Abstract
Gibberellic acid-stimulated transcript (GAST) proteins are widely distributed among plant species and are involved in various aspects of plant growth, plant responses to stress, and hormone crosstalk. However, in moso bamboo (Phyllostachys edulis), bioinformatics data and functional information on the GAST family are limited. Here, a total of eight GAST genes were first identified in the whole genome of moso bamboo and then clustered into three groups based on phylogenetic analysis. Each clade is well supported by the conserved intron/exon structures and motifs, suggesting that the members within the same subfamily perform similar functions. A number of development-, hormone-, and stress-related elements are randomly distributed in the promoter sequences of PheGASTs. Tissue expression analysis indicates that most PheGASTs might be associated with moso bamboo flower development and rapid shoot growth. Furthermore, the PheGASTs display phytohormone- and stress-inducible expression patterns in response to various circumstances (abscisic acid, gibberellins, cold, drought, and salinity). Among these, PheGASR1 was assumed to have a distinct regulatory role in rapid shoot growth and abiotic stress responses involving the ABA pathway. Collectively, our results provide extensive insights into the GAST gene family, thereby contributing to the screening of additional candidate genes in moso bamboo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibberellic acid-stimulated transcript (GAST) proteins are widely distributed among plant species and comprise three distinct domains: (1) a putative signal peptide of 18–29 residues; (2) a variable region displaying high levels of divergence among family members in terms of amino acid composition and sequence length; and (3) a C-terminal region of approximately 60 amino acids containing 12 cysteine residues at conserved positions, which is also known as the GASA domain (Aubert et al. 1998). To date, our understanding of the mode of action of GAST proteins is limited, although studies have suggested that the cysteine residues play a central role in their antioxidant activity (Ben-Nissan et al. 2004; Rubinovich and Weiss 2010; Rubinovich et al. 2014).
Most GAST genes are regulated by gibberellins (GAs) as well as by other hormones, such as abscisic acid (ABA), auxins (IAA), and brassinosteroids (BRs) (Zhang and Wang 2008; Wang et al. 2009; Liu et al. 2013; Sun et al. 2013; Lee et al. 2015; Jie et al. 2016). The GAST genes are also involved in various biological processes, including plant development and abiotic stress responses. For example, in strawberry, FaGAST is upregulated in the fruits and roots but downregulated in leaves and stolons, and its overexpression in transgenic Fragaria vesca causes a delay in growth and a reduction in fruit size (de la Fuente et al. 2006). Moreover, ectopic expression of FsGASA4 enhances salt, oxidative, and heat stress tolerance during seed germination in transgenic Arabidopsis thaliana by increasing salicylic acid (SA) biosynthesis (Alonso-Ramirez et al. 2009). Expression analyses indicate that the spatial and temporal regulation of GAST genes is highly specific, each displaying a distinct expression pattern, and their expression is mostly correlated with young tissues and actively growing organs (Zhang and Wang 2008; Zimmermann et al. 2010; Nahirñak et al. 2016). Overall, the GAST family participates in hormone signaling pathways that modulate hormonal responses and levels, as well as in a variety of plant developmental processes and environmental responses, thereby suggesting that these play essential functions in plants (Nahirñak et al. 2012).
China has extensive bamboo resources and a large bamboo industry, with a natural bamboo forest area of 6,010,000 ha. The total area of moso bamboo forest accounts for about 74% of the total area of bamboo forests in China, reaching about 4,430,000 ha. The annual felling of moso bamboo is 500 million–6 billion trees, and the output of various bamboo woods is about 4,500,000 tons, thereby providing valuable materials for the development of industrial and agricultural production, as well as cultural events in China (Wu et al. 2015). Additionally, moso bamboo is characterized by a prolonged vegetative phase before it flowers and a rapid shoot growth rate (as fast as 1 m/day at peak growth, reaching a height of 20 m in 45–60 days) (Peng et al. 2013a). GAs are essential endogenous hormones that play vital roles in controlling growth and a wide variety of developmental processes in plants. Treatment of moso bamboo seedlings with exogenous GA3 induces elongation of the culm internode and an increase in plant height, blade quality, and cell length (Jiang et al. 2016). Furthermore, during the rapid shoot growth phase, the shoot apical meristem (SAM) region produces high amounts of total GAS, and temporal changes in concentrations of endogenous GA3 are generally bimodal in different parts of the shoots (bottom, middle, and top) (Gamuyao et al. 2017; Li et al. 2017). A total of 57 genes that are involved in the GA pathway are differentially expressed during rapid shoot growth (Peng et al. 2013b). During the reproductive growth phase, GA metabolism genes related to flower development, such as GAMYB, GID1, and GID2, are upregulated at four different periods of flower development (Gao et al. 2014; Ge et al. 2017). The GAST family may act as downstream members of the DELLA protein family in the GA pathway (Zhang and Wang 2008). Our team has been investigating the functional roles of some GA-related genes (unpublished data), but the role of the GAST family is still not well understood. The present study performed genome-wide identification of the GAST family, followed by detailed bioinformatics analysis. Moreover, expression patterns of the PheGAST genes were also investigated during flower and shoot development, as well as in response to hormones (GA and ABA) and abiotic stresses (salt, drought, and cold). Thus, the goal of this study was to elucidate the role of the GAST gene family in moso bamboo tissue development, as well as in response to abiotic stress.
Materials and Methods
Identification of Putative Genes That Encode GAST Proteins
The GAST genes were searched in the bamboo genome database (BambooGDB, http://www.bamboogdb.org/) (Peng et al. 2013a) using known plant GAST sequences of a nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore) as query. The aligned result was filtered by searching Conserved Domain Database (https://www.ncbi.nlm.nih.gov/guide/domains-structures/). Only the gene models that contained intact GASA domains and hits with E values < 0.001 were identified as members of the PheGAST family. We also referred to the database of full-length non-chimeric (FLNC) reads (http://www.forestrylab.org/db/PhePacBio/), searching putative novel or mis-annotated GAST genes. To obtain a more comprehensive overview on PheGASTs, the GAST genes of Zea mays, Oryza sativa, A. thaliana, and Brachypodium distachyon were identified based on previous studies (Zhang and Wang 2008; Zimmermann et al. 2010; Nahirñak et al. 2016) and downloaded from Phytozome v11.0 (http://www.phytozome.net/). The sequences of Petunia hybrida GIP1, wheat TaGASR7-A1, and potato GAST1 were obtained from the protein database (https://www.ncbi.nlm.nih.gov/protein/308535464).
