Abstract
Purpose
The purpose of this study is to evaluate the potential association between genetic variants in genes encoding the components of RNA-induced silencing complex and prostate cancer (PCa) risk. Genetic variants chosen for this study are rs3742330 in DICER1, rs4961280 in AGO2, rs784567 in TARBP2, rs7813 in GEMIN4 and rs197414 in GEMIN3.
Methods
The study involved 355 PCa patients, 360 patients with benign prostatic hyperplasia and 318 healthy controls. For individuals diagnosed with PCa, clinicopathological characteristics including serum prostate-specific antigen level at diagnosis, Gleason score (GS) and clinical stage were determined. Genotyping was performed using high-resolution melting analysis, PCR–RFLP, TaqMan SNP Genotyping Assay and real-time PCR-based genotyping assay using specific probes. Allelic and genotypic associations were evaluated by unconditional linear and logistic regression methods.
Results
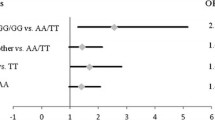
The study provided no evidence of association between the analyzed genetic variants and PCa risk. Nevertheless, allele A of rs784567 was found to confer the reduced risk of higher serum PSA level at diagnosis (P = 0.046; Difference = −66.64, 95 % CI −131.93 to 1.35, for log-additive model). Furthermore, rs4961280, as well as rs3742330, were shown to be associated with GS. These variants, together with rs7813, were found to be associated with the lower clinical stage of PCa. Also, rs3742330 minor allele G was found to be associated with lower PCa aggressiveness (P = 0.036; OR 0.14, 95 % CI 0.023–1.22, for recessive model).
Conclusions
According to our data, rs3742330, rs4961280 and rs7813 qualify for potentially protective genetic variants against PCa progression. These variants were not shown to be associated with PCa risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent statistics on prostate cancer (PCa) show that this malignancy is the second most commonly diagnosed cancer among males. Furthermore, it contributes substantially to cancer-related death rates, ranking fifth on mortality scales in global male population [1]. Among the newly diagnosed, latent PCa that remains indolent during the life time represents a significant percent. According to estimations based on clinical reports from developed countries, as much as a half of patients are overdiagnosed with PCa [2], which leads to unnecessary morbidity due to application of invasive therapeutic procedures [3]. Therefore, among prevailing aims in modern scientific research on PCa is to identify genetic markers potentially significant for constructing reliable algorithms applicable for assessing the risk of disease progression to a more aggressive form [4].
Regulatory activities of microRNA molecules have been extensively studied in the last decade [5]. During this period, numerous lines of evidence have been found to support the involvement of deregulations in RNA interference process in carcinogenesis in various tissues, including prostatic [6–9]. These data mostly consist of the observed differences in microRNA and their targets’ expression between normal and malignant cells [8, 10]. Furthermore, forced or silenced expression of numerous microRNAs was found to be correlated with presentation of various aspects of PCa malignant phenotype in in vitro systems [10].
Due to their potential effect on microRNA biogenesis and function, components of RNA-induced silencing complex (RISC) could also be involved in molecular pathogenesis of PCa. This hypothesis is augmented by the recent data indicating the aberrant expression of several components of this protein complex in malignant, compared to normal prostatic tissue [9, 11]. Also, downregulated or increased expression of several RISC proteins has been associated with cancer growth, apoptosis, as well as with the development of metastases [11, 12].
Based on these data, genetic variants potentially influencing the biogenesis and function of microRNAs qualify for candidates in case–control studies on PCa risk and progression [13–16]. Among these genetic variants are, therefore, those located in genes encoding the components of RISC: DICER1, AGO2, TARBP2, GEMIN3 and GEMIN4 [17].
To date, microRNA genetic variants have been evaluated for their potential association with PCa in both Asian and European populations [13–16]. Still, those located in RISC genes have been analyzed in a single study in Han Chinese [18]. The only component of RISC that was a subject of this study is GEMIN4. Evidence was obtained supporting the association of two GEMIN4 variants (rs2740348 and rs7813) with PCa risk. Furthermore, three genetic variants were found to be associated with the increased stage of localized PCa [18].
