Abstract
Practical design of the XOR gate is an important milestone in the field of DNA computing. In this study, we aim to develop an enzyme-free XOR gate driven by a toehold-mediated strand displacement mechanism possessing the true detection property. The advantages of our design are as follows: dual-rail logic is not required, the explicit use of the NOT gate is avoided, the circuit structure is simple, and the design is achievable with fewer DNA strands than that designed by the combination of four NAND gates. A rational circuit design is performed and the dynamic behaviors of the biochemical reaction and the secondary structures of DNA strands are confirmed by computer simulation. In particular, both the domain-level design technique with G-T mismatched base pairs and base sequence-level fine-tuning are successfully achieved to alleviate the performance degradation arising from unintended and leaky reactions present in the circuit. The validity of the XOR gate design is confirmed by experimental studies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Molecular programming using DNA strand displacement (DSD) has a long and rich history in the field of DNA dynamic nanotechnology [12]. Design concepts enabling bottom-up, rational design of DNA molecular circuits have permitted the development of DNA circuits for various molecular machines, such as molecular robots [10]. Based on the beneficial characteristics of DSD, i.e., that we can consider a DSD reaction as an basic element with input and output channels in the circuit design, it has been proven that arbitrary systems of chemical equations are realizable in vitro by suitably connecting an output strand of a DSD reaction to an input strand of another DSD element to successfully create an ensemble of connected DSD elements (hereafter referred to as the DSD circuit) [18]. The study of DSD circuits has moved beyond proof-of-principle studies to relatively small circuits [1] and the major issue is the development of large-scale circuits comprised of a collection of some basic logical elements. To this end, the combinational circuit design must be established using a “complete set” for logic functions, where AND, OR, and NOT gates are a typical solution for the complete set in the sense that arbitrary Boolean functions are constructible in a combinational structure with these three gates [13]. In fact, this design principle provides a crucial cornerstone for electric circuit design.
Although AND, OR, and NOT gates have been already realized as a DSD circuit in many studies [14, 17], it is unclear whether middle- or large-scale DSD circuits can be developed with this complete set for practical applications and the circumstances are quite complex. As reported in previous studies [13, 14], the use of the NOT gate frequently causes problems because NOT gates generate a high concentration of output strand while consuming substrate or fuel strands, even before circuit operation. Thus, substrate and fuel strands required to drive the NOT gate are depleted. In addition to this dysfunction of the NOT gate, downstream gates could also react with the output of the NOT gate while consuming fuel strands before circuit operation. Because it would still be difficult to overcome the problems associated with the use of the NOT gate in a DNA combinational circuit, the main strategy for combinational circuit design is to utilize (i) dual-rail logic [9, 17] or (ii) another complete set; indeed, in the field of Boolean algebra, various complete sets are known to exist [22].
For the former (i), Qian et al. proposed a complete set with only AND and OR gates by applying dual-rail logic, allowing them to avoid the explicit use of the NOT gate in the design of NAND, NOR, and XOR gates [13, 14]. However, dual-rail logic requires two types of subsignals (\(x^0\) and \(x^1\)) for defining a main signal (x) in a DNA circuit, where \(x^0\) and \(x^1\) correspond to the logical “low” and “high”, respectively. In this context, for example, the logical low of x is given by a combination of the high level of \(x^0\) and low level of \(x^1\); similarly, the logical high is given by a combination of the low level of \(x^0\) and high level of \(x^1\). Although dual-rail logic is a powerful tool to construct large-scale DNA circuits [21], “true detection of the absence of input” (hereafter referred to as the true detection property), a major property of Boolean circuits cannot be handled [6], where a logic circuit is said to have the property of “true detection of the absence of input” if the logical high and low of the input signal correspond to the high and low of the concentration of the input molecule, respectively [4]. As for the latter (ii), Soloveichik and colleagues established a method for constructing arbitrary chemical reaction networks by introducing unimolecular (\(X \rightarrow Y + Z\)) and bimolecular (\(X+Y \rightarrow Z\)) reactions [18], where X, Y, and Z denote single-stranded DNA molecules, which corresponds to a complete set with “implication” [22]. This remarkable method encourages many researchers to develop “analog” DNA circuits with specific functions for various application purposes [11, 16, 19].