Phylogenetic Reconstruction
Multiple sequence alignment of the full-length protein sequences of PheGASTs was performed using Clustal X 2.1, and neighbor-joining (NJ) and maximum likelihood (ML) phylogenetic trees were constructed using MEGA 7.0 with 1000 bootstrap replicates, respectively. To compare the evolutionary relationship and the predicted functional roles of PheGASTs, phylogenetic trees were also constructed using the amino acid sequences of moso bamboo and other species using the same method. To support subgroup designations during phylogenetic analysis, the intron pattern of moso bamboo GAST family was displayed using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/index.php) by aligning the cDNA sequences to their corresponding genomic DNA sequences. The conserved motifs of PheGAST proteins were defined by Multiple Em for Motif Elicitation (MEME, http://meme-suite.org/) (Bailey et al. 2009). The following parameter settings were used: distribution of motifs, zero or one per sequence; maximum number of motifs to find, 50; minimum width of motif, six; and maximum width of motif, 100. Only motifs with an E value of < 1e−20 were retained for further analysis. Subsequently, the MAST program (http://meme.nbcr.net/meme/cgi-bin/mast.cgi) was used to search detected motifs in protein databases (Bailey and Gribskov 1998).
Calculation of Non-synonymous (K a)/Synonymous (K s) Values
To further analyze gene duplication events, paralogous and orthologous gene pairs were aligned using Clustal X 2.1 and analyzed using DnaSP software to calculate the non-synonymous (Ka) and synonymous (Ks) substitution rates (Rozas 2009). The divergence time (T) for moso bamboo, rice, and B. distachyon was calculated using the following equation: T = Ks/(2 × 6.5 × 10−9) MYA (Wu et al. 2016). Sliding window analysis of Ka/Ks ratios was performed using SWAKK (http://ibl.mdanderson.org/swakk/), with the following parameters: window size 60 and 90 bp.
In Silico Analysis of Cis-Acting Elements of PheGASTs
The elements in the promoter fragments (from −1500 bp to the transcription start site) of the PheGASTs genes were analyzed using the online program PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant Materials and Treatments
Moso bamboo seeds used in our experiments were obtained from Guilin City, Guangxi Province, China. These were germinated on moist germination paper in culture dishes in the dark at room temperature (25 °C), and the seedlings were transferred into a greenhouse at 25 °C under a 16/8-h light/dark cycle as previously described (Chen et al. 2017). These were planted in vermiculite and watered with 1/2 strength Hoagland’s nutrient medium once a week. Until 2 months of age, the young seedlings were subject to the hormones and abiotic stresses, and GAST gene expression was assayed.
For the hormone treatments, the seedlings were watered with GA3 (100 µM; Sigma) and ABA (100 µM; Sigma) solution as previously described (Nahirñak et al. 2016), respectively. For low-temperature stress, the plants were transferred to a growth chamber at 4 °C under the same light and photoperiod conditions (16 h light/8 h dark). For salinity and drought stress, the seedlings were watered with 1/2 Hoagland’s solution with 200 mM NaCl and 20% polyethylene glycol 6000 (PRG 6000), respectively. The samples at low-temperature and normal environment were also treated simultaneously with 1/2 strength Hoagland’s solution. The second or third young leaves were collected at different time points (0, 1, 6, and 9 h for hormone treatments; 0, 3, 12, 48 h, and 6 days for abiotic stress treatments; and 0, 1, 3, 6, 9, 12, 48 h and 6 days for mock-treated samples), with three biological replicates for RNA preparation. All leaf samples were immediately frozen in liquid nitrogen and stored at − 80 °C until further analysis.
For tissue-specific analysis, the Illumina RNA-seq data (Peng et al. 2013b; Gao et al. 2014; Li et al. 2017) of developing flowers and shoots of moso bamboo were used for expression profiling of the PheGASTs. Four stages of flowering moso bamboo were defined based on the anatomical structure of the floral organs (F1, F2, F3, and F4). At stage F1, the floral buds begin to form. At stage F2, the floral organs gradually mature but do not undergo flowering. At stage F3, the flowers are in full blossom. At stage F4, pistils and stamens have withered, and embryo formation commences (Gao et al. 2014). Seven developmental stages of moso bamboo shoots were selected, including winter shoots (underground period) and six shoot heights (0.5, 1, 3, 6, 9, and 12 m), which were designated as S1, S2, S3, S4, S5, S6, and S7, respectively (Li et al. 2017).
Gene Expression Analysis
Previously generated and processed transcriptome data of PheGASTs in developing flowers and shoots were used in transcriptomics analysis. The reads per kilobase of exon model per million mapped reads (RPKM) units representing the expression in developing flowers were obtained from the NIH Short Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/?term=SRR1187864). For developing shoots, the FPKM data of different stages were provided by Li et al. (2017). The RPKM/FPKM values of all GAST genes were retrieved and displayed in Table S5 as previously described (Gao et al. 2014; Li et al. 2017).
