibrio parahaemolyticus. Appl Environ Microbiol 79:7305–7312. https://doi.org/10.1128/aem.01723-13
Islam M, Doyle MP, Phatak SC, Millner P, Jiang XP (2004) Persistence of enterohemorrhagic Escherichia coli O157 : H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J Food Prot 67:1365–1370. https://doi.org/10.4315/0362-028x-67.7.1365
Jager T, Alexander J, Kirchen S, Dotsch A, Wieland A, Hiller C, Schwartz T (2018) Live-dead discrimination analysis, qPCR assessment for opportunistic pathogens, and population analysis at ozone wastewater treatment plants. Environ Pollut 232:571–579. https://doi.org/10.1016/j.envpol.2017.09.089
Jamal Z, Miot-Sertier C, Thibau F, Dutilh L, Lonvaud-Funel A, Ballestra P, Le Marrec C, Dols-Lafargue M (2013) Distribution and functions of phosphotransferase system genes in the genome of the lactic acid bacterium Oenococcus oeni. Appl Environ Microbiol 79:3371–3379. https://doi.org/10.1128/AEM.00380-13
Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L (2010) Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol 6:364. https://doi.org/10.1038/msb.2010.18
Kim SH, Lee SR, Kim KS, Ko A, Kim E, Kim YH, Chang KT (2010) Shiga toxin A subunit mutant of Escherichia coli O157:H7 releases outer membrane vesicles containing the β-pentameric complex. FEMS Immunol Med Microbiol 58:412–420. https://doi.org/10.1111/j.1574-695X.2010.00654.x
Kim E-H, Nies DH, McEvoy MM, Rensing C (2011a) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 193:2381–2387. https://doi.org/10.1128/JB.01323-10
Kim YM, Cho HS, Jung GY, Park JM (2011b) Engineering the pentose phosphate pathway to improve hydrogen yield in recombinant Escherichia coli. Biotechnol Bioeng 108:2941–2946. https://doi.org/10.1002/bit.23259
Kim Y, Wegner CE, Werner L (2014) Soil metatranscriptomics. In: Nannipieri P, Pietramellara G, Renella G (eds). Omics in Soil Science. Caister Academic Press, Norfolk, pp 63–93
Kojima S, Blair DF (2004) The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233:93. https://doi.org/10.1016/S0074-7696(04)33003-2
Krebs SJ, Taylor RK (2011) Nutrient-dependent, rapid transition of Vibrio cholerae to coccoid morphology and expression of the toxin co-regulated pilus in this form. Microbiology 157:2942–2953. https://doi.org/10.1099/mic.0.048561-0
Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, Karp PD, Larsson E, Liu Y, Michell S, Prior J, Prior R, Malfatti S, Sjostedt A, Svensson K, Thompson N, Vergez L, Wagg JK, Wren BW, Lindler LE, Andersson SG, Forsman M, Titball RW (2005) The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet 37:153–159. https://doi.org/10.1038/ng1499
Lemke JJ, Durfee T, Gourse RL (2009) DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74:1368–1379. https://doi.org/10.1111/j.1365-2958.2009.06939.x
Liang J, Cao RX, Zhang YJ, Xia Y, Zheng YH, Li XJ, Wang LW, Yang WW, Lu ZM (2016) PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis. Nat Commun 7. https://doi.org/10.1038/ncomms12431
Lothigius A, Sjoling A, Svennerholm AM, Bolin I (2010) Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J Appl Microbiol 108:1441–1449. https://doi.org/10.1111/j.1365-2672.2009.04548.x
Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, Smith DGE, Low JC, Gally DL (2006) Analysis of firnbrial gene clusters and their expression in enterohaernorrhagic Escherichia coli O157 : H7. Environ Microbiol 8:1033–1047. https://doi.org/10.1111/j.1462-2920.2006.00995.x
Magnusson LU, Farewell A, Nystrom T (2005) ppGpp: a global regulator in Escherichia coli. Trends Microbiol 13:236–242. https://doi.org/10.1016/j.tim.2005.03.008
Monahan LG, Harry EJ (2016) You are what you eat: metabolic control of bacterial division. Trends Microbiol 24:181–189. https://doi.org/10.1016/j.tim.2015.11.007
Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T (2010) Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol 78:1117–1129. https://doi.org/10.1111/j.1365-2958.2010.07391.x
Munson GP, Lam DL, Outten FW, O’Halloran TV (2000) Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol 182:5864–5871. https://doi.org/10.1128/JB.182.20.5864-5871.2000
Nannipieri P, Ascher-Jenull J, Ceccherini MT, Pietramellara G, Renella G, Schloter M (2020) Beyond microbial diversity for predicting soil functions: a mini review. Pedosphere 30:5–17. https://doi.org/10.1016/s1002-0160(19)60824-6