Since the association between variants located within genes encoding the components of RISC and PCa risk has not been analyzed in European populations, we conducted a case–control study in Serbian population. Potentially functional genetic variants chosen for this study are rs3742330 in DICER1, rs4961280 in AGO2, rs784567 in TARBP2, rs7813 in GEMIN4 and rs197414 in GEMIN3. Furthermore, we have evaluated the possible association of selected genetic variants with standard prognostic parameters of PCa progression, as well as with the risk of PCa progression. Genetic variants included in this study were selected based on their potential functional significance and also for their previously found association with PCa and/or other malignant diseases.
Materials and methods
The study used peripheral blood samples obtained from patients treated in the period between 2009 and 2013 at Clinical Centre “Dr Dragiša Mišović Dedinje”, Belgrade, Serbia. Research was conducted with the approval of ethics committee of this medical institution. Written informed consents were obtained from participants before their inclusion in the study. Experiments are in accordance with the Helsinki Declaration of 1964.
Three hundred and fifty-five samples of peripheral blood were obtained from patients with PCa and 360 samples from patients with benign prostatic hyperplasia (BPH). The control group comprised 318 healthy volunteers who gave samples of buccal swabs. The exclusion criteria for potential controls were the presence of any self-reported diseases and family history of PCa. Controls were recruited after passing standard annual physical examination. Mean ages for PCa, BPH patients and controls were 69.91, 68.10 and 69.11 years, respectively. Diagnoses of PCa and BPH were made by using standard clinical procedure which included digital rectal examination, transrectal ultrasonography, abdominal and pelvic ultrasound, bone scintigraphy and radiography, serum prostate-specific antigen (PSA) level and biopsy of the prostate. Serum PSA levels were determined by Hybritech method of monoclonal immunoassay. Clinical stage of cancer was determined according to TNM classification system. H&E-stained slides of paraffin-embedded prostate biopsy material were used to determine histological type of cancer and Gleason score (GS).
Patients with PCa were selected into groups based on the values of standard prognostic parameters—PSA at diagnosis (PSA < 10 ng/ml; 10 ng/ml ≤ PSA ≤ 20 ng/ml; PSA > 20 ng/ml), Gleason score (GS < 7; GS = 7; GS > 7) and clinical stage (T1; T2; T3/T4). Two groups of patients were formed based on the presence of distant metastases. Based on the risk for localized cancer progression, three groups of patients were formed, according to D’Amico criteria and as recommended by European Association of Urology (EAU). Groups were defined as low-risk (PSA < 10 ng/ml, GS < 7, and clinical stage T1–T2a), intermediate-risk (PSA 10–20 ng/ml or GS = 7 or clinical stage T2b–T2c) and high-risk (PSA > 20 ng/ml or GS > 7 or stage T3/T4) [19]. Since patients with metastases were included in the study, the criteria were modified to include this subset into high-risk group. Patients were also selected into low-risk (Gleason score <7 and stage T1–T2) and high-risk (Gleason score ≥7 or stage T3/T4 or bone metastases) groups according to Medeiros et al. [20].
Genomic DNA was isolated from peripheral blood and buccal swab samples using the QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturers’ protocol.
Genotyping of rs3742330 was performed by high-resolution melting analysis (HRMA). Primers used to amplify 198-bp-long segment of DNA surrounding this genetic variant were designed by using Primer3 software [21, 22]: 5′-CAAAGTCTTCACTTCCCTGCCA-3′ and 5′-GATGTTTAACTCCTCTCCACGTGATC-3′. The amplifications were performed in 10 μl volumes containing 10–20 ng of genomic DNA, 1X MeltDoctor™ HRM Master Mix (Applied Biosystems, Foster City, CA, USA), 0.3 μM of both primers (Invitrogen, Life Technologies, Grand Island, NY, USA) and nuclease-free water (Serva, Westbury, NY, USA). PCR cycling included an initial denaturation and enzyme activation at 95 °C for 10 min followed by 40 cycles of 15 s at 95 °C and 15 s at 60 °C. The heteroduplex formation step included denaturation at 95 °C for 10 s, followed by 30 s at 40 °C, while the melt curve run included hold at 60 °C for 1 min, 15 s at 95 °C with ramp rate of 0.3 % during which fluorescence was acquired and 15 s at 60 °C. The HRMA was performed by using High Resolution Melting Software version 3.0.1 (Applied Biosystems, Foster City, CA, USA).
