Abstract
FT homologs are crucial for the flowering process, playing a vital role as ‘florigens’ in plants. In this study, we isolated and characterized an FT homolog, FaFT1, from cultivated strawberry. Nucleotide sequence analysis revealed a 531-bp open reading frame in FaFT1, encoding a putative protein with typical DPDxP and GxHR motifs belonging to the PEBP family proteins. For vegetative tissues or organs, qRT-PCR revealed that FaFT1 was primarily expressed in leaves. Notably high expression levels were detected in flowers and fruits, including the pith and cortex of the receptacle. Analysis of potential putative cis-acting regulatory elements (CREs) in this gene promoter indicated that many of them are associated with plant hormonal responses and abiotic stress responses. Expression detection confirmed that GA3 treatment enhanced the expression of FaFT1. When ectopically expressed in Arabidopsis, FaFT1 could promote the flowering process under SD conditions. These results suggested the possible role of FaFT1 in the regulation of reproductive development in cultivated strawberry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flowering is regulated by different cues, such as hereditary characteristics or plant hormone levels, the endogenous factors, temperature or day length, the environmental conditions. There are six defined distinct genetic pathways involved in flowering induction, including the autonomous, photoperiod, vernalization, gibberellin, thermosensory, and age pathways (Srikanth and Schmid 2011). In Arabidopsis, it has been reported that more than 300 genes are implicated in flowering-time regulation (Bouché et al. 2016). Such signaling pathways responding to endogenous or environmental signals converge on key regulators, such as FLOWERING LOCUS T (FT), which then activate other floral homeotic genes. FT proteins contain the phosphatidylethanolamine-binding (PEBP) domain (Pin and Nilsson 2012; Putterill and Varkonyi-Gasic 2016) and belong to the PEBP protein family. In the LD (long-day) plant Arabidopsis, FT is induced by photoperiod and other activators in leaves and then moves from leaves to the shoot apex through the phloem (Krzymuski et al. 2015). In the shoot apex, together with the bZIP transcriptional factor FD (FLOWERING LOCUS D), FT promotes the activation of other key flowering regulation genes, such as LEAFY (LFY) and APETALA1 (AP1), to induce flowering (Abe et al. 2005; Pin and Nilsson 2012; Song et al. 2013).
The functions of FT-like proteins in flowering regulation are largely conserved in different photoperiodic sensitive plants. In rice, which flowers earlier in SD (short-day) conditions, the flowering process is also induced by two FT homologs, HEADING DATE 3a (Hd3a) and Rice FT 1 (RFT1), under SD or LD conditions, respectively (Komiya et al. 2008). However, there are also other cases. In sugar beets, two FT homologs, BvFT1 and BvFT2, play antagonistic roles in flowering regulation by acting as inhibitor or activator, respectively (Pin et al. 2010). Similar results were also found in poplar (Hsu et al. 2011) and soybean (Zhai et al. 2014). Considering the importance of flowering regulation in crop production, the function of FT homologs in different economically significant crops needs to be verified separately.
Cultivated strawberry (Fragaria × ananassa) is one of the most economically important berry crops worldwide. Most cultivars of strawberry are Junebearing SD plants, which means that they are induced to flower in a decreased photoperiod and lower temperature in autumn (Stewart and Folta 2010). There are also some strawberry cultivars that flower perpetually and do not require SDs or low-temperature for flowering, known as Everbearing types (EB). Flowering habits are an important agricultural trait for most crops, and different flowering times or flower numbers lead to varied marketing times or yields in crops. Understanding the molecular regulation mechanism controlling strawberry flowering is of crucial importance for the breeding or production of this important berry crop.
The FT homolog of Fragaria was first reported in Fragaria vesca, the diploid wild strawberry (2n = 2x = 14). The expression of FvFT1 is detected in leaf tissues exclusively under LDs (Koskela et al. 2012). Further studies found that FvFT1 could promote the expression of FvSOC1, which then activates the expression of FvTFL1, the floral inhibitor in F. vesca (Rantanen et al. 2014). It seems that the expression of FvFT1 correlates negatively with flowering in SD genotypes of F. vesca. However, knockdown of FvFT1 in perpetual flowering F. vesca accession H4 by RNAi produced a clearly late flowering phenotype under LD conditions (Koskela et al. 2012), which implies that FvFT1 functions as the floral promoter at least when the function of TFL1 is absent.
Nakajima et al. (2014) first reported the Fragaria ananassa FT homologue FaFT1 (AB840273) which was isolated from cultivated strawberry cv. Tochimotome. It was highly expressed in leaves under non-inductive long-day and high-temperature (LDHT) condition, but not expressed in leaves under inductive short-day and low-temperature condition (SDLD). Further study by Koembuoy et al. (2017) revealed that ectopic expression of this gene in Arabidopsis couldn’t cause an early flowering phenotype.