Other promising selections for complete sets without the NOT gate include those containing the XOR gate, such as \(\{ \mathrm{ XOR, AND} \}\) and \(\{ \mathrm{XOR, OR} \}\), because the XOR gate essentially contains the characteristics of the NOT gate; indeed, the XOR function can be theoretically assembled by the NOT function along with the OR or AND functions. However, the realization of an ideal XOR gate possessing the true detection property is still challenging in the field of DNA computing, although some studies have reported the XOR gates using dual-rail logic [3, 15] or enzymatic mechanisms [8, 20].
In this study, we develop an enzyme-free XOR gate possessing the true detection property for rational combinational circuit design by the complete set with the XOR gate. Our design has the following advantage: (i) dual-rail logic is not used, (ii) explicit use of the NOT gate is avoided, and (iii) because of the circuit size, the number of strands in the DNA circuit is considerably small. Inspired by the operation principle of the XOR gate based on pseudorotaxane formed by self-assembly of a wire-type electron acceptor and a macrocyclic electron donor [5], we establish different approaches to suppress the output status to logical low when the two input statuses are both logical high. Rational circuit design is performed and the dynamic behaviors of the biochemical reaction and secondary structures of DNA strands are determined by computer simulation. Technically, both the domain-level design technique with G-T mismatched base pairs [7] and base sequence-level fine-tuning are successfully achieved to alleviate the performance degradation arising from unintended and leaky reactions present in the circuit. The validity of the XOR gate design is then confirmed by experimental studies.
New XOR Gate Design
Principle of Circuit Operation
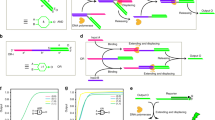
An overview of our two-input XOR gate is depicted in Fig. 1a, where the inputs (\(I_1\) and \(I_2\)) and output (O) strands are single-stranded DNA (ssDNA) and the predefined strands (\(G_1\), \(G_2\), \(F_1\), and \(F_2\)) are double-stranded DNA (dsDNA) with initial concentrations. The truth table of the XOR operation is given in Fig. 1b, where an input status of a high level of \(I_1\) or \(I_2\) leads to a high level of O; otherwise, a low output is obtained.
Figure 1c shows the detailed reaction processes in the circuit operation. In the case (1), i.e., the input status with a logical high \(I_1\) and low \(I_2\) (denoted as \((I_1,I_2)=(\mathrm H, \mathrm L)\)), the input strand \(I_1\) binds to the gate \(G_1\) at the toehold domain and then released both the signal strand \(S_1\) and the terminator strand \(T_1\) via toehold-mediated strand displacement (TMSD), while generating a waste strand (rxn\(\alpha _1\) in Fig. 1c). Subsequently, \(S_1\) in turn binds to the fuel strand \(F_1\), generating both the output strand O and a waste via TMSD (rxn\(\beta _1\)); \(T_1\) and \(F_2\) form a waste complex (rxn\(\gamma _1\)). Case (2) can be explained by the same mechanism as case (1), alternating the subscript numbers from “1” to “2”, and vice versa. In case (3), it is obvious that no output is generated because no reactions occur without no inputs. In case (4), the administration of both \(I_1\) and \(I_2\) leads to the generation of \(T_1\), \(S_1\), \(T_2\), and \(S_2\) via TMSD (rxns\(\alpha _1\) and \(\alpha _2\)). Because the fuel strand binds to the terminator strand much faster than the signal strand, no output is generated because the subsequent reaction after rxns\(\alpha _1\) and \(\alpha _2\) is dominated by fuel–terminator binding, resulting in the formation of a waste complex (rxns\(\gamma _1\) and \(\gamma _2\)). Taken together, the circuit operation determined by cases (1)–(4) coincides with the truth table shown in Fig. 1b, which indicates that the XOR gate is designed to have the true detection property.