Genotyping of rs4961280 was performed by custom-designed real-time PCR-based genotyping assay using specific probes (PrimerDesign Ltd, Southampton, UK). The assay was carried out using the standard method recommended by the manufacturer.
HRMA was used to genotype rs784567. Primers used to amplify 198-bp-long segment of DNA surrounding this genetic variant were designed by using Primer3 software [21, 22]: 5′-AGCCCTGCGGAAACAGAG-3′ and 5′-GTCGGATCCTGGCTCTTTG-3′. PCR amplification, melting curve run and the analysis of the obtained results were conducted as described for rs3742330.
Genotyping of rs7813 was performed using TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA). The assay was carried out using the standard method recommended by the manufacturer.
Genetic variant rs197414 was genotyped by using PCR–RFLP method. Primers used to amplify a segment of DNA surrounding rs197414 were designed by using Primer3 software [21, 22]: 5′-TCTTCCCAGAGCAAAGGAAA-3′ and 5′-TGGTGGTTGTTCCAAAGAAA-3′. The 15-µl PCR reaction mixture contained approximately 50 ng of genomic DNA, 0.3 μM of both primers (Invitrogen, Life Technologies, Grand Island, NY, USA), 200 μM of each dNTP (Fermentas, Hanover, MD, USA), 1.5 μl of 10X PCR buffer A (containing 15 mM MgCl2, Kapa Biosystems, Woburn, MA, USA), 0.04 U/μl of Taq DNA polymerase (Kapa Biosystems, Woburn, MA, USA) and nuclease-free water (Serva, Westbury, NY, USA). After initial denaturation at 97 °C for 3 min, PCR reactions were run for 35 cycles: 95 °C for 60 s, 62 °C for 60 s and 72 °C for 60 s, while final extension was at 72 °C for 10 min. The amplified fragments were separated by 1.5 % agarose gel electrophoresis and stained with ethidium bromide. Their expected length was 111 bp. Ten microliters of PCR products were digested at 37 °C overnight with 1 U of MboI enzyme (Fermentas, Hanover, MD, USA) per single reaction (15-μl reaction mixture). Digested products were separated by 3 % agarose gel electrophoresis. The expected lengths of fragments resulting from restriction digest were 84 and 27 bp for CC genotype, 111, 84 and 27 bp for CA genotype and 111 bp for AA genotype.
Statistical analysis of SNP association was done using SNPStats software [23, 24]. Hardy–Weinberg equilibrium was assessed using exact test implemented in SNPStats software. Allelic and genotypic associations were evaluated by unconditional linear (for serum PSA level in PCa patients) and logistic regression method with adjustment for age. Separate comparisons were done for five different genetic models: allelic (log-additive), codominant, dominant, recessive and overdominant. Odds ratio (OR) and its 95 % confidence intervals (95 % CI) were used as risk estimates. The best-fitting models were determined by using Akaike information criterion (AIC).
Results
Clinical and pathological characteristics of PCa patients are presented in Table 1. According to available data, 15.8 % of PCa patients had distant metastases at diagnosis, while the most frequently determined serum PSA scores were higher than 20 ng/ml. Also, the majority of PCa patients had GG = 6 (53.8 %) as well as clinical stage T2 of primary cancer (55 %). Not all of the PCa patients were included in the tests of association of the analyzed genetic variants with the values of standard prognostic parameters and PCa progression risk due to the lack of data on initial serum PSA, GS or clinical stage in patients’ records. Also, for some patients, clinical stage was initially assessed according to different classification systems which could not be reliably converted to TNM stages. Furthermore, instead of GS, histological grades were obtained for several patients in the initial period of the collection of data.