Another two strawberry FT homologues, FaFT1 (KP184716) and FaFT1 (LC017713), were reported by Lei et al. (2015) and Nakano et al. (2015) individually in the cultivated strawberry cv. Camarosa and cv. Nyoho, respectively. Although FaFT1 (KP184716) exhibited a diurnal circadian rhythm both under SD and LD conditions, its expression level under SD is lower. While ectopic expression of FaFT1 (KP184716) in Arabidopsis caused an early flowering phenotype in LD. FaFT1 (LC017713) was expressed with a much higher level in the leaves under long-day condition than in SD condition. Wang et al. (2017) also reported a FT homologue of cultivated strawberry, FaFT (FR729042). Ectopic expression of this gene in tobacco produced a moderately late flowering time.
Considering the crucial role played by FT homologs in the flowering regulation process in plants and the inconsistent results about FaFTs in different reports, detailed information about the FT homolog in cultivated strawberry, the octoploid species (2n = 8x = 56) of Fragaria is needed.
In this work, FaFT1 (GenBank: JQ364958), an FT homolog of cultivated strawberry cv. Huaji was isolated and characterized. Based on the promoter information analysis, its expression in response to gibberellin was further investigated. Finally, expression profile and ectopic expression in Arabidopsis indicated its possible role not only in flowering time but also in fruit development. Our data provide more insights about the molecular information of FT homologs in cultivated strawberry.
Materials and Methods
Plant Materials and Nucleic Acid Extraction
F. ananassa cv. Huaji, a Junebearing cultivar, was used in this study. Strawberry plants were grown under natural conditions in a solar greenhouse at Shenyang Agricultural University. The plants were planted in September and blossomed and set fruits in December to April of the next year. The temperature in the greenhouse in this period was approximately 2–10 °C at night and 10–30 °C at daytime. To keep the solar greenhouse inside warmer, the blanket was also used at night in addition to the PVC plastic sheet at the top of the solar greenhouse from November to March. The average sunshine time in this period was approximately 9 h per day, which is an SD condition for strawberry. After March, the blanket was no longer used and it turns to LD conditions after March.
Tissues and flower organs, including leaf, stolon stem, stolon bud, flower, fruit and separated flower organs, were separated individually from the plants and then immediately frozen in liquid nitrogen for RNA extraction. Flowers were collected at 3–5 days after anthesis, and young leaves were approximately 80–100 mm in length. Total RNA of the relative tissues or organs was extracted by the modified CTAB method as described (Chang et al. 2007). DNA was extracted from young leaves. All the tissues or organs used for expression detection were collected on April 20, 2019 at 9:00 AM (4 h after dawn), which is an LD condition for strawberry with approximately 13 h of sunshine per day.
Wild-type Arabidopsis of Columbia (Col) was used in the ectopic expression analysis.
Cloning of the FaFT1 Gene
Gene-specific primers FaFTfwd and FaFTrev were designed based on the reported diploid strawberry (F. vesca) FT homolog FvFT1 (JN172098). Primer information and the PCR program are listed in Table 1. Leaf cDNA and DNA were used for cDNA fragment and gDNA fragment amplification, respectively. LA Taq (TaKaRa) was used in the PCR. First-strand cDNA was synthesized using a PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa) following the instruction manual.
The amplified fragments were cloned into the pGEM-T easy vector (Promega) individually and then sequenced with the Sanger method.
Isolation of the Promoter Sequence
Compared to diploid strawberry, genomic information of octoploid strawberry is insufficient. A forward primer PFF (Table 1) was designed based on the FvFT1 promoter detected in the diploid strawberry genome data at GDR (Genome Database for Rosaceae) website: https://www.rosaceae.org/species/fragaria/all. The reverse primer PFR was designed based on the FaFT1 DNA sequence. PCR was performed using leaf DNA as the template.
Bioinformatic Analysis
The amino acid sequences were aligned with Clustal W. A phylogenetic tree was generated using the neighbor-joining method. Boot strap values were derived from 1000 replicate runs. The phylogenetic tree was constructed by MEGA 7.0 software (Kumar et al. 2016).
Cis-acting regulatory elements (CREs) distributed in the putative FaFT1 promoter were analyzed by PlantCARE online software (Lescot et al. 2002): http://bioinformatics.psb.ugent.be/webtools/plantcare/html/.
Expression Detection
Vegetative tissues/organs, such as leaf, stolon stem and stolon bud, and reproductive tissues/organs, such as flower, inflorescence stem and fruits, were used to detect the gene expression profile. Four whorls of flower organs were also included in the expression detection. For expression detection in fruit tissues, green fruit without seeds was cut in half and the pith and cortex were separated individually for RNA extraction (Fig. S1).