To confirm the circuit operation principle, we perform numerical simulations based on chemical kinetics [24]. Our XOR gate can be mathematically described by the following ordinary differential equations:
where bracket \([\cdot ]\) denotes the concentration of a strand and the parameters \(k_{f_a}\) and \(k_{f_b}\) are association rate constants. We assume that the terminator strands are generated at more than twice the abundance of signal strands as described by the coefficient “2” in the right-hand sides of \(T_1\) and \(T_2\); this permits the terminator strands to suppress the output generation in case (4). Moreover, based on the specifications of our XOR gate with \(k_{f_a} \gg k_{f_b}\), we set these values as \(k_{f_a} = 3\times 10^6\) (M\(^{-1}\)sec\(^{-1}\)) and \(k_{f_b} = 5\times 10^3\) (M\(^{-1}\)sec\(^{-1}\)) according to the feasibility of implementation of DNA base sequences (see Table 2 in Sect. 2.2.4 and Discussion section for details). The simulation results are shown in Fig. 2, where the initial concentrations are given in Table 1, and the output concentration is normalized according to the maximum value in time-course data for all four cases. Then, we confirm that our XOR gate has the true detection property and works successfully as expected in all four cases, indicating that the operation principle is theoretically adequate.
Implementation Using DNA Base Sequences
The design procedure for the DNA circuit is summarized as follows. First, for each of the symbolic notations of DNA strands (\(I_1\), \(F_1\), etc.), its domain structure is designed such that the main reactions shown in Fig. 1 occur. Second, if harmful crosstalk that can ruin the normal circuit operation arises from complementary domains, the mismatch technique [2] is applied to the controversial domains to alleviate crosstalk binding. Finally, for each of the designed domains, the base sequence is designated while avoiding harmful secondary structures and inserting a short “clamp” domain to reduce leaky reactions [17].
Domain-Level Representation
The domain structure of each DNA strand in the XOR gate is designed as shown in Fig. 3, where the DNA strand is indicated by an arrow and the \(3^\prime\) end is indicated by an arrowhead. The region \(t_i\) (\(i=1,\ldots ,6\)) denotes a toehold domain that serves as a starting point for TMSD, \(s_i\) (\(i=1,\ldots ,7\)) denotes a branch migration domain, and \(t_i^*\) (\(i=1,\ldots ,6\)) and \(s_i^*\) (\(i=1,\ldots ,7\)) denote the complementary domains of \(t_i\) and \(s_i\), respectively. The binding domain forming the double helix structure is depicted by hatching. For better visibility in the figure, a small space is inserted between domains, which are connected with a thin line.
In accordance with the specifications of the XOR circuit, the terminator strands \(T_1\) and \(T_2\) need to have sufficient ability to suppress the output generation in case (4), as modeled by the coefficient 2 in the right-hand sides of the differential equation for \(T_1\) and \(T_2\) in Eq. (1). Hence, gates \(G_1\) and \(G_2\) are designed to contain double \(T_1\) and \(T_2\) strands in their structures, respectively. In addition to structural considerations, the differential equation (1) has two types of association rate constants with the relationship \(k_{f_a} \gg k_{f_b}\), enabling more rapid binding of the terminator strand to the fuel strand than to the signal strand. Because the association rate constant of TMSD is tunable by changing the length of the relevant toehold domain [24], the toehold domains of the terminator strands \(T_1\) and \(T_2\) are given by \(t_2-t_3\) and \(t_6-t_4\) domains, respectively, and those of the signal strands \(S_1\) and \(S_2\) are given by \(t_4\) and \(t_3\) domains, respectively. This design rationally ensures the condition \(k_{f_a} \gg k_{f_b}\), where \(k_{f_a}\) is determined by the length of the toehold \(t_2-t_3\) (or \(t_6-t_4\)), and \(k_{f_b}\) is determined by the length of the toehold \(t_4\) (or \(t_3\)) (also see Table 2 in Sect. 2.2.4 for a specific example).
