Abstract
Dicer, Argonaute (AGO), and RNA-dependent RNA polymerase (RDR) comprise the core components of RNA-induced silencing complexes, which trigger RNA silencing. Here, we performed a complete analysis of the cucumber Dicer-like, AGO, and RDR gene families including the gene structure, genomic localization, and phylogenetic relationships among family members. We identified seven CsAGO genes, five CsDCL genes, and eight CsRDR genes in cucumber. Based on phylogenetic analysis, each of these genes families was categorized into three or four clades. The orthologs of CsAGOs, CsDCLs, and CsRDRs were identified in apple, peach, wild strawberry, foxtail millet, and maize, and the evolutionary relationships among the orthologous gene pairs were investigated. We also investigated the expression levels of CsAGOs, CsDCLs, and CsRDRs in various cucumber tissues. All CsAGOs were relatively higher upregulated in leaves and tendrils than in other organs, especially CsAGO1c, CsAGO1d, and CsAGO7. All CsDCL genes were relatively higher upregulated in tendrils, with almost no expression detected for CsDCL1, CsDCL4a, or CsDCL4b in other organs. In addition, CsRDR1a, CsRDR2, CsRDR3, and CsRDR6 had relatively higher upregulation in tendrils, whereas almost all CsRDRs were downregulation in other organs. The results of this study will facilitate further studies of gene silencing pathways in cucumber.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA silencing is a process triggered by 21–24 nt small RNAs (such as microRNAs [miRNAs] and short-interfering RNAs [siRNAs]) that represses gene expression and regulates development and physiology to maintain genome stability (Ding 2010; Bai and others 2012). In plants, the generation of small RNAs mainly depends on proteins encoded by members of the Dicer-like (DCL), Argonaute (AGO), and RNA-dependent RNA polymerase (RDR) gene families. Recent studies have revealed that the plant Dicer-like protein, Argonaute, and RNA-dependent RNA polymerase gene families usually comprise multiple members and are involved in different RNAi pathways. The structures and functions of these core proteins have also recently been clarified (Pattanayak and others 2013; Yang and others 2013; Shao and Lu 2013; Liu and others 2014). The Argonaute proteins belong to the core components of RNAi effector complexes, which play central roles in RNA silencing (Moazed 2009). AGO proteins are evolutionarily highly conserved in eukaryotes and can be subdivided into three groups (Hutvagner and Simard 2008). These proteins contain several functional domains, including the DUF1785, PAZ, MID, and PIWI domains (Kapoor and others 2008; Hutvagner and Simard 2008). Based on sequence comparisons, DCL proteins have six domains, namely DEAD-helicase, helicase C, Duf283, PAZ, RNase III, and double-stranded RNA binding (dsRB) (Margis and others 2006). Among these RNAi machinery components, plant DCL proteins mainly process long double-stranded RNAs into mature small RNAs (Bernstein and others 2001; Chapman and Carrington 2007). The third major type of RNAi protein is RDR proteins, which are necessary for the initiation and amplification of silencing signals (Kapoor and others 2008). RDR proteins contain a unique conserved RNA-dependent RNA polymerase (RdRP) domain. RDR proteins are required for RNAi in fungi, nematodes, and plants, but they have not been identified in insects or vertebrates (Djupedal and Ekwall 2009).
Dicer, Argonaute, and RNA-dependent RNA polymerase comprise the core components of RNA-induced silencing complexes, which trigger RNA silencing and are implicated in the initiation and maintenance of the mechanism that is central to this mode of gene regulation (Kapoor and others 2008). Functional analysis of DCL, AGO, and RDR genes has revealed that different genes play multiple roles in regulating growth and development. For example, 4 DCL, 10 AGO, and 6 RDR genes have been identified in Arabidopsis (Fang and Spector 2007; Vaucheret 2008; Xie and others 2004). Among these genes, AtDCL1 mainly contributes to the production of miRNAs from noncoding, imperfect stem-loop precursor RNAs (Voinnet 2009). AtDCL2 is associated with viral defense, whereas AtDCL3 and AtAGO4 are required for RNA-directed DNA methylation (Zilberman and others 2003; Henderson and others 2006), and AtDCL4 regulates vegetative phase change (Margis and others 2006). Qu and others examined the role of four DCLs, two AGOs, and one RDR in controlling viral accumulation in infected Arabidopsis plants, revealing that all four DCLs contribute to antiviral RNA silencing. DCL1 represses antiviral RNA silencing through negatively regulating the expression of DCL3 and DCL4 (Qu and others 2008). Argonautes (AGOs) play crucial roles in RNAi and related pathways in several species, and they regulate plant growth and development. Yang and others focused on the expression patterns and co-expression profiles of 19 OsAGO genes in rice and found that most OsAGOs are expressed specifically and preferentially during various stages of reproductive development, and they are preferentially upregulated at the panicle stages (Yang and others 2013). Ten SmAGO genes were identified in Salvia miltiorrhiza. Analysis of their expression levels in various tissues revealed that some SmAGOs play similar roles to those of their counterparts in Arabidopsis, whereas other SmAGOs might be more species specialized. This study also confirmed that SmAGO1 and SmAGO2 are targeted by S. miltiorrhiza miR168a/b and miR403, respectively (Shao and Lu 2013). Furthermore, RDRs might be involved in several types of gene silencing in plants, including cosuppression (Dalmay and others 2000). Among the six Arabidopsis RDR genes, AtRDR1, 2, and 6 function in distinct and overlapping processes such as viral resistance, chromatin silencing, and PTGS (Donaire and others 2008; Kapoor and others 2008; Curaba and Chen 2008; Vaistij and Jones 2009).