To investigate the regulatory effect of hormones on FaFT1 expression, GA3 solution was sprayed onto the surface of strawberry leaves. The treated plants were at flowering and were grown in the same solar greenhouse. The treatment started on April 25, 2019, which is an LD condition for strawberry. A stock solution of 100 mM GA3 (BBI) was made in ethanol and diluted with water before application. Based on the previous reports (Thompson and Guttridge 1959; Kang et al. 2013), the final treatment concentration of GA3 in this study was 500 μM. The GA3 solution was sprayed every 2 days. Water was sprayed onto the mock control. Considering that frequent leaf removal of the strawberry plant may affect its vegetative/reproductive development balance, which then should affect the expression of FaFT1 in the samples, we sprayed GA3 or water at different times before sampling ( − 144 h, − 96 h and − 48 h). All the samples were collected at the same time, including the mock control. For the mock control, water was sprayed at each spraying time point but was only sampled on the last day. Treated leaf samples were collected at 9:00 AM, which is about 4.5 h after dawn.
Quantitative PCR (qRT-PCR) was conducted using SYBR® Premix Ex Taq™ II (TaKaRa) with the gene-specific primer sets FaFTF and FaFTR on an ABI 7500 Real-Time PCR system (Applied Biosystems). Three biological and three technical replicates were performed to calculate the relative expression level using the comparative Ct(2−ΔΔCt) method after normalization to the 26S rRNA internal transcript control.
Ectopic Expression of FaFT1 in Arabidopsis
The FaFT1 overexpression construct was made by inserting a full-length cDNA clone of FaFT1 in sense orientation into the Nco I and Bgl II sites of the plasmid pCAMBIA1304 under the control of the CaMV 35S promoter (Fig. S2). The target plasmid was then introduced into the disarmed strain of Agrobacterium tumefaciens GV3101.
Arabidopsis plants (Col) were used for transformation with floral-dip method (Clough and Bent 1998). After hygromycin selection and PCR confirmation, the transformants were transplanted into moistened potting soil consisting of vermiculite and perlite [1:1 (V:V)] after they developed 2–5 adult leaves. Both the transformants and the WT were all grown in a chamber with a short-day (SD) treatment (8 h light/16 h dark). The chamber temperature was set at 23 °C. The phenotypes of 12 plantlets in each typical independent T3 generation line were analyzed. The day of sowing was counted as day 0.
Flowers and siliques from different plants of the lines were collected for qRT-PCR detection of FaFT1. Leaves were collected for PCR confirmation of the FaFT1 insertion. Primers for both qRT-PCR and PCR were FaFTF and FaFTR. The amplified FaFT1 segment was 261 bp in length.
Results
Isolation and Characterization of FaFT1
Based on the sequence information of FvFT1 in diploid strawberry, the gene-specific primers FaFTfwd and FaFTrev were used to isolate the FT homolog in cultivated strawberry. One cDNA fragment which is about 500 bp was amplified and then cloned into the T-vector. Sequencing results showed that it is 531 bp in length and encodes a putative protein with 176 amino acid residues. This FT homolog was named as FaFT1 (GenBank: JQ364958). Sequence alignment indicated that there are 3 nucleotide changes in FaFT1 and FvFT1, resulting in one amino acid difference between their coding proteins.
Alignment of FaFT1 with other FT-like proteins showed that they share the typical D-P-D-x-P and G-x–H-R motifs (Fig. 1), which are identical to PEBP family proteins (Banfield and Brady 2000). Multiple sequence alignment also revealed that FaFT1 contains the conserved key amino acid residue Tyr84 (Y) in the position corresponding to Tyr85 in AtFT, which has been reported to be the crucial position for differentiating FT from TFL1 (TERMINAL FLOWER 1) (Hanzawa et al. 2005).
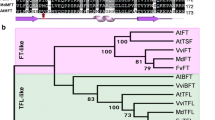
Phylogenetic analysis indicated that FaFT1 falls into a small clade with other FaFT1s (KP184716, AB840273 and LC017713) and the F. vesca FvFT1, distinctly separated from TFL1-like proteins (Fig. 2A). FaFT (FR729042), together with the other two FT homologs of F. vesca, FvFT2 and FvFT3, falls into another small clade.