Technical Problems Caused by Crosstalk
Although the domain structures designed in Fig. 3 realize the intended reactions rxns\(\alpha _1\), \(\alpha _2\), \(\beta _1\), \(\beta _2\), \(\gamma _1\), and \(\gamma _2\), two unintended reactions are also induced by domain complementarity, as shown in Fig. 4. The intended reaction is the main signal flow, whereas the unintended reaction is a “crosstalk” between signals. This could then ruin the normal operation of a circuit. Fig. 4a explains the crosstalk reaction between input \(I_1\) and fuel \(F_2\), mediated by toehold \(t_2-t_3\) (rxn\(\delta _1\)). In this case, this crosstalk is inevitable; in principle, \(I_1\) and \(T_1\) partially contain the same domain structures \(t_2-t_3-s_2\) because rxn\(\alpha _1\) is a TMSD to exchange \(I_1\) for \(T_1\). However, according to our specifications, \(T_1\) is designed to bind to \(F_2\), which is why the crosstalk occurs and is unavoidable. Similarly, Fig. 4b illustrates a similar mechanism of crosstalk between input \(I_2\) and fuel \(F_1\), mediated by the toehold \(t_6-t_4\) (rxn\(\delta _2\)). In practical applications, the harmful effects of crosstalk on the main signal flow are circuit-dependent. Indeed, we experimentally confirmed that the XOR circuit could not operate correctly (data not shown). Therefore, some kind of measure is needed to reduce the effects of crosstalk.
Redesign Using the Mismatch Technique
One effective measure is to suppress crosstalk reactions by breaking the complementarity of the toehold domain involved in the crosstalk reaction. The redesign strategy for the XOR gate can be outlined as follows. First, for rxn\(\delta _1\), two toehold domains \(t_2^*\) and \(t_3^*\) of \(F_2\) are replaced with \(\hat{t}_2^*\) and \(\hat{t}_3^*\) not to bind to their counterparts \(t_2\) and \(t_3\) of \(I_1\) (Fig. 5a). However, these replacements destroy the intended reactions rxn\(\gamma _1\) between \(F_2\) and \(T_1\) because the relevant toeholds involved in the TMSD become incompatible. Therefore, in turn, two toehold domains \(t_2\) and \(t_3\) of \(T_1\) are replaced with \(\hat{t}_2\) and \(\hat{t}_3\) to bind to their counterparts \(\hat{t}_2^*\) and \(\hat{t}_3^*\) of \(F_2\) for rxn\(\gamma _1\) (Fig. 5a). Again, these replacements ruin the double helix structure of \(G_1\) because of a lack of complementarity between the two domains \(\hat{t}_2-\hat{t}_3\) and \(t_2^*-t_3^*\). To recover the double helix structure of \(G_1\), noncomplementary base pairs between \(\hat{t}_2-\hat{t}_3\) and \(t_2^*-t_3^*\) are repaired using G-T mismatches, which exhibit the strongest binding affinity of all mismatched base pairs (Fig. 5b). It is reasonably expected that the double helix structure of \(G_1\) can be formed by appropriately selecting the base sequences because this dsDNA is created by annealing (see Fig. 7 for an example). Likewise, a similar redesign is applicable to \(F_1\), \(T_2\), and \(G_2\) to suppress rxn\(\delta _2\). As a result, by going through a series of modifications of the base sequences while making the use of G-T mismatching, it would be possible to suppress the unintended reactions rxns\(\delta _1\) and \(\delta _2\) while maintaining the intended reactions rxns\(\alpha _1\), \(\alpha _2\), \(\beta _1\), \(\beta _2\), \(\gamma _1\), and \(\gamma _2\).
Redesign by making use of G-T mismatch base pairs is generalized as follows. For simplicity, we consider a simple (toy) case. Suppose that we want to design a series of reactions such that strand 1 binds to strand 2, strand 2 binds to strand 3, strand 3 binds to strand 4, and but strand 4 does not bind to strand 1. However, if we design their base sequences just based on complementarity, it is unavoidable that strand 1 would bind to strand 4 (Fig. 5c). In contrast, if it is acceptable to use G-T mismatched base pairs between strands 2 and 3, strand 1 would not bind to strand 4 (Fig. 5d). Typically, such G-T mismatches are applicable when bonds of strands 2 and 3 are prepared by annealing.