The expression profiles of PheGASTs in different hormones and abiotic stresses treatments were analyzed by quantitative RT-PCR (qRT-PCR). Each 20-µL reaction contained 0.4 µL (10 µM) of each primer, 1.5 µL (30 ng) of cDNA, 7.7 µL H2O, and 10 µL of SYBR Green I master mix (Roche, Mannheim, Germany), according to the manufacturer’s instructions. The qRT-PCR parameters were as follows: 95 °C for 5 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s. The gene-specific primers were designed using Primer 3.0, and their specificity was checked with BLAST using information provided by the local CDS database that was downloaded from BambooGDB (http://www.bamboogdb.org/page/microrna.jsp) (Chen et al. 2017). The primers used for qRT-PCR and the melting curves of the PCR products are shown in Online Resources 1 and 2, respectively. The tonoplast intrinsic protein 41 gene (TIP41) was used as reference gene under different stress treatments (Fan et al. 2013; Qi et al. 2013; Wu et al. 2015, 2016). To ensure the reliability of the data, another reference of nucleotide tract-binding protein gene (NTB) was also used to validate the expression patterns (Fan et al. 2013; Sun et al. 2016). The qPCR assays were performed with three biological and four technical replicates, and quantitative analysis was performed using the \({2^{ - \Delta \Delta {C_T}}}\) method, whereas the untreated control (0 h) were normalized to 1 as described elsewhere (Chen et al. 2017). Finally, the statistical analyses were performed using SPSS 19.0 software.
Results
Identification, Scaffold Location, and Structural Analysis of Moso Bamboo GAST Genes.
A total of eight GAST genes in moso bamboo were finally identified based on homology to members of the GASA family from other dicots or monocots, which were further confirmed by the Conserved Domain Database for a complete GASA domain. Moreover, no novel and mis-annotated genes were detected in the FLNC database (Wang et al. 2017). To obtain a more comprehensive overview on the PheGAST genes, we also searched for GAST homologs in rice, maize, A. thaliana, B. distachyon and P. hybrida GIP1, wheat TaGASR7-A1, and potato GAST1. The moso bamboo GAST genes were named according to their closest rice and maize relatives (PheGAST1, PheGASR2, PheGSL9, PheGASR1, and PheGSL8), and others were numbered in sequence when no ortholog was available (PheGAST2, PheGAST3, and PheGAST4). All non-redundant PheGASTs were mapped and found to be distributed among eight scaffolds of the P. edulis genome (Table 1). Each scaffold only harbored one gene, unlike those in maize and rice (Zimmermann et al. 2010).
Similar to other species (Zhang and Wang 2008; Zimmermann et al. 2010; Nahirñak et al. 2016), most PheGASTs contain an individual stretch of 19–29 amino acids at the N terminus and a conserved GASA domain with 12 cysteines (Cys, C) at its C-terminal (Fig. 1a). However, a base substitution was detected at the first three C residues in PheGAST2 and the ninth C residue in PheGASR2, respectively. For PheGAST2, the first C residue was deleted, and the other two positions were substituted to alanine (Ala, A). For PheGASR2, the ninth C residue was substituted by glycine (Gly, G).
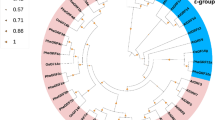
Sequences alignment and structure analysis of PheGASTs. a Multiple alignment of deduced amino acid sequences of PheGAST proteins. Amino acid sequences are aligned using Clustal W software. Color shading shows amino acid identities; the putative signal peptide and conservative cysteine-rich C-terminal domain are labeled. The highly conserve C residues in C-terminal are shaded in purple. b Phylogenetic relationships among GAST proteins in moso bamboo. An unrooted phylogenetic tree constructed using the neighbor-joining method. The branches are reconfirmed by 1000 bootstrap replicates. The bootstrap value of each branch is indicated. c Exon–intron organization of the corresponding genes. The exons and introns are represented by yellow boxes and lines, respectively. d Distribution of conserved motifs of corresponding genes. Using MEME, the conserved motifs within the protein sequence are represented by boxes. The box size indicates the length of the motif. Gray lines represent the length of the proteins
To better mine the gene structure and conserved motifs among moso bamboo GASTs, we constructed NJ and ML phylogenetic trees using amino acid sequences (Fig. 1b and Online Resource 3), respectively. These showed good agreement with the same classification, dividing PheGASTs into three subfamilies with at least 50% bootstrap support. Among these, the PheGASR2 was grouped alone in both phylogenetic trees. Figure 1c shows that all PheGAST genes are disrupted by one (PheGAST2, PheGSL8, PheGSL9, and PheGASR1) to three (PheGAST4) introns. Remarkably, genes in the same subfamily generally show the similar intron pattern; for example, PheGSL9 and PheGASR1 contain a single intron (that is, intron 1), whereas PheGAST1 and PheGSL8 lack intron 2. Ten conserved motifs were also detected within the PheGAST proteins (Fig. 1d), with the exception of a putative signal peptide and GASA domain (motifs 1, 2, 3, and 4), and some PheGASTs have the same motif in their variable regions, but the functional roles of these motifs remain unclear.
Phylogenetic Relationship of PheGAST Proteins
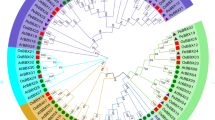
To investigate the evolutionary relationship among members of the GASA family, NJ and ML phylogenetic trees were constructed based on the full-length protein sequences of all known GASA members in P. edulis, Z. mays, O. sativa, A. thaliana, and B. distachyon, as well as the TaGASR7-A1 from wheat, the GIP1 from a P. hybrid, and the StGAST1 from potato (Fig. 2 and Online Resource 4). The resulting phylogenetic tree is organized into four subfamilies, namely, PheGASR2 in subfamily 1, PheGAST1 and PheGSL8 in subfamily 2, PheGSL9 in subfamily 3, and PheGASR1 in subfamily 4, with PheGAST2, PheGAST3, and PheGAST4 forming a subfamily specific to moso bamboo (Fig. 1). The subfamilies 1–4 comprise P. edulis, Z. mays, O. sativa, and B. distachyon, forming a distinct branch and are thus likely to represent monocot-specific functions. Similar to ZmGASTs (Zimmermann et al. 2010), it was also difficult to assign the PheGASTs to distinct homologs from A. thaliana, indicating that some functional roles might have been lost after the divergence of monocots and dicots.