A Phylogenetic analysis of FaFT1 with other FT/TFL1 homologous proteins. FaFT1(JQ364958) is indicated with a black circle, other FaFT1s and FaFT are indicated with a white circle. Three FT homologs in Fragaria vesca are indicated with triangles. B The intron (line) distributions of several FT-like genes. Sequence length is shown at the end of each gene. Proteins used in phylogenetic tree construction are: Fragaria ananassa FaFT1(AB840273), BAP18900, FaFT1(LC017713), BAR63708, FaFT1(KP184716), AKO90119, FaFT1(JQ364958), AFA42331, FaFT(FR729042), CBY25183; Fragaria vesca FvFT1, AEP23098, FvFT2, XP_004297273, FvFT3, XP_004295610; Arabidopsis thaliana AtFT, NP_176726; Malus domestica MdFT1, NP_001280791, MdFT2, NP_001280810; Allium cepa AcFT1 AGO81838; Beta vulgaris BvFT1 XP_010690385, BvFT2, XP_010673871; Dimocarpus longan DlFT1, AEZ63949, DlFT2, AEZ63950; Nicotiana tomentosiformis NtFT1, XP_009592530, NtFT2, XP_016459944; Fragaria ananassa FaTFL1, BAO03909; Fragaria ananassa FaTFL1-like, AFA42328, Pyrus bretschneideri PcTFL1-2, NP_001289244; Eriobotrya japonica EjTFL1-1, BAD10966; Pyrus bretschneideri PcTFL1-1, XP_009374732; Malus domestica MdTFL1a, NP_001280794, MdTFL1, NP_001280887; Vitis vinifera VvTFL1B, NP_001267931; Citrus sinensis CsTFL, NP_001275848; Cydonia oblonga CoTFL1-2, BAD10970; Arabidopsis thaliana AtTFL1, NP_196004
The genomic DNA sequence of FaFT1, gFaFT1, was also isolated, and sequencing results showed that it is 2104 bp in length. Alignment of gFaFT1 with its cDNA sequence revealed that there are three introns with nucleotide lengths of 238 bp, 1233 bp and 102 bp, respectively (Fig. 2B). The number of introns is similar to that of other FT-like genes, such as MdFT2 of apple and PtFT of poplar. However, the position and length of their introns differ. gFaFT1 and gFvFT1 shared not only the same exon number but also the same length for each exon. However, the introns of these genes also differ from each other, especially for their second introns.
Cis-Acting Regulatory Element Analysis of the FaFT1 Promoter
Based on the primer pair PFF and PFR, a DNA fragment of 1,588 bp was isolated. Compared with the complete cDNA sequence of another reported octoploid strawberry FT homolog Flowering Locus T-like 1 (LC017713), the 5′UTR of FaFT1 is 53 bp in length and the promoter we isolated is 1,481 bp in length.
Sequence analysis showed that the FaFT1 promoter contained several typical TATA-boxes, the core promoter elements, and CAAT-boxes, the common cis-acting elements. In addition to these fundamental elements, many plant hormone-responsive elements were detected, including two ABA-responsive elements, three GA-responsive elements and one MeJA-responsive element (Fig. 3, Table 2). We also identified two CREs that are associated with low-temperature in the promoter region of FaFT1.
In the photoperiod flowering pathway, transcription of FT is normally activated by CONSTANS (CO) by binding at the CO response element (CORE). However, in the isolated 1481-bp FaFT1 promoter, no typical CORE motif (TGTG(N2-3)ATG) was found. While three NF-Y complex binding sites (CCAAT-box), which are also possible CO binding sites, were detected.
Expression Profile of FaFT1 in Strawberry Tissues and Organs
Expression detection of FaFT1 in vegetative tissue/organs showed that a higher expression level was found in leaves than in stolon stems and stolon buds (Fig. 4A). This result is consistent with the expression profile of other FT homologs, such as the AtFT of Arabidopsis, whose expression is also activated in leaves.
Expression detection of FaFT1. A Expression in different tissues/organs. L: leaf, SS: stolon stem, SB: stolon bud, IS: inflorescence stem, Fl: flower, Fr: fruit. B Expression in floral organs. L: leaf, Se: sepal, P: pistol, Stm: stamen, Stg: stigma. C Expression in fruit. GF: green fruit, P: pith, C: cortex. The average expression level of three biological replicates is shown for each sample, all normalized to the expression level of Fragaria × ananassa 26S rRNA. Error bars indicate ± SD
Interestingly, unexpectedly high expression levels were also detected in most of the detected reproductive tissues/organs. FaFT1 showed notably high expression levels in the flowers and the fruits of strawberry, with approximately 12-fold and 20-fold higher expression levels compared with that in leaves, respectively. For this unexpected expression result, we further detected its expression in flower and fruit in detail.
For the four whorls of floral organs, FaFT1 showed a similar transcription pattern in sepals as in leaves, while a considerably lower expression level was detected in the other three whorls of floral organs (Fig. 4B).
Meanwhile, we detected the expression of this gene in green fruit and separated fruit tissues, including the pith and cortex. The results showed that green fruit indeed showed considerably stronger FaFT1 transcription with an approximately 20-fold higher expression level than that in the leaf. Stronger expression was also detected in pith and cortex tissues of the fruit individually. In addition, our results also indicated that much higher FaFT1 expression level was found in the pith than in the cortex (Fig. 4C).
GA3 Regulates the Expression of FaFT1
CRE analysis of the FaFT1 promoter suggested that the expression of this gene might be regulated by plant hormones. To test this hypothesis, we also detected changes in FaFT1 expression after treatment with GA3.
The expression of FaFT1 in leaves was significantly upregulated at 48 h after GA3 spraying with approximately 1.7-fold expression change compared to the mock (Fig. 5). Furthermore, FaFT1 expression was detected to be continuously upregulated following the repetitive spraying of GA3. After 6 days, at which point GA3 had been sprayed three times, the expression level of FaFT1 in leaves was ninefold higher than that in the mock.