DNA Base Sequence Design
DSD circuits frequently suffer from some types of leaky reactions that do not appear in the ideal reaction scheme expected by the domain structures of strands. In this case, a “fuel-gate leak” that contaminates the reaction scheme depicted in Fig. 1 may exist. Figure 6a shows the leaky reaction (rxn\(\lambda _1\)) between fuel \(F_1\) and gate \(G_1\), where zero-toehold strand displacement may occur [13]. Such leaky reactions can be eliminated by inserting a clamp domain in the original domain structure [17]. Hence, the clamp domains \(s_{4a}\) and \(s_{4a}^*\) are inserted next to domains \(t_4\) and \(t_4^*\) for \(G_1\), respectively, as shown in Fig. 6b; this could suppress rxn\(\lambda _1\). In response, input \(I_1\), which is a counterpart of \(G_1\), also needs to be added to the clamp domain \(s_{4a}\), as depicted in Fig. 6c. Similar insertion of clamp domains is applicable to \(I_2\), \(G_2\), and \(F_2\).
Finally, Tables 2 and 3 summarize the length of each domain and the base sequence of each strand. Figure 7 demonstrates the results of analysis of nucleic acid structures by NUPACK [23] to examine the validity of G-T mismatched base pairs between \(G_1\) (under) and \(T_1\) and between \(G_2\) (under) and \(T_2\), respectively, indicating that double helix structures are successfully formed.
Analysis of nucleic acid structures (NUPACK[23])
Experimental Verification
Materials and methods are summarized in the Appendix. Figure 8 shows the experimental results of time-course responses of output O under four types of input statuses, where the input concentrations of the logical low and high are 0 and 100 nM, respectively. Other initial concentrations are summarized in Table 1. Because output O is mounted on both \(F_1\) and \(F_2\), as shown in Fig. 3, Cy5 and FAM fluorophores are labeled for output strands mounted on \(F_1\) and \(F_2\), respectively, to distinguish which fuel strand is the source of the output (see Table. 3). In addition, simulation results are included as a comparison. To make the comparison easier in terms of the transient property, the simulation curve is normalized to match at the final time point (135 min) of the experimental data in Figs. 8b, c.
For the input status \((I_1 , I_2) = (\)L, L), both Cy5- and FAM-labeled outputs remained low, although the data tended to increase slightly over time owing to leaky reactions. For the input status \((I_1 , I_2) = (\)H, L), only Cy5-labeled output increased to a high level, whereas FAM-labeled output did not appear. Inversely, for the input status \((I_1 , I_2) = (\)L, H), only FAM-labeled output increased to a high level, whereas Cy5-labeled output did not appear. For the input status \((I_1 , I_2) = (\)H, H), no output responses are observed, indicating that terminator strands suppressed the generation of output by the fuel strand by binding to the fuel strand much faster than binding between fuel and signal strands, as shown in Fig. 1. Taken together, our experimental results confirmed that the designed circuit successfully functioned in accordance with the truth table for the XOR operation with the true detection property.
Discussion: Reaction Rate Constants
The reaction rate constants of TMSD can be “estimated” reasonably based on the lengths of toehold sequences in accordance with the calculus given in a previous study [24]. In our simulation, we assume that the lengths of toehold sequences related to the reaction rate constants \(k_{f_a}\) and \(k_{f_b}\) are the same between \(I_1\)- and \(I_2\)-induced reaction pathways. Therefore, we believe that our parameter setting would be reasonable from the perspective of simulation techniques in the field of DNA computing. On the other hand, the “actual” reaction rate constant is also affected by the type and combination of base sequences, particularly for GC content. For \(k_{f_a}\), the association rate constant of TMSD saturates if the corresponding toehold length is greater than six [24]. In our design shown in Table 2, the toehold lengths related to \(k_{f_a}\) are seven (\(t_1\) for the \(I_1-G_1\) association, and \(t_5\) for the \(I_2-G_2\) association) and eight (\(t_2-t_3\) for the \(T_1-F_2\) association, and \(t_6-t_4\) for the \(T_2-F_1\) association), therefore, \(k_{f_a}\) is saturated beyond the influence of differences in base sequences among toehold domains. For \(k_{f_b}\), the GC contents of toehold sequences related to \(k_{f_b}\) are the same (e.g., “ATCG” for \(t_4\) and “GCTT” for \(t_3\)), therefore, \(k_{f_b}\) may be a reasonable parameter in our validation simulation. In addition, the experimental results shown in Fig. 8 also demonstrate the validity of our design, even in cases of (high, high).