Basic data of the genetic variants selected for the analysis in this study are presented in Table 2. Genotyping of all five genetic variants analyzed was successful for more than 99 % of subjects (Table 3). Genotype distributions, presented in Table 3, were not found to significantly deviate from HWE (Table 2).
By comparing genotype distributions among PCa patients and healthy controls, as well as among PCa and BPH patients, no evidence of association between the analyzed genetic variants and PCa was found (Table 3). Nevertheless, a statistical trend of significance was reached for association between rs784567 and the risk of developing PCa (P = 0.055, for dominant model). Also, rs4961280 minor allele A was found to confer the reduced risk of developing BPH (P = 0.03; OR 0.74, 95 % CI 0.56–0.97, for log-additive model), while the opposite direction of association was found for rs784567 minor allele A (P = 0.013; OR 1.52, 95 % CI 1.09–2.11, for dominant model; results not shown).
Allele A of rs784567 was found to confer the reduced risk of higher serum PSA level at diagnosis (P = 0.046; Difference = −66.64, 95 % CI −131.93 to 1.35, for log-additive model). Besides for log-additive genetic model, statistical significance was obtained for dominant model (P = 0.05; Table 4). The analysis of association between other analyzed genetic variants and the serum PSA level at diagnosis did not show statistical significance for any genetic model tested (results not shown).
When genotype frequencies among PCa patients with GS < 7 and GS > 7 were compared, rs3742330 minor allele G was found to confer the decreased risk for high GS (P = 0.027; OR 0.40, 95 % CI 0.16–1.00, for log-additive model; Table 5). Furthermore, the comparison of genotype distributions among PCa patients with GS > 7 and GS = 7 showed the statistical trend for the association between this variant and GS (P = 0.059). Also, the same comparison showed that rs4961280 minor allele A confers the decreased risk of high GS (P = 0.045; OR 0.45, 95 % CI 0.20–1.00, for overdominant model). Conversely, opposite direction of association was observed in comparison of rs4961280 genotype distributions among patients with GS < 7 and GS = 7 (P = 0.033; OR 1.64, 95 % CI 1.04–2.58, for log-additive model).
By comparing genotype frequencies among subgroups of PCa patients with primary tumor clinical stages T1 and T2, rs4961280 and rs7813 minor alleles were found to be associated with the lower stage disease (P = 0.0013; OR 0.34, 95 % CI 0.18–0.66, for dominant model; and P = 0.035; OR 0.46, 95 % CI 0.22–0.97, for dominant model, respectively; Table 6). The results for rs4961280 were also confirmed by the comparison involving PCa patients with T3 and T1 stages (P = 0.037; OR 0.47, 95 % CI 0.23–0.96, for dominant model). DICER1 genetic variant rs3742330 was also shown to be associated with the decreased risk for high clinical stage, when comparing genotype distributions among PCa patients stratified into groups with T3 and T2 stages (P = 0.022). The exact OR could not be calculated, since there were no PCa patients with T3 stage who had GG genotype.
The genetic variants tested were not shown to be associated with the presence of bone metastases among PCa patients (results not shown).
When classifying PCa patients according to D’Amico criteria for disease aggressiveness, tests for genetic associations yielded no statistically significant results (results not shown). Conversely, when PCa patients were selected according to different criteria for progression risk assessment, rs3742330 minor allele G was found to be associated with the lower PCa aggressiveness (P = 0.036; OR 0.14, 95 % CI 0.023–1.22, for recessive model; Table 7). The same comparison showed statistical trend of association between rs7813 and the risk of PCa progression (P = 0.074, for overdominant model; results not shown).