Expression of FaFT1 in Fragaria × ananassa leaves in response to GA3 treatment. Changes in FaFT1 expression after every 2 days of spraying with 500 μM GA3 are shown. The average expression level of three biological replicates is shown for each time point, all normalized to the expression level of Fragaria × ananassa 26S rRNA. Error bars indicate ± SD
Phenotypic Analysis of FaFT1 Transgenic Arabidopsis
The FT-like gene generally promotes the flowering process of transformants overexpressing it for its well-known function as ‘Florigen’. To evaluate the function of FaFT1, we generated transgenic Arabidopsis plants carrying FaFT1 driven by the CaMV35S promoter. The T3 generation transgenic lines were used for further analysis in this study.
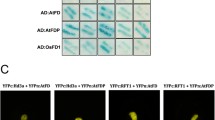
When grown in the noninductive SD conditions, most of the Arabidopsis transformants showed an early flowering phenotype compared with the wild type. Three typical independent T3 lines (T3-1, T3-2 and T3-3) were selected for further analysis. Insertion of FaFT1 in the genomic DNA was confirmed in the selected lines by PCR using primers FaFTF and FaFTR (Table 1, Fig. S3). Although the flowering times of these lines were not consistent with each other, all of them showed early flowering compared with the WT line (Figs. 6A, 7A).
Phenotypes of FaFT1 transgenic plants. A Early flowering phenotypes of three lines (T3-1, T3-2 and T3-3) in the T3 generation are shown, while WT were still in vegetative growth development. B Siliques of the WT (left) and the transformants (T3-1, right). C Flowers of the WT (left) and the transformants (T3-1, right) (Color figure online)
Promoted flowering process and reduced vegetative growth of the transformants. A Average days needed for WT and transformants to be flowering. B Number of rosette leaves when flowering in WT plants and transgenic lines. Error bars indicate ± SD (n = 13). Asterisks indicate significant differences between WT and transgenic lines (Student’s t-test, *P < 0.05, **P < 0.01)
Plantlets of line T3-1 showed the most severe early flowering phenotype compared with the WT, exhibiting flowering approximately 11 days earlier than the control. At the same time, the decreased rosette leaf numbers and the reduced leaf size at flowering also indicated shortened vegetative development (Figs. 6A, 7B). The other two lines also showed early flowering and shortened vegetative development, although their phenotypes were not as severe as those in line T3-1. Even in line T3-3, the latest flowering line among the three lines, plantlets flowered 2 days earlier on average than the control.
High FaFT1 expression levels were detected in the reproductive tissues of strawberry, such as fruit and flower. However, phenotypes of the siliques and flowers of the transformants were similar to those in WT (Fig. 6B, C), while notable FaFT1 expression was detected in flowers (Fig. 8A) and siliques (Fig. 8B) of three lines by qRT-PCR.
Discussion
FaFT1 is a Typical FT-Like Homolog of Strawberry
The physiology underlying flowering in cultivated strawberry has been extensively studied for nearly a century. However, due to the complex genetic background of this berry, studies aimed at elucidating the genetic basis of the flowering character via genetic mapping have led to varying results (Ahmadi et al. 1990; Weebadde et al. 2008). The closely related diploid woodland strawberry F. vesca has been widely used in functional genetic studies to elucidate the molecular pathways controlling flowering in Fragaria. Considerable progress has been made on the strawberry TERMINAL FLOWER1 gene (TFL1) (Koskela et al. 2016, 2017; Iwata et al. 2012; Rantanen et al. 2015), the competitor of FT in binding with FD (Hanano and Goto 2011).
However, the role of the FT homologue in flowering regulation of strawberry was not well elucidated as the TFL1 homolog. However, in F. vesca, FvFT1 was found to be expressed specifically under LDs and negatively correlated with the flower induction in SD genotypes of F. vesca (Koskela et al. 2012). A similar result was also found in the octoploid Junebearing strawberry (Nakano et al. 2015). Moreover, Koskela et al. (2012) reported that the FvFT1 RNAi lines, which were produced in an everbearing H4 background, grown in LD conditions showed a clearly late flowering phenotype. These results indicated that the role of FT homologs in Fragaria is more complex than that of the relevant genes in Arabidopsis or other plants. Characterization of FT homologs in cultivated strawberry is strongly warranted.
In this study, we isolated and characterized an FT-like gene, FaFT1, from cultivated strawberry. Sequence analysis showed that FaFT1 is a typical FT-like PEBP family member. In addition to the conserved key amino acid residue Tyr84 (Y), several other important amino acid residues that have been reported to be responsible for the FT-like function are also detected, such as Tyr133(Y) and Trp137(W) (Klintenäs et al. 2012; Wickland and Hanzawa 2015). The genomic sequence of FaFT1 consisted of four exons, which resembled the genomic structure of other FT genes. Such information about the FaFT1 sequence suggested its potential role in flowering regulation.