Conclusion
In this study, we developed an enzyme-free XOR gate made from a combination of TMSD reactions possessing the true detection property. Our XOR gate does not require dual-rail logic and can be realized with fewer types of DNA strands than that designed by a combination of four NAND gates. Circuit design is carried out based on a series of reasonable procedures and the dynamic behaviors of the biochemical reaction and the secondary structures of DNA strands are confirmed by computer simulation. In DNA circuit design, unintended and leaky reactions significantly affect the success or failure of the design at the experimental level. Domain-level redesign using G-T mismatched base pairs is performed to mitigate unintended reactions and sequence-level fine-tuning is achieved, along with using the clamp domain, to mitigate leaky reactions. Overall, our experimental results showed that the circuit design is effective.
References
Berleant, J., Berlind, C., Badelt, S., Dannenberg, F., Schaeffer, J., Winfree, E.: Automated sequence-level analysis of kinetics and thermodynamics for domain-level dna strand-displacement systems. J. R. Soc. Interface 15, 20180107 (2018)
Broadwater, D.W.B.J., Kim, H.D.: The effect of basepair mismatch on dna strand displacement. Biophys. J. 110(7), 1476–1484 (2016)
Cardelli, L., Kwiatkowska, M., Whitby, M.: Chemical reaction network designs for asynchronous logic circuits. Nat. Comput. 17(1), 109–130 (2018)
Chiniforooshan, E., Doty, D., Kari, L., Seki, S.: Scalable, time-responsive, digital, energy-efficient molecular circuits using dna strand displacement. arXiv:1003.3275 [cs.OH] (2010)
Credi, A., Balzani, V., Langford, S.J., Stoddart, J.F.: Logic operations at the molecular level. an xor gate based on a molecular machine. J. Am. Chem. Soc. 119(11), 2679–2681 (1997)
Garg, S., Shah, S., Bui, H., Song, T., Mokhtar, R., Reif, J.: Renewable time-responsive dna circuits. Small 14(1801470), 1–9 (2018)
Jian, Y.S., Bhadra, S., Li, B., Ellington, A.D.: Mismatches improve the performance of strand-displacement nucleic acid circuits. Angew. Chem. Int. Ed. Engl. 53(7), 1845–1848 (2014)
Lederman, H., Macdonald, J., Stefanovic, D., Stojanovic, M.N.: Deoxyribozyme-based three-input logic gates and construction of a molecular full adder. Biochemistry 45(4), 1194–1199 (2006)
Muller, D.E.: Asynchronous logics and application to information processing. Proc. Symp. Switching Theory in Space Technology, pp. 289–297 (1963)
Murata, S., Konagaya, A., Kobayashi, S., Saito, H., Hagiya, M.: Molecular robotics: a new paradigm for artifacts. N. Gener. Comput. 31(1), 27–45 (2013)
Oishi, K., Klavins, E.: Biomolecular implementation of linear i/o systems. IET Syst. Biol. 5(4), 252–260 (2011)
Padirac, A., Fujii, T., Rondelez, Y.: Nucleic acids for the rational design of reaction circuits. Curr. Opin. Biotechnol. 24, 575–580 (2013)
Qian, L., Winfree, E.: Scaling up digital circuit computation with dna strand displacement cascades. Science 332(6034), 1196–1201 (2011)
Qian, L., Winfree, E.: A simple dna gate motif for synthesizing large-scale circuits. J. R. Soc. Interface 8(62), 1281–1297 (2011)
Qian, L., Winfree, E., Bruck, J.: Neural network computation with dna strand displacement cascades. Nature 475, 368–372 (2011)
Sawlekar, R., Montefusco, F., Kulkarni, V.V., Bates, D.G.: Implementing nonlinear feedback controllers using dna strand displacement reactions. IEEE Trans. NanoBiol. 15(5), 443–454 (2016)
Seelig, G., Soloveichik, D., Zhang, D.Y., Winfree, E.: Enzyme-free nucleic acid logic circuits. Science 314(5805), 1585–1588 (2006)