Phylogenetic analysis of predicted GAST proteins in different species. The unrooted neighbor-joining (NJ) tree was constructed based on the amino acids of moso bamboo (8), Arabidopsis (14), rice (9), maize (10), B. distachyon (12) P. hybrida GIP1, wheat TaGASR7-A1, and potato GAST1 using MEGA7.0 with 1000 bootstrap replicates. Branches with less than 50% bootstrap support are collapsed. The names of the subfamilies (S1, S2, S3, and S4) are shown outside the circle. Branch lines of subtrees are colored based on GAST subfamilies
Identification of GAST Orthologs and Paralogs in Moso Bamboo, Rice, and B. distachyon
Peng et al. (2013a) earlier showed that bamboo, B. distachyon, and rice from the BEP clade (Bambusoideae, Ehrhartoideae, and Pooideae) are more closely related than those belonging to the Panicoideae clade (maize, sorghum, and foxtail millet) (Peng et al. 2013a). To better understand the evolutionary relationship of the members of the GAST gene family of this branch, the time of gene duplication and environmental selection pressure of the three species were further investigated. Phylogenetic reconstruction and BLASTP analysis identified two paralogous gene pairs (Phe–Phe) and two orthologous gene pairs (Os–Phe). Moreover, two orthologous gene pairs (Os–Bd) were also observed (Table 2; Fig. 3). Bootstrap support for the internal nodes was relatively lower, which was mainly due to the variable N-terminal region of the GASTs.
Subsequently, Ka/Ks ratios were assessed to verify Darwinian positive selection (Table 2). Interestingly, the Ka/Ks ratios of the two Phe–Phe pairs differed, with Ka/Ks above 1 for PheGAST4/PheGAST2 and Ka/Ks below 1 for PheGAST1/PheGSL8. The Ks values of PheGAST4/PheGAST2 and PheGAST1/PheGSL8 were 0.55 and 0.75, respectively, indicating that their divergence time was about 42 and 57 million years ago (MYA), respectively. Additionally, the Ks values of the orthologous pairs of Phe–Os were both less than 1, and their divergence time was about 29 and 55 MYA, with a mean of 42 MYA. Afterwards, the divergence time for the Os–Bd pairs was also calculated based on the Ks values, with a mean of about 40 MYA.
Furthermore, sliding window analysis of the Ka/Ks ratios was conducted for pairwise comparison (Fig. 4a). The motifs representing a cleavable signal sequence (motif 4) and the conserved C-terminal region with a GAST domain (motifs 1, 2, and 3) were described, respectively (Fig. 4b). Regardless of the paralogous pairs of Phe–Phe or orthologous pairs of Os–Phe, the Ka/Ks ratios were less than 1. It gradually decreased from the N-terminal to the C-terminal, and roughly remained the same at the motif 2 region.
Sliding window plots of representative duplicated PheGAST genes and orthologous of Phe–Os. a The characterized motifs are noted by different colored lines. The window sizes were 90 and 60 bp in length, respectively. b The logo of each motif. The motifs, numbered 1–4, are displayed in different colored lines
Promoter Analysis of PheGASTs
Although the exact role played by the Snakin/GAST proteins remains elusive, these have been associated with different aspects of plant development and stress tolerance (Nahirñak et al. 2016). To identify the cis-acting elements in the promoters of moso bamboo GAST genes, we searched the 1.5-kb upstream sequences from the translation start sites using the PlantCARE database. Hormone-, stress-, and development-related elements were randomly distributed upstream of the PheGASTs (Fig. 5a, b), which may be responsible for the wide range of functions of this protein family. Among these, the jasmonic acid (CGTCA-motif)-, abscisic acid (ABRE)-, gibberellin (P-box and GARE-motif)-responsive elements were found within the promoter regions of 6, 6, and 3 PheGASTs, respectively. These were all present within the promoters of four genes, namely, PheGASR1, PheGASR2, PheGAST1, and PheGAST4. The element involved in anoxic-specific inducibility (GC-motif) was abundant and mainly distributed upstream of the four genes, whereas the other stress-related elements such as defense- and stress-responsive elements (TC-rich repeats), heat-responsive elements (HSE), low-temperature-responsive elements (LTR), and drought-inducible elements (MBS) were also detected. Moreover, with the exception of PheGAST2, all the other PheGASTs have at least three stress-related elements within their promoters. Additionally, five PheGAST promoters also had binding sites for transcription factors that were related to tissue development, such as the elements involved in seed-specific regulation (RY-element), endosperm expression (Skn-1_motif), and meristem-specific activation (CCGTCC-box).
The cis-elements distributed in the promoters of PheGASTs. a The number of cis-acting elements in the promoter region of each PheGAST gene. Annotation of cis-elements: ERE ethylene-responsive element, AuxRR-core auxin-responsive element, TCA-element involved in salicylic acid responsiveness, CGTCA-motif involved in MeJA responsiveness, ABRE involved in abscisic acid responsiveness, GARE-motif and P-box gibberellin-responsive element, W box binds WRKY type transcription factors and wounding and pathogen responsiveness. ARE essential for the anaerobic induction, Box-W1 fungal elicitor responsive element, GC-motif enhancer-like element involved in anoxic-specific inducibility, HSE cis-acting element involved in heat stress responsiveness, LTR involved in low-temperature responsiveness, MRE, MBS, and MYB binding sites involved in light responsiveness and drought inducibility, TC-rich repeats involved in defense and stress responsiveness, MSA-like cis-acting element involved in cell cycle regulation, O2-site cis-acting regulatory element involved in zein metabolism regulation, motif I, cis-acting regulatory element root specific, RY-element cis-acting regulatory element involved in seed-specific regulation, circadian involved in circadian control, CAT-box related to meristem expression, Skn-1 motif and GCN4_motif cis-regulatory element involved in endosperm expression, MBSIMYB-binding site involved in flavonoid biosynthetic gene regulation, CCAAT-box MYBHv1-binding site, CCGTCC-box related to meristem-specific activation. b The number of cis-elements and the PheGAST genes containing cis-acting elements
Expression Patterns of PheGAST Genes During Shoot and Flower Development
We analyzed the expression patterns of all PheGAST genes in the flowers and shoots based on publicly available RNA-seq data (Peng et al. 2013b; Li et al. 2017) and a heatmap was generated (Fig. 6 and Online Resource 5). We also selected four genes from different groups (PheGAST2, PheGSL8, PheGASR1, and PheGASR2) to verify transcripts levels using qPCR (Online Resource 6). These genes exhibit similar expression patterns similar to previous RNA-seq data, thereby suggesting that our transcriptome results are highly reliable. It shows that all PheGAST genes present similar expression patterns during flower development in moso bamboo (Fig. 6a). These are upregulated during the earlier stages (F1 and F2) and downregulated in the later stages (F3 and F4). Among these, the expression level of PheGASR2 and PheGSL8 decreases by more than 10-fold in the F4 period, and PheGAST2 is hardly detected in the last stage (Table S5). For shoot growth, most PheGAST genes show twin peaks during the early stages (S2 and S3) and late stages (S6, S7), respectively (Fig. 6b). Interestingly, PheGASR1 is highly expressed throughout the whole growth process of the bamboo shoots (FPKM > 60), which peaks at periods S1 and S4, respectively (Fig. 6b).