Several plant hormone-responsive and low-temperature responsive elements were detected in the promoter region of FaFT1, suggesting that FaFT1 expression may be affected by phytohormones and temperature. Further study (Fig. 5) confirmed that GA3 treatment significantly upregulated the expression of FaFT1. A previous study reported that treatment with bioactive GAs inhibited the flowering process of strawberry, while runner production was enhanced (Thompson and Guttridge 1959; Tafazoli and Vince-Prue 1978; Choma and Himelrick 1984). Further work is needed to clarify the possible role of FaFT1 in the GA regulation of flowering or runner formation in strawberry.
The CO-FT module plays central, crucial roles in the photoperiodic flowering pathway (Turck et al. 2008). Although the typical CORE motif was not found in the identified FaFT1 promoter, several CCAAT boxes (NF-Y complex binding sites) were detected, indicating that the expression of FaFT1 may also be regulated by the CONSTANS homolog of strawberry. In F. vesca, it was reported that FvCO up-regulates FvFT1 in light (Kurokura et al. 2017).
Expression Profile of FaFT1 Implies Its Function in Reproductive Development
Normally, florigen is synthesized in leaves (Turck et al. 2008). For the vegetative organs, FaFT1 showed a higher expression level in leaves than in others, such as stolon stems or stolon buds. Interestingly, compared with the expression in vegetative organs, its expression in the reproductive organs was found to be considerably higher (Fig. 4A). There are also some other similar reports indicating that FT-like genes are abundantly expressed in reproductive organs, such as flowers and pods in soybean (Sun et al. 2011), flowers and berries in grape (Sreekantan and Thomas 2006), and fruit skin and fruit pulp in banana (Chaurasia et al. 2017). Such findings suggest that FaFT1 may also play roles in reproductive development processes other than flowering.
Although the expression of FaFT1 is considerably higher in an integrated flower, its expression in each of the four whorls of flower organs individually is not higher than in the leaf (Fig. 4B). As a kind of accessory fruit, in addition to the four whorls of flower organs, the receptacle is also an integral part of the flower. The existence of the receptacle may be the reason that FaFT1 expression in an integrated flower is considerably higher than that in the leaf.
We further detected the expression of FaFT1 in the fruit, which developed from the receptacle. Gene expression results (Fig. 4C) confirmed our hypothesis, and its expression is considerably higher in the green fruit than in the leaf. Notably higher expression levels were also detected in the pith and cortex. For the receptacle tissue, the expression of FaFT1 is considerably higher in the pith than in the cortex. Considering the vital role of the receptacle in the development of strawberry fruit, this expression profile of FaFT1 implies its possible function in the regulation of strawberry fruit development.
FaFT1 Activates the Flowering Process of Arabidopsis
There are some different reports about the function of the Fragaria FT homolog in the regulation of the flowering process. In F. vesca, downregulation of FvFT1 in Hawaii-4 by RNAi resulted in a clearly late flowering phenotype, which indicated that FvFT1 functions as a flowering activator in F. vesca LD accessions (Koskela et al. 2012). In this study, when ectopically expressed in Arabidopsis, FaFT1 could promote the Arabidopsis flowering process under noninductive SD conditions, indicating that it could promote the flowering process in Arabidopsis.
While when FaFT (FR729042), another reported FT homolog of cultivated strawberry, ectopically expressed in tobacco, the transformants showed a moderately late flowering phenotype (Wang et al. 2017). Koembuoy et al. (2017) also reported that ectopic expression of FaFT1 (AB840273) in Arabidopsis only caused the aerial rosette phenotype under SD, and they deduced that FaFT1 (AB840273) acts at the vegetative propagation stage.
For those confusing results, we further checked the deduced protein sequence similarity coded by five reported FaFTs (JQ364958, AB840273, LC017713 and FR729042). The alignment results (Fig S4) showed that protein sequence of the FaFT1 (JQ364958) we isolated is same as the FaFT1 (KP184716) reported by Lei et al (2015), while FaFT1 (AB840273) and FaFT1 (LC017713) are the same one. Early flowering phenotype of the transgenic Arabidopsis in both our finding and Lei et al. (2015) indicate that FaFT1 (JQ364958/KP184716) could function as a flowering activator when ectopically expressed in Arabidopsis.
FaFT1 (JQ364958/KP184716) and FaFT1 (AB840273/LC017713) showed a high similarity with only two amino acid variations. In the study reported by Koembuoy et al. (2017), the role of FaFT1 (AB840273) in flowering induction was evaluated by counting Arabidopsis leaf numbers at the flowering stage, not only the rosette leaf number but also the cauline leaf number. The flowering time data of transgenic Arabidopsis was not shown. Different evaluation methods may result in the different conclusions deduced by our study and that study.
While sequence of FaFT (FR729042) owns much more variation compared with the other two FaFT1 sequences. Considering the late flowering phenotype in tobacco caused by ectopic expression of this gene, FaFT (FR729042) may function as a flowering inhibitor, just as the different roles of BvFT1 and BvFT2 in sugar beets (Pin et al. 2010).