Soloveichik, D., Seelig, G., Winfree, E.: Dna as a universal substrate for chemical kinetics. PNAS 107(12), 5393–5398 (2010)
Song, T., Garg, S., Mokhtar, R., Bui, H., Reif, J.: Analog computation by dna strand displacement circuits. ACS Synt. Biol. 5, 898–912 (2016)
Stojanovic, M.N., Mitchell, T.E., Stefanovic, D.: Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 124(14), 3555–3561 (2002)
Thubagere, A.J., Thachuk, C., Berleant, J., Johnson, R.F., Ardelean, D.A., Cherry, K.M., Qian, L.: Compiler-aided systematic construction of large-scale dna strand displacement circuits using unpurified components. Nat. Commun. 8(14373), 1–12 (2017)
Wernick, W.: Complete sets of logical functions. Trans. Am. Math. Soc. 51, 117–132 (1942)
Zadeh, J.N., Steenberg, C.D., Bois, J.S., Wolfe, B.R., Pierce, M.B., Khan, A.R., Dirks, R.M., Pierce, N.A.: Nupack: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2010)
Zhang, D.Y., Winfree, E.: Control of dna strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009)
Acknowledgement
This work was supported by JSPS KAKENHI (Grant No. 17K06500).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Materials and Methods
Materials and Methods
System Specifications
All DNA oligos in Table 3 are synthesized and purified by Nippon Gene, Co., Ltd (Tokyo, Japan). Each DNA oligo is diluted with 1\(\times\)TE buffer (Nippon Gene, Co., Ltd.) with 12.5 mM MgCl\(_2\) added at concentrations shown in Table 1. The dsDNAs \(G_1\), \(G_2\), \(F_1\), and \(F_2\) are prepared by annealing from 90 to \(20\ ^\circ\)C with a temperature gradient of \(-1\ ^\circ\)C per min after heating at \(90\ ^\circ\) for 15 min with a thermal cycler (GeneAtlas ASTEC325 by Astec Co., Ltd; Fukuoka, Japan).
Signal intensity measurements
After annealing, all samples are held for 30 min and are then mixed in test tubes, except for samples with input strands. Signal intensity measurements are performed with a fluorescence spectrophotometer (FP-8300; JASCO Co., Ltd.; Tokyo Japan) at a temperature of \(20\ ^\circ\)C, where the \(5^\prime\) end of the output O mound on \(F_1\) is labeled with Cy5, the \(5^\prime\) end of the output O mound on \(F_2\) is labeled with FAM, and the \(3^\prime\) ends of fuels \(F_1\) and \(F_2\) are labeled with the black fluorescence quencher BHQ (see Table 3). Input strands required for each experimental condition are added 15 min after the start of measurement and observation of signal intensity is performed for 120 min. For FAM measurement, the excitation wavelength is 496 nm and the fluorescence wavelength is 524 nm; for Cy5 measurement, the excitation wavelength is 643 nm and the fluorescence wavelength is 667 nm.
Normalization of measured time-course data
For each experimental condition, three independent time series measurements are taken to determine the mean and standard deviation. The fluorescence intensity before input administration is set to the minimum value. In the case of status \((I_1,I_2)=(\mathrm L, \mathrm L)\), after 2 h of measurement, excess amounts of \(I_1\) and \(I_2\) are added and the average of the last five time-course points at which the fluorescence intensity converged is taken as the maximum value [13]. Using these minimum and maximum values, the experimental data are normalized to be in the range of 0–1.
About this article
Cite this article
Nishijima, K., Nakakuki, T. XOR Gate Design Toward a Practical Complete Set for DNA Computing. New Gener. Comput. 38, 285–301 (2020). https://doi.org/10.1007/s00354-020-00090-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00354-020-00090-3