Expression pattern of PheGASTs genes in developing flowers and shoots of moso bamboo. a The expression profile of PheGASTs in different stages of flowering, F1 The floral bud formation stage. F2 The inflorescence growing stage. F3 The flowers with both pistils and stamens emerging from glumes at bloom stage. F4 The flowers are withered. b The expression profile of PheGASTs at different shoot growth stages. S1–S7 represent winter bamboo shoot and different heights of shoots (50, 100, 300, 600, 900, and 1200 cm)
Regulation of the PheGAST Genes Expression
As earlier mentioned, most GAST genes are regulated by plant hormones and external factors. Here, in silico analyses also revealed that the upstream sequences of most PheGAST genes carry a variety of hormones and stresses response elements (Fig. 5). Compared to the untreated samples, the expression of the PheGAST genes significantly changes with hormone and abiotic stress treatment (Figs. 7, 8). We initially analyzed the effect of GA3 and ABA on the expression pattern of the PheGASTs (Fig. 7). We found that all PheGASTs, except for PheGAST3, are upregulated by GA3 treatment. Some show similar expression profiles (PheGAST1, PheGAST4, and PheGSL8), which were induced by exposure to GA3 for 1 and 6 h and repressed after 9 h. However, the paralogous pair PheGSL9 and PheGASR1 presents double peaks and is upregulated by 1 and 9 h of GA3 treatment, respectively. For ABA treatment, five PheGAST genes were upregulated, and most of their expressions were initially downregulated (1 and 6 h) and then upregulated after 9 h of exposure. Interestingly, PheGASR1 showed a specific expression pattern. It was significantly induced at 1 h after ABA treatment and upregulated during the rest of the time points.
Expression analysis of the PheGAST genes in the leaves of moso bamboo under mock, GA, and ABA treatments. Bars represent standard deviations (SDs) of three technical replicates. *Represents a significant difference relative to the control (0 h). The X axes show the time points of GA and ABA treatments for each gene (0–9 h). The Y axes show the relative expression levels (fold-change). The control expression at 0 h was standardized by 1, which is represented by broken lines. The TIP41 gene is used as reference
Expression analysis of the PheGAST genes in the leaves of moso bamboo under mock and different abiotic stress treatments. Bars represent standard deviations (SDs) of three technical replicates. *Represents a significant difference relative to the control (0 h). The X axes show the time points of cold, drought, and salt treatments for each gene (0–6 days). The Y axes show the relative expression levels (fold-change). The control expression at 0 h was standardized by 1, which is represented by red lines. The TIP41 gene is used as reference
We further analyzed the expression profiles of the PheGASTs during different abiotic stress conditions (that is, salt, drought, and cold; Figs. 8, 9). Except for PheGAST4, the expression of all PheGAST genes was induced by at least one stress treatment to varying degrees (Fig. 8). We found most PheGASTs (PheGAST1, PheGAST2, PheGAST3, PheGSL8, and PheGASR2) share a similar expression pattern regardless of cold, drought, or salt condition (Fig. 9). For example, these were upregulated at 3 h, but gradually downregulated after 48 h and 6 days of cold treatment (Fig. 9a). With drought treatment, these genes were initially upregulated (after 3 h), subsequently rapidly downregulated (48 h), and then significantly upregulated again after 6 days (Fig. 9b). Their expression was downregulated with salt treatment (Fig. 9c). However, PheGASR1 exhibits a unique expression pattern under all stress treatments; it is upregulated under low temperature and downregulated under drought conditions. It also peaked at 48 h after salt treatment. This characteristic response model under abiotic stress distinguishes PheGASR1 from the other PheGAST genes.
Additionally, all PheGAST genes were validated using another reference gene, NTB. The expression levels of some genes varied, but the similar expression patterns were observed with hormone and stress treatment (Online Resources 7 and 8). The results on the DEGs further strengthens the data generated using TIP41.
Discussion
In the present study, a total of eight PheGAST genes were identified and characterized for the first time from the moso bamboo genome database (Peng et al. 2013a; Zhao et al. 2014). Recently, Wang et al. (2017) used a combination of single-molecule long-read sequencing technology and polyadenylation site sequencing (PAS-seq) to reannotate the bamboo genome, resulting in the correction of 2241 mis-annotated genes and identification of 8091 previously unannotated loci, significantly improving current annotations (Wang et al. 2017). We further consulted and searched for GAST genes based on the FLNC reads database; however, no novel or erroneous genes were identified. Therefore, at least eight PheGAST genes were finally established, forming a GAST family in moso bamboo, and their details are described here for the first time, including their gene structure, phylogeny, conserved motifs, promoter cis-elements, and regulation of expression.