However, further work should be performed to clarify the detailed function of FaFTs in strawberry. The expression of FaFT1 (AB840273) was induced in strawberry grown under noninductive LD and high-temperature conditions (Nakajima et al. 2014). Thompson and Guttridge (1959) reported that GA treatment on strawberry could suppress flower initiation and activate runner initiation, which normally occurs in strawberry plants grown under LD conditions. In this study, we found that GA3 treatment could activate the expression of FaFT1 (Fig. 5). All these findings imply the function of FaFT1 not as a flowering inducer in strawberry, at least in SD genotypes of strawberry. However, when ectopically expressed in Arabidopsis, FaFT1 promoted the flowering process of Arabidopsis, even in the non-inductive photoperiod. Such different results regarding the function of FaFT1 obtained from its expression character and its ectopic expression in Arabidopsis may derive from the varied flowering regulation mechanisms in strawberry and Arabidopsis. In the meantime, the extremely high expression level in strawberry fruit tissue (Fig. 4) implies that it should be involved in strawberry fruit development. However, Arabidopsis owns the silique fruit, which is different from the berry fruit of strawberry that developed from the expanded receptacle. The possible role of FaFT1 in strawberry fruit development cannot be verified by this work for the different fruit development mechanisms between Arabidopsis and strawberry. Further analysis of the possible function of FaFT1 in flowering and fruit development by overexpression or knockout in strawberry is required.
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Ahmadi H, Bringhurst RS, Voth V (1990) Modes of inheritance of photoperiodism in Fragaria. J Am Soc Hortic Sci 115:146–152
Banfield MJ, Brady RL (2000) The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J Mol Biol 297:1159–1170
Bouché F, Lobet G, Tocquin P, Périlleux C (2016) FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res 44:D1167–D1171
Chang L, Zhang Z, Yang H, Li H, Dai H (2007) Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J Phytopathol 155:431–436
Chaurasia AK, Patil HB, Krishna B, Subramaniam VR, Sane PV, Sane AP (2017) Flowering time in banana (Musa spp.), a day neutral plant, is controlled by at least three FLOWERING LOCUS T homologues. Sci Rep 7:5935
Choma ME, Himelrick DG (1984) Responses of day-neutral, June-bearing and everbearing strawberry cultivars to gibberellic acid and phthalimide treatments. Sci Hortic 22:257–264
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcription repression. Plant Cell 23:3172–3184
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102:7748–7753
Hsu CY, Adams JP, Kim H, No K, Ma C, Strauss SH, Drnevich J, Vandervelde L, Ellis JD, Rice BM, Wickett N, Gunter LE, Tuskan GA, Brunner AM, Page GP, Barakat A, Carlson JE, DePamphilis CW, Luthe DS, Yuceer C (2011) FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA 108:10756–10761
Iwata H, Gaston A, Remay A, Thouroude T, Jeauffre J, Kawamura K, Oyant LH, Araki T, Denoyes B, Foucher F (2012) The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J 69:116–125
Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25(6):1960–1978
Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O (2012) Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol 196:1260–1273
Koembuoy K, Nakajima R, Otagaki S, Kurokura T, Takahashi H, Nakazono M, Shiratake K, Matsumoto S (2017) Functional analyses of cultivated strawberry FT and TFL1 homologs. Acta Hortic 1156:95–102
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135:767–774
Koskela EA, Mouhu K, Albani MC, Kurokura T, Rantanen M, Sargent DJ, Battey NH, Coupland G, Elomaa P, Hytönen T (2012) Mutation in TERMINAL FLOWER 1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol 159:1043–1054
Koskela EA, Anita S, Flachowsky H, Heide OM, Magda-Viola H, Elomaa P, Hytönen T (2016) TERMINAL FLOWER1 is a breeding target for a novel everbearing trait and tailored flowering responses in cultivated strawberry (Fragaria × ananassa Duch.). Plant Biotechnol J 14:1852–1861
Koskela EA, Kurokura T, Toivainen T, Sønsteby A, Heide OM, Sargent DJ, Isobe S, Jaakola L, Hilmarsson H, Elomaa P, Hytönen T (2017) Altered regulation of TERMINAL FLOWER 1 causes the unique vernalisation response in an arctic woodland strawberry accession. New Phytol 216:841–853
Krzymuski M, Andrés F, Cagnola JI, Jang S, Yanovsky MJ, Coupland G, Casal JJ (2015) The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J 83:952–961
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kurokura T, Samad S, Koskela E, Mouhu K, Hytönen T (2017) Fragaria vesca CONSTANS controls photoperiodic flowering and vegetative development. J Exp Bot 68:4839–4850
Lei H, Guo X, Wang Y, Yao L, Wang S, Li T (2015) Identification and characterization of FaFT1: a homolog of Flowering Locus T from strawberry. Adv J Food Sci Technol 8(3):180–188