Nicolas C, Kiefer P, Letisse F, Kromer J, Massou S, Soucaille P, Wittmann C, Lindley ND, Portais JC (2007) Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett 581:3771–3776. https://doi.org/10.1016/j.febslet.2007.06.066
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. https://doi.org/10.1128/mmbr.67.4.593-656.2003
Nilsson AI, Berg OG, Aspevall O, Kahlmeter G, Andersson DI (2003) Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother 47:2850–2858. https://doi.org/10.1128/AAC.47.9.2850-2858.2003
Nyberg ET, White SA, Jeffers SN, Bridges WC (2014) Removal of plant pathogen propagules from irrigation runoff using slow filtration systems: quantifying physical and biological components. Water Air Soil Pollut 225. https://doi.org/10.1007/s11270-014-1999-5
Patrone V, Campana R, Vallorani L (2013) CadF expression in Campylobacter jejunistrains incubated under low-temperature water microcosm conditions which induce the viable but non-culturable (VBNC) state. Anton Leeuw 103:979–988. https://doi.org/10.1007/s10482-013-9877-5
Pinto D, Santos MA, Chambel L (2015) Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit Rev Microbiol 41:61–76. https://doi.org/10.3109/1040841x.2013.794127
Pu YY, Li YX, Jin X, Tian T, Ma Q, Zhao ZY, Lin SY, Chen ZH, Li BH, Yao G, Leake MC, Lo CJ, Bai F (2019) ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell 73:143–156. https://doi.org/10.1016/j.molcel.2018.10.022
Qian RH, Zhao HX, Chong WZ, Wu YC, Lian SW, Hong HZ, Yong wW, Lian Y (2008) A conserved outer membrane protein as an effective vaccine candidate from Vibrio alginolyticus. Aquaculture 278:0-9. https://doi.org/10.1016/j.aquaculture.2008.03.010
Radzikowski JL, Vedelaar S, Siegel D, Álvaro Dario O, Schmidt A, Heinemann M (2016) Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol Syst Biol 12:882. https://doi.org/10.15252/msb.20166998
Randa MA, Polz MF, Lim E (2004) Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469–5476. https://doi.org/10.1128/aem.70.9.5469-5476.2004
Renella G, Ogunseitan O, Giagnoni L, Arenella M (2014) Environmental proteomics: a long march in the pedosphere. Soil Biol Biochem 69:34–37. https://doi.org/10.1016/j.soilbio.2013.10.035
Rosche TM, Yano Y, Oliver JD (2005) A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol Immunol 49:381–389. https://doi.org/10.1111/j.1348-0421.2005.tb03731.x
Sandoval JM, Arenas FA, Vasquez CC (2011) Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS One 6. https://doi.org/10.1371/journal.pone.0025573
Schauer K, Rodionov DA, de Reuse H (2008) New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci 33:330–338. https://doi.org/10.1016/j.tibs.2008.04.012
Schimel J, Balser TC, Wallenstein M (2007) Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. https://doi.org/10.1890/06-0219
Schloter M, Nannipieri P, Sorensen SJ, van Elsas JD (2018) Microbial indicators for soil quality. Biol Fertil Soils 54:1–10. https://doi.org/10.1007/s00374-017-1248-3
Semenov AV, Van Bruggen AHC, Van Overbeek L, Termorshuizen AJ, Semenov AM (2007) Influence of temperature fluctuations on Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in cow manure. FEMS Microbiol Ecol 60:419–428. https://doi.org/10.1111/j.1574-6941.2007.00306.x
Semenov AV, van Overbeek L, van Bruggen AH (2009) Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar typhimurium in soil amended with contaminated dairy manure or slurry. Appl Environ Microbiol 75:3206–3215. https://doi.org/10.1128/AEM.01791-08
Strauss B, Kelly K, Dincman T, Ekiert D, Biesieda T, Song R (2004) Cell death in Escherichia coli dnaE(Ts) mutants incubated at a nonpermissive temperature is prevented by mutation in the cydA gene. J Bacteriol 186:2147–2155. https://doi.org/10.1128/JB.186.7.2147-2155.2004
Sun DS, Bi QF, Li KJ, Zhu J, Zhang QC, Jin CW, Lu LL, Lin XY (2018) Effect of soil drying intensity during an experimental drying-rewetting event on nutrient transformation and microbial community composition. Pedosphere 28:644–655. https://doi.org/10.1016/s1002-0160(17)60450-8
Tilden J Jr, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, Morris JG Jr (1996) A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health 86:1142–1145. https://doi.org/10.2105/AJPH.86.8_Pt_1.1142