Discussion
Dysregulation of microRNA-based regulatory mechanisms was found to be involved in the pathogenesis of PCa. These observations mainly refer to microRNAs, but similar findings were obtained for components of microRNA machinery [8, 25]. To date, evidence was found to support the association of genetic variants in microRNA genes with PCa risk and/or progression in Asians, as well as in Europeans, as found in our previous studies in Serbian population [13–16]. Nevertheless, a single case–control study on PCa analyzed the variants within microRNA machinery genes [18]. The mentioned study provided evidence of association between two genetic variants in GEMIN4 and PCa risk and progression [18]. Still, these results needed to be replicated in other populations in order to validate the observed effects of GEMIN4 variants on PCa risk/progression. Therefore, we decided to conduct the first case–control study on a GEMIN4 variant and PCa in a European population.
The exonic genetic variant in GEMIN4 chosen for this study is rs7813, for which it was previously found that genotype TT confers the increased risk of developing PCa [18]. The same genotype was also found to be associated with the higher clinical stage of PCa [18]. Our result did not support the association of this genetic variant with the risk of developing PCa. The discordance with the previous results can be explained by the differences in ethnic backgrounds, since they were obtained in a European and an Asian population. Also, the potential reasons for differences in the observed effect of rs7813 on PCa could be unmatched subject recruitment procedure. Still, our results show similarities with previously obtained regarding PCa progression, even though patient classifications differed between these two studies. Findings from both studies qualify rs7813 allele G for protective allelic variant against disease aggressiveness.
Other genetic variants selected for the present study were never previously analyzed for association with PCa. DICER1 variant rs3742330, located in the 3′UTR, was found to be associated with both GS and clinical stage of primary PCa. Also, this genetic variant was shown to be related to disease aggressiveness. These findings, even though unique for PCa, are consistent with previously obtained suggesting the association with osteosarcoma prognosis, with minor allele G being protective [26]. Similarly, minor allele of rs784567 located in TARBP2 promoter was found to be associated with the lower serum PSA level. Still, these results are in contrast to those obtained for renal cell carcinoma outcome [27].
Genetic variant rs4961280 located in AGO2 promoter showed opposing direction of association with GS when different groups of PCa patients were compared. This can be explained by the relatively small number of patients within some GS-based groups. More significant are results regarding the association with the clinical stage of PCa, suggesting the protective effect of minor allele A. The observed results are concordant with the potential biological function of rs4961280 as a repressive promoter variant, since AGO2 was found to be upregulated in PCa [28].
Our study provided no evidence of association between the analyzed genetic variants in RISC genes and PCa susceptibility. Nevertheless, the protective effect of rs3742330, rs4961280 and rs7813 on PCa progression risk was shown. The most significant results obtained in multiple comparisons are those implying the association of rs3742330 located in DICER1 with both GG and clinical stage of PCa, as well as with cancer aggressiveness. Still, in order to make further conclusions about the association between the analyzed genetic variants and the risk of PCa progression, additional studies in European and non-European populations are required.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65:87–108
Mühlberger N, Kurzthaler C, Iskandar R, Krahn MD, Bremner KE, Oberaigner W, Klocker H, Horninger W, Conrads-Frank A, Sroczynski G, Siebert U (2015) The ONCOTYROL prostate cancer outcome and policy model: effect of prevalence assumptions on the benefit-harm balance of screening. Med Decis Mak 35:758–772
Klotz L (2012) Active surveillance for favorable-risk prostate cancer: background, patient selection, triggers for intervention, and outcomes. Curr Urol Rep 13:153–159
Goh CL, Eeles RA (2014) Germline genetic variants associated with prostate cancer and potential relevance to clinical practice. In: Cuzick J, Thorat MA (eds) Prostate cancer prevention. Springer, Heidelberg, pp 9–26 (Recent Results in Cancer Research; vol 202)
Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13:271–282
Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T (2011) MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 59:671–681
Zimmerman AL, Wu S (2011) MicroRNAs, cancer and cancer stem cells. Cancer Lett 300:10–19
Patil PA, Magi-Galluzzi C (2015) MicroRNA in prostate cancer: practical aspects. Histol Histopathol 30(12):11647
Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R (2006) Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol 169:1812–1820
Wang YL, Wu S, Jiang B, Yin FF, Zheng SS, Hou SC (2015) Role of MicroRNAs in prostate cancer pathogenesis. Clin Genitourin Cancer 13:261–270
Bian XJ, Zhang GM, Gu CY, Cai Y, Wang CF, Shen YJ, Zhu Y, Zhang HL, Dai B, Ye DW (2014) Down-regulation of Dicer and Ago2 is associated with cell proliferation and apoptosis in prostate cancer. Tumour Biol 35:11571–11578
Zhang B, Chen H, Zhang L, Dakhova O, Zhang Y, Lewis MT, Creighton CJ, Ittmann MM, Xin L (2014) A dosage-dependent pleiotropic role of Dicer in prostate cancer growth and metastasis. Oncogene 33:3099–3108
Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang ZD, Tong N, Wang JF, Song NH, Zhang W, Hua LX, Wu HF (2010) A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate 70:467–472
George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD (2011) Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep 38:1609–1615
Nikolić ZZ, Pavićević DLS, Vukotić VD, Tomović SM, Cerović SJ, Filipović N, Romac SP, Brajušković GN (2014) Association between genetic variant in hsa-miR-146a gene and prostate cancer progression: evidence from Serbian population. Cancer Causes Control 25:1571–1575
Nikolić Z, Savić Pavićević D, Vučić N, Cidilko S, Filipović N, Cerović S, Vukotić V, Romac S, Brajušković G (2015) Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol 99:145–150
Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640
Liu J, Liu J, Wei M, He Y, Liao B, Liao G, Li H, Huang J (2012) Genetic variants in the microRNA machinery gene GEMIN4 are associated with risk of prostate cancer: a case-control study of the Chinese Han population. DNA Cell Biol 31:1296–1302
D’Amico AV, Whittington R, Malkowicz B, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A (1998) Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 280:969–974
Medeiros RM, Morais A, Vasconcelos A, Costa S, Pinto D, Oliveira J, Ferreira P, Lopes C (2002) Outcome in prostate cancer: association with endothelial nitric oxide synthase Glu-Asp298 polymorphism at exon 7. Clin Cancer Res 8:3433–3437
Primer3 software. http://primer3.ut.ee/
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3: new capabilities and interfaces. Nucleic Acids Res 40:e115
SNPStats Prevention Program. http://bioinfo.iconcologia.Net/SNPstats
Solé X, Guinó E, Valls J, Iniesta R, Moreno V (2006) SNPstats: a web tool for the analysis of association studies. Bioinformatics 22:1928–1929
Huang JT, Wang J, Srivastava V, Sen S, Liu SM (2014) MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol 4:113
Weng Y, Chen Y, Chen J, Liu Y, Bao T (2016) Common genetic variants in microRNA processing machinery genes are associated with risk and survival in patients with osteosarcoma. Mol Genet Genom 291:511–511
Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X (2010) Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 31:1805–1812
Fu X, Xue C, Huang Y, Xie Y, Li Y (2010) The activity and expression of microRNAs in prostate cancers. Mol BioSyst 6:2561–2572
Acknowledgments
The research was supported by the Ministry of Education, Science and Technological Development of Serbia (Project No. 173016). The scientific work of Zorana Nikolic was supported by the PhD student scholarship provided by the Ministry of Education, Science and Technological Development of Serbia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Authors’ contribution
Protocol development, data collection, data analysis and manuscript writing were done by Z Nikolić. Protocol development, data analysis and manuscript editing were carried out by D Savić Pavićević. Data collection and data analysis were performed by N Vučić, S Cerović and V Vukotić. Protocol development, data collection, data analysis and manuscript writing/editing were done by G Brajušković.
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments.
Rights and permissions
About this article
Cite this article
Nikolić, Z., Savić Pavićević, D., Vučić, N. et al. Genetic variants in RNA-induced silencing complex genes and prostate cancer. World J Urol 35, 613–624 (2017). https://doi.org/10.1007/s00345-016-1917-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-016-1917-0