All moso bamboo GAST family members have structural characteristics typical of secreted polypeptide molecules. Each putative protein contains an N-terminal hydrophobic region consisting of 18–29 amino acids that could act as a cleavable signal sequence (Nahirñak et al. 2016). Downstream of the putative signal peptide, a variable region and a conserved C-terminal domain were defined. Generally, the cysteines of the C-terminus are essential to their biochemical activity and are responsible for their protein structure (Zimmermann et al. 2010; Liu et al. 2013; Nahirñak et al. 2016). Interestingly, different mutations in the moso bamboo PheGAST2 and PheGASR2 domains were detected (Fig. 1a). Previous studies have suggested that these proteins play a role in reactive oxygen species scavenging due to the presence of these redox-active cysteines (Wigoda et al. 2006; Rubinovich and Weiss 2010). In plants and bacteria, the replacement of four of the conserved cysteines with alanine residues in AtGASA4 results in the inhibition of antioxidant activity (Rubinovich and Weiss 2010). This suggests that the redox activity of PheGASR2, particularly PheGAST2, is probably limited by mutations; however, other factors influencing this particular biological activity remain unclear.
Similar to the findings of a previous study (Peng et al. 2013a), phylogenetic reconstruction reveals a higher degree of homology among moso bamboo, rice, and B. distachyon GAST family proteins. Selection plays an important role in shaping gene families, resulting in different evolutionary patterns among gene families or even within a single gene family (Zhang et al. 2016). Ka/Ks ratios reflect the selection pressure that is experienced by a gene pair. A Ka/Ks ratio of less than 1 is indicative of purifying or negative selection, Ka/Ks = 1 suggests neutral selection, and Ka/Ks greater than 1 reflects positive selection (Lynch and Conery 2000). The findings of the present study indicate that the evolution of the PheGAST family may have initially involved positive selection, followed by purifying selection. Additionally, Peng et al. (2013a) reported a putative recent whole genome duplication (WGD) event occurring 7–12 MYA based on analyzing the evolutionary history of P. edulis, and the estimated times for the divergence of bamboo from B. distachyon and rice are approximately 46.9 and 48.6 MYA, respectively (Peng et al. 2013a). The time of division for GAST genes in rice and moso bamboo was established at a mean of around 42 MYA, suggesting that the PheGAST family underwent further evolution after its divergence from rice. However, the duplication event involving GASTs in moso bamboo occurred between around 42 and 57 MYA, long before the recent WGD event, indicating that the recent WGD event is not the main reason for the expansion of the PheGAST family in moso bamboo (Peng et al. 2013a). Together, we propose that the PheGAST family in moso bamboo completed its expansion following the divergence of other grasses at about 50 MYA, and the recent WGD event is not the primary cause of the duplication of PheGAST genes (Kellogg 1998; Yu et al. 2002; Guyot and Keller 2004; Gaut 2010).
Moso bamboo is famous for its prolonged vegetative growth period and rapid shoot growth rates. Plant hormones, including IAA, cytokinins (CK), ABA, and GAs (Peng et al. 2013b; Gao et al. 2014; Ge et al. 2017; Gamuyao et al. 2017; Li et al. 2017), and their related genes are essential to these developmental processes. GAST proteins act as integrators of internal and environmental cues and participate in hormone homeostasis to modulate plant development and stress tolerance. These are also involved in plant developmental processes and stress responses that regulate redox homeostasis through their conserved cysteine-rich domains (Székely 2009; Almasia et al. 2010; Nahirñak et al. 2012; Sun et al. 2013). Therefore, we examined the expression patterns of PheGASTs in moso bamboo flowers and in shoot growth processes. For flower development, all PheGAST genes are upregulated during the early stages (F1 and F2) and downregulated during the later stages (F3 and F4). A number of GAST genes are correlated with young tissues and actively growing flowers, including AtGASA1, 4, 6, and 10 in A. thaliana (Aubert et al. 1998; Trapalis et al. 2017), whereas GASA4 and GASA6 also affect flowering time (Jie et al. 2016). This indicates that PheGAST genes, particularly PheGAST2, PheGSL8, and PheGASR2, which are highly expressed at F1 or F2 (RPKM value > 100), might be associated with floral meristem differentiation and are probably involved in a GA-signaling pathway in regulating flowering time in moso bamboo. However, additional investigations verifying our hypothesis are warranted. Our team has been investigating the phenomenon of rapid shoot growth for several years, accumulating valuable information on spatiotemporal changes in morphology, the production of endogenous hormones, and differentially expressed genes (DEGs) (Peng et al. 2013b; Li et al. 2016, 2017). Endogenous hormones involved in moso bamboo shoot growth display a wide range of variations in expression levels (Li et al. 2017). IAA and BR contents increase from S1 and continue to increase with growth, and the expression level of IAA remains high from S5 to S7 (> 150 ng/g FW). The temporal changes of GA3 were generally bimodal, occurring as double peaks at the early (S2 and S3) and late (S6) stages (Li et al. 2017). The inhibitory factor ABA is upregulated in S1 (> 150 ng/g FW), transiently decreases, and then increases again from S5 to S7. In our study, the expression patterns of most PheGASTs show twin peaks that coincide with variations in endogenous GA3 levels (Li et al. 2017), thereby suggesting that shoot growth involves a GA-dependent pathway. Gene expression analysis of PheGASTs in response to hormones (GA and ABA) further supports our viewpoint. We found that most PheGASTs respond to GA3 signaling in a significant and earlier (1 h) manner. Although these are also upregulated by ABA signaling, induction was observed later (after 9 h of treatment). A similar complex regulation using hormones has been previously reported. For example, the expression of the potato Snakin-2 gene in shoot apices was induced by GA after 12 and 24 h of treatment and is repressed by GA 24 h after treatment in leaves. In addition, Snakin-2 gene expression in the shoot apices was induced after 12 h of ABA treatment (Nahirñak et al. 2016). Comparatively, it is more important to evaluate the regulation of PheGASA genes at earlier time points because if these are early responsive genes, then more drastic expression changes might be observed. Thus, PheGSL8, PheGSL9, and PheGASR2 are upregulated by more than dozens or hundreds of times after 1 h of GA3 treatment. We also noticed that PheGAST3 expression was not induced by GA treatment; however, this may be an earlier response factor that was induced before 1 h of treatment. In summary, the results of the present study suggest that GA signaling plays a direct regulatory role in early responsive GAST genes, possibly involving moso bamboo tissue growth and development. Finally, we also investigated the effect of abiotic stresses on the expression of GAST genes and observed that all PheGAST genes, except for PheGAST4, are significantly upregulated in moso bamboo seedlings when subjected to stress, particularly PheGAST1 and PheGAST3. These results also coincide with the participation of the GAST genes in biotic or abiotic stress tolerance (Ko et al. 2007; Alonso-Ramirez et al. 2009; Sun et al. 2013; Nahirñak et al. 2016), thereby suggesting their possible positive roles in moso bamboo stress resistance.