van Elsas JD, Semenov AV, Costa R, Trevors JT (2010) Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J 5:173–183. https://doi.org/10.1038/ismej.2010.80
Vives-Rego J, Lebaron P, Nebe-von Caron G (2000) Current and future applications of flow cytometry in aquatic microbiology. FEMS Microbiol Rev 24:429–448. https://doi.org/10.1016/s0168-6445(00)00033-4
Werts C, Charbit A, Bachellier S, Hofnung M (1992) DNA-sequence analysis of the lamb gene from klebsiella-pneumoniae - implications for the topology and the pore functions in maltoporin. Mol Gen Genet 233:372–378. https://doi.org/10.1007/bf00265433
Xie J, Dai C, Hu X (2016) Evidence that does not support pyruvate kinase M2 (PKM2)-catalyzed reaction as a rate-limiting step in cancer cell glycolysis. J Biol Chem 291:8987–8999. https://doi.org/10.1074/jbc.M115.704825
Xing J, Jia X, Wang H, Ma B, Falcão Salles J, Xu J (2020) The legacy of bacterial invasions on soil native communities. Environ Microbiol. https://doi.org/10.1111/1462-2920.15086
Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR (1982) Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 8:313–323. https://doi.org/10.1007/BF02010671
Xu CG, Wang SY, Ren HX, Lin XM, Wu LN, Peng XX (2005) Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 5:3142–3152. https://doi.org/10.1002/pmic.200401128
Yamasaki R, Song S, Benedik MJ, Wood TK (2020) Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience 23:100792. https://doi.org/10.1016/j.isci.2019.100792
Yaron S, Matthews KR (2002) A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: investigation of specific target genes. J Appl Microbiol 92:633–640. https://doi.org/10.1046/j.1365-2672.2002.01563.x
Yoshida T, Qin L, Egger LA, Inouye M (2006) Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J Biol Chem 281:17114–17123. https://doi.org/10.1074/jbc.M602112200
Zhang HL, Huang FC, Cai GZ, Li YT, Lin JH (2018a) Rapid and sensitive detection of Escherichia coli O157:H7 using coaxial channel-based DNA extraction and microfluidic PCR. J Dairy Sci 101:9736–9746. https://doi.org/10.3168/jds.2018-14730
Zhang NW, Gao RX, Yang JY, Zhu YY, Zhang Z, Xu XL, Wang JF, Liu XK, Li ZL, Li ZH, Gong DX, Li J, Bi JB, Kong CZ (2018b) Quantitative global proteome and lysine succinylome analyses reveal the effects of energy metabolism in renal cell carcinoma. Proteomics 18. https://doi.org/10.1002/pmic.201800001
Zhang YQ, Wu QP, Zhang JM, Yang XH (2019a) Effects of ozone on membrane permeability and ultrastructure in Pseudomonas aeruginosa. J Appl Microbiol 111:1006–1015. https://doi.org/10.1111/j.1365-2672.2011.05113.x
Zhang YX, Zhang Y, Yu JJ, Zhang H, Wang LY, Wang SN, Guo SY, Miao YC, Chen SX, Li Y, Dai SJ (2019b) NaCl-responsive ROS scavenging and energy supply in alkaligrass callus revealed from proteomic analysis. BMC Genomics 20. https://doi.org/10.1186/s12864-019-6325-6
Zhao F, Wang Y, An H, Hao Y, Hu X, Liao X (2016) New insights into the formation of viable but nonculturable Escherichia coli O157:H7 induced by high-pressure CO2. mBio 7. https://doi.org/10.1128/mBio.00961-16
Zhao XH, Zhong JL, Wei CJ, Lin CW, Ding T (2017) Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol 8. https://doi.org/10.3389/fmicb.2017.00580
Zhong Q, Tian J, Wang J, Fang X, Liao Z (2018) iTRAQ-based proteomic analysis of the viable but nonculturable state of Vibrio parahaemolyticus ATCC 17802 induced by food preservative and low temperature. Food Control 85:369–375. https://doi.org/10.1016/j.foodcont.2017.10.011
Funding
This study was financially supported by grants from the National Key Research and Develop Program of China (2016YFD0800200) and the Fundamental Research Funds for the Central Universities (2019QNA6013) and was supported by the Zhejiang University Education Foundation Global Partnership Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 1368 kb)
Rights and permissions
About this article
Cite this article
Se, J., Fu, Y., Xie, Y. et al. Proteomic changes of viable but nonculturable (VBNC) Escherichia coli O157:H7 induced by low moisture in an artificial soil. Biol Fertil Soils 57, 219–234 (2021). https://doi.org/10.1007/s00374-020-01520-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-020-01520-6