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Nakajima R, Otagaki S, Yamada K, Shiratake K, Matsumoto S (2014) Molecular cloning and expression analyses of FaFT, FaTFL, and FaAP1 genes in cultivated strawberry: their correlation to flower bud formation. Biol Plant 58:641–648
Nakano Y, Higuchi Y, Yoshida Y, Hisamatsu T (2015) Environmental responses of the FT/TFL1 gene family and their involvement in flower induction in Fragaria × ananassa. J Plant Physiol 177:60–66
Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35:1742–1755
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400
Putterill J, Varkonyi-Gasic E (2016) FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol 33:77–82
Rantanen M, Kurokura T, Mouhu K, Pinho P, Tetri E, Halonen L, Palonen P, Elomaa P, Hytönen T (2014) Light quality regulates flowering in FvFT1/FvTFL1 dependent manner in the woodland strawberry Fragaria vesca. Front Plant Sci 5:271
Rantanen M, Kurokura T, Jiang P, Mouhu K, Hytönen T (2015) Strawberry homologue of TERMINAL FLOWER1 integrates photoperiod and temperature signals to inhibit flowering. Plant J 82:163–173
Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18:575–583
Sreekantan L, Thomas MR (2006) VvFT and VvMADS8, the grapevine homologues of the floral integrators FT and SOC1, have unique expression patterns in grapevine and hasten flowering in Arabidopsis. Funct Plant Biol 33:1129–1139
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Stewart PJ, Folta KM (2010) A review of photoperiodic flowering research in strawberry (Fragaria spp.). Crit Rev Plant Sci 29:1–13
Sun H, Jia Z, Cao D, Jiang B, Wu C, Hou W, Liu Y, Fei Z, Zhao D, Han T (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6:e29238
Tafazoli E, Vince-Prue D (1978) A comparison of the effects of long days and exogenous growth regulators on growth and flowering in strawberry, Fragaria × ananassa, Duch. J Hortic Sci 53:255–259
Thompson PA, Guttridge CG (1959) Effect of Gibberellic acid on the initiation of flowers and runners in the strawberry. Nature 184:BA72–BA73
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594
Wang Z, Yang R, Devisetty UK, Maloof JN, Zuo Y, Li J, Shen Y, Zhao J, Bao M, Ning G (2017) The divergence of flowering time modulated by FT/TFL1 is independent to their interaction and binding activities. Front Plant Sci 8:697
Weebadde CK, Wang D, Finn CE, Lewers KS, Luby JJ, Bushakra J, Sjulin TM, Hancock JF (2008) Using a linkage mapping approach to identify QTL for day-neutrality in the octoploid strawberry. Plant Breed 127:94–101
Wickland DP, Hanzawa Y (2015) The FLOWERING LOCUS T/TERMINAL FLOWER1 gene family: functional evolution and molecular mechanisms. Mol Plant 8:983–997
Zhai H, Lü S, Liang S, Wu H, Zhang X, Liu B, Kong F, Yuan X, Li J, Xia Z (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9:e89030
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 31872073), the Liaoning Key R&D Program (2020JH2/10200032) and the Natural Science Foundation of Liaoning Province (No. 2014027015, 20180550431). Our sincere thanks go to Dr. Junhui Zhou of UMD for grammar checking. We also thank the editor and the reviewers for their valuable comments and suggestions.
Author information
Authors and Affiliations
Contributions
YL designed the research. WC, HL, DZ, YY, CL and AY performed the experiments. YL, WC, HL and ZZ wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Karine David.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2021_10409_MOESM1_ESM.tif
Supplementary file1 Fruit without seed cross-section showing pith (center) and cortex (flanking) tissues; the red line indicates the boundary of two tissues (TIF 91 kb) (Color figure online)

344_2021_10409_MOESM2_ESM.jpg
Supplementary file2 Vector construction. FaFT1 was inserted into the plasmid pCAMBIA1304. 35S promoter: the cauliflower mosaic virus (CaMV) 35S promoter (JPG 43 kb)
344_2021_10409_MOESM3_ESM.tif
Supplementary file3 PCR detection of FaFT1 in Arabidopsis T3 generation lines. WT: nontransgenic Arabidopsis as a negative control, 1–9: PCR detection results for T3 generation plantlets (TIF 51 kb)
344_2021_10409_MOESM4_ESM.tif
Supplementary file4 Sequence alignment of five reported FaFTs. Black and blue colors indicate higher and lower amino acid residue conservation, respectively. Red and blue boxes indicate the same protein sequences with different accession numbers (TIF 102 kb) (Color figure online)
Rights and permissions
About this article
Cite this article
Chen, W., Li, H., Zou, D. et al. Expression Profile of FaFT1 and Its Ectopic Expression in Arabidopsis Demonstrate Its Function in the Reproductive Development of Fragaria × ananassa. J Plant Growth Regul 41, 1687–1698 (2022). https://doi.org/10.1007/s00344-021-10409-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10409-z