The present study assessed the expression pattern of the PheGASR1 gene in response to ABA signaling as well as in relation to shoot growth and abiotic stress resistance. Our findings may be used in discriminating it from other GAST genes. During shoot development, the PheGASR1 gene shows its highest expression levels at the S1 stage; it then subsequently decreases and peaks again at the S4 stage and remains upregulated until S6 (FPKM > 100). Previous research has shown that the expression pattern of PheGASR1 is apparently regulated by both endogenous ABA and GA3 (Li et al. 2017). Moreover, the PheGASR1 gene rapidly responds to ABA treatment and is upregulated at least eightfold higher after 1 h of ABA treatment. The PheGASR1 gene also exhibits a distinct expression pattern in response to abiotic stresses, particularly under cold and drought treatment. Phylogenetic reconstruction suggests that rice OsGASR1 is the ortholog of PheGASR1 (Fig. 2). OsGASR1 is related to cell proliferation in meristem and panicle development (Furukawa et al. 2006); moreover, it plays important role in seedling growth as well as in salt and ABA stress resistance (Lee et al. 2015, 2017). The function of OsGASR1 homologs have also been functionally investigated in other species, such as GhGAST3 in cotton and TaGASR1 in wheat, which are involved in fiber elongation and heat stress resistance, respectively (Liu et al. 2013; Zhang et al. 2017). More importantly, PheGASR1 might be involved in regulating the morphological development of rods of moso bamboo and its mutant lines (unpublished data), which is rarely studied in other species. Above all, the distinct expression pattern and related literature indicate the involvement of PheGASR1 in growth and stress responses through both GA and ABA pathways. However, the correlation between the transcript regulation and protein expression levels remains unclear. Finally, although our study only indicates the particular regulatory role of PheGASR1 under limited hormones and stresses treatments, it lays the foundation for future functional research investigations.
References
Almasia NI, Narhirñak V, Hopp HE, Vazquez-Rovere C (2010) Isolation and characterization of the tissue and development-specific potato snakin-1 promoter inducible by temperature and wounding. Electron J Biotechnol 13:8–9. https://doi.org/10.2225/vol13-issue5-fulltext-12
Alonso-Ramirez A, Rodríguez D, Reyes D, Jiménez JA, Nicolás G, López-Climent M, Gómez-Cadenas A, Nicolás C (2009) Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol 150:1335–1344. https://doi.org/10.1104/pp.109.139352
Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M (1998) Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol 36:871–883. https://doi.org/10.1023/A:1005938624418
Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54. https://doi.org/10.1093/bioinformatics/14.1.48
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Ben-Nissan G, Lee JY, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37:229–238. https://doi.org/10.1046/j.1365-313X.2003.01950.x
Chen D, Chen Z, Wu M, Wang Y, Wang Y, Yan H, Xiang Y (2017) Genome-wide identification and expression analysis of the HD-zip gene family in moso bamboo (Phyllostachys edulis). J Plant Growth Regul 36:323–337. https://doi.org/10.1007/s00344-016-9642-x
de la Fuente JI, Amaya I, Castillejo C, Sanchez-Sevilla JF, Quesada MA, Botella MA, Valpuesta V (2006) The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot 57:2401–2411. https://doi.org/10.1093/jxb/erj213
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 8:e56573. https://doi.org/10.1371/journal.pone.0056573
Furukawa T, Sakaguchi N, Shimada H (2006) Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst 81:171
Gamuyao R, Nagai K, Ayano M, Mori Y, Minami A, Kojima M, Suzuki T, Sakakibara H, Higashiyama T, Ashikari M, Reuscher S (2017) Hormone distribution and transcriptome profiles in bamboo shoots provide insights on bamboo stem emergence and growth. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcx023
Gao J, Zhang Y, Zhang C, Qi F, Li X, Mu S, Peng Z (2014) Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis. PLoS ONE 9:e98910. https://doi.org/10.1371/journal.pone.0098910
Gaut BS (2010) Evolutionary dynamics of grass genomes. New Phytol 154:15–28. https://doi.org/10.1046/j.1469-8137.2002.00352.x
Ge W, Zhang Y, Cheng Z, Hou D, Li X, Gao J (2017) Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol J 15:82–96. https://doi.org/10.1111/pbi.12593
Guyot R, Keller B (2004) Ancestral genome duplication in rice. Genome 47:610. https://doi.org/10.1139/g04-016
Jiang X, Lou C, Yuan N, Yue JJ, Gu XP (2016) Effects of exogenous GA3 on culm form and culm fiber quality of Phyllostachys edulis seedlings neonatal tiller. J Nanjing For Univ 40:121–126
Jie Q, Kang SG, Hah C, Jang JC (2016) Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci 246:1–10. https://doi.org/10.1016/j.plantsci.2016.01.009
Kellogg EA (1998) Relationships of cereal crops and other grasses. Proc Natl Acad Sci USA 95:2005–2010
Ko CB, Woo YM, Lee DJ, Lee MC, & Kim CS (2007) Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol Biochem 45(9):722–728. https://doi.org/10.1016/j.plaphy.2007.07.010
Lee SC, Han SK, Kim SR (2015) Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J Plant Biol 58:96–101. https://doi.org/10.1007/s12374-014-0497-z
Lee SC, Kim SJ, Han SK, An G, Kim SR (2017) A gibberellin-stimulated transcript, OsGASR1, controls seedling growth and α-amylase expression in rice. J Plant Physiol 214:116–122. https://doi.org/10.1016/j.jplph.2017.04.010
Li L, Hu T, Li X, Mu S, Cheng Z, Ge W, Gao J (2016) Genome-wide analysis of shoot growth-associated alternative splicing in moso bamboo. Mol Genet Genom 291:1695–1714. https://doi.org/10.1007/s00438-016-1212-1
Li L, Cheng Z, Ma Y, Bai Q, Li X, Cao Z, Wu Z, Gao J (2017) The association of hormone signalling genes, transcription and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol J 16(1):72–85. https://doi.org/10.1111/pbi.12750
Liu ZH, Zhu L, Shi HY, Chen Y, Zhang JM, Zheng Y, Li XB (2013) Cotton GASL genes encoding putative gibberellin-regulated proteins are involved in response to GA signaling in fiber development. Mol Biol Rep 40:4561. https://doi.org/10.1007/s11033-013-2543-1
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151. https://doi.org/10.1126/science.290.5494.1151
Nahirñak V, Almasia NI, Hopp HE, Vazquezrovere C (2012) Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav 7:1004–1008. https://doi.org/10.4161/psb.20813
Nahirñak V, Rivarola M, Urreta MGD, Paniego N, Hopp HE, Almasia NI, Vazquez-Rovere C (2016) Genome-wide analysis of the snakin/GASA gene family in Solanum tuberosum cv. Kennebec Am J Potato Res 93:172–188. https://doi.org/10.1007/s12230-016-9494-8
Peng Z et al (2013a) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45:456–461. https://doi.org/10.1038/ng.2569
Peng Z, Zhang C, Zhang Y, Hu T, Mu S, Li X, Gao J (2013b) Transcriptome sequencing and analysis of the fast growing shoots of moso bamboo (Phyllostachys edulis). PLoS ONE 8:e78944. https://doi.org/10.1371/journal.pone.0078944
Qi F, Hu T, Peng Z, Gao J (2013) Screening of reference genes used in qRT-PCR and expression analysis of PheTFL1 gene in moso bamboo. Acta Bot Boreali-Occident Sin 33:48–52
Rozas J (2009) DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol 537:337
Rubinovich L, Weiss D (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64:1018–1027. https://doi.org/10.1111/j.1365-313X.2010.04390.x
Rubinovich L, Ruthstein S, Weiss D (2014) The Arabidopsis cysteine-rich GASA5 is a redox-active metalloprotein that suppresses gibberellin responses. Mol Plant 7:244–247. https://doi.org/10.1093/mp/sst141
Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, Wang X (2013) GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot 64:1637–1647. https://doi.org/10.1093/jxb/ert021
Sun H, Li L, Lou Y, Zhao H, Gao Z (2016) Genome-wide identification and characterization of aquaporin gene family in moso bamboo (Phyllostachys edulis). Mol Biol Rep 43:437–450. https://doi.org/10.1007/s11033-016-3973-3
Székely G (2009) Oxidative stress in plants. Stud Univ Babes-Bolyai Biol 10:19–19
Trapalis M, Li SF, Parish RW, Li SF (2017) The Arabidopsis GASA10 gene encodes a cell wall protein strongly expressed in developing anthers and seeds. Plant Sci 260:71. https://doi.org/10.1016/j.plantsci.2017.04.003
Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57:498–510. https://doi.org/10.1111/j.1365-313X.2008.03707.x
Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, Lin C, Ma L, Gu L (2017) Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J 91(4):684–699. https://doi.org/10.1111/tpj.13597
Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D (2006) The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J 48:796–805. https://doi.org/10.1111/j.1365-313X.2006.02917.x
Wu H, Lv H, Li L, Liu J, Mu S, Li X, Gao J (2015) Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB Genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 10:e0126657. https://doi.org/10.1371/journal.pone.0126657
Wu M, Li Y, Chen D, Liu H, Zhu D, Xiang Y (2016) Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci Rep 6:24520. https://doi.org/10.1038/srep24520
Yu J, Hu S, Wang J et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:79–92. https://doi.org/10.1126/science.1068037
Zhang S, Wang X (2008) Expression analysis on GASA genes downstream of DELLA in Arabidopsis. Sci Bull 53:2760–2767
Zhang J, Jia H, Li J, Li Y, Lu M, Hu J (2016) Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci Rep 6:30050. https://doi.org/10.1038/srep30050
Zhang LY, Geng XL, Zhang HY, Zhou CL, Zhao AJ, Wang F, Zhao Y, Tian XJ, Hu ZR, Xin MM, Yao YY, Ni ZF, Sun QX, Peng HR (2017) Isolation and characterization of heat-responsive gene TaGASR1 from wheat (Triticum aestivum L.). J Plant Biol 60:57–65. https://doi.org/10.1007/s12374-016-0484-7
Zhao H, Peng Z, Fei B, Li L, Hu T, Gao Z, Jiang Z (2014) BambooGDB: a bamboo genome database with functional annotation and an analysis platform Database. Database 2014:bau006. https://doi.org/10.1093/database/bau006
Zimmermann R, Sakai H, Hochholdinger F (2010) The gibberellic acid stimulated-like gene family in maize and its role in lateral root development. Plant Physiol 152:356–365. https://doi.org/10.1104/pp.109.149054
Funding
Fundamental Research Funds of ICBR (Grant Number 1632018006) and the National Natural Science Foundation of China (Grant Number 31570673) supported this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hou, D., Bai, Q., Li, J. et al. The Gibberellic Acid-Stimulated Transcript Gene Family in Moso Bamboo: A Genome-Wide Survey and Expression Profiling During Development and Abiotic Stresses. J Plant Growth Regul 37, 1135–1147 (2018). https://doi.org/10.1007/s00344-018-9805-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-018-9805-z