Recently, RNA silencing components in soybean (7 GmDCL, 7 GmRDRs, and 21 GmAGOs), sorghum (5 SbDCLs, 7 SbRDRs, and 14 SbAGOs), rice (8 DCL, 19 AGO, and 5 RDR), maize (5 DCL, 18 AGO, and 5 RDR), and grape (4 VvDCLs, 13 VvAGOs, and 5 VvRDRs) were identified (Liu and others 2014; Zhao and others 2014; Kapoor and others 2008; Qian and others 2011). Meanwhile, 7 Dicer-like (SIDCL), 15 Argonaute (SIAGO), and 6 RNA-dependent RNA polymerase (SIRDR) genes were identified in tomato (Solanum lycopersicum), and comprehensive analyses of gene structure, expression patterns, genomic localization, and similarity among these genes have revealed that the DCL2 family has played an important role in the evolution of tomato (Bai and others 2012). Moreover, an analysis of the stress-induced transcription patterns of seven duplicated GmDCL gene pairs involved in RNAi and DNA methylation processes in soybean (Glycine max) has revealed that the Dicer-like 2 (DCL2) gene pair exhibits the strongest response to stress and has the most highly conserved co-expression pattern (Curtin and others 2012). In addition, 8 SiDCL, 19 SiAGO, and 11 SiRDR genes were identified in foxtail millet (Setaria italica), and the expression profiling revealed the differential expression pattern of the candidate genes at different time points of stresses, which provides insights into the putative roles of these genes in abiotic stresses (Yadav and others 2015).
In cucumber, few RNAi machinery components have been characterized to date. In this study, we analyzed the gene structures, protein motifs, phylogenetic relationships, and gene expression patterns of members of the DCL, RDR, and AGO gene families. We identified 20 core components of RNAi genes belonging to these gene families. The results of this study provide basic genomic information about these gene families, and they provide a basis for further, more detailed investigations aimed at understanding the contributions of individual components of RNA silencing machinery to plant growth and development.
Materials and Methods
Identification of Dicer-Like, Argonaute, and RDR Genes in Cucumber
To identify all Dice-like (DCL), Argonaute (AGO), and RNA-dependent RNA polymerase (RDR) genes in the cucumber genome, the annotated cucumber database was searched using the following sequences as queries: six types of conserved DEAD/DEAH box helicase (DEAD) domains, Helicase conserved C-terminal (Helicase C) domain, Dicer dimerization domain (Dicer dimer), PAZ domain (PAZ), Ribonuclease III domain (Ribonuclease 3), and double-stranded RNA-binding domain from DE (DND1 DSRM) from the putative polypeptide sequence in CsDCL proteins; three types of conserved domains of unknown function (DUF1785), PAZ domain (PAZ), and Piwi domain (Piwi) from the putative polypeptide sequence in CsAGO proteins; and one type of conserved domain of RNA-dependent RNA polymerase (RdRP) from the putative polypeptide sequences in CsRDR proteins generated from the HMM profile in the Pfam program (http://pfam.xfam.org/search/sequence) (Finn and others 2014). First, for the CsDCLs, CsAGOs, and CsRDRs, all predicted CsDCL, CsAGO, and CsRDR protein sequences were used as query sequences to search against the Cucumber Genome Database (http://cucumber.genomics.org.cn/page/cucumber/index.jsp) using the BLASTP program (with a P value = 0.001 to avoid false positives). The sequences of DCL, AGO, and RDR genes in Arabidopsis were used as queries to search against the DATF (Database of Arabidopsis Transcription Factor, http://datf.cbi.pku.edu.cn/browsefamily.php?fn=Dicer-like, Argonaute and RNA-dependent RNA polymerase). Finally, the Pfam and SMART (Simple Model Architecture Research Tool, http://smart.embl-heidelberg.de) (Letunic and others 2004) databases were used to determine whether any candidate CsDCL, CsAGO, and CsRDR protein sequences were members of the Dicer-like, Argonaute, and RNA-dependent RNA polymerase gene families, respectively. To exclude any overlapping genes, all of the candidate DCLs, AGOs, and RDRs were aligned using Clustal W (Larkin and others 2007) and the sequences were checked manually. All non-overlapping DCL, AGO, and RDR genes were subjected to further analysis.
Structural Analysis of Dicer-Like, Argonaute, and RDR Genes
Information about the CsDCL, CsAGO, and CsRDR genes was retrieved from the Cucumber Genome Database, including their sequence IDs, chromosomal locations, and deduced polypeptide sequences. The position of each CsDCL, CsAGO, and CsRDR gene on cucumber chromosomes was determined by BLAST searching against the genomic sequences of each cucumber chromosome. Molecular weights (MWs) and isoelectric points (PIs) were determined using the Protparam program on the Expasy website (http://au.expasy.org/tools/protparam.html).
To predict the exon–intron structures of the Dicer-like, Argonaute, and RDR genes, a comparison of the genomic sequences and their predicted coding sequences (CDS) was performed using GSDS (http://gsds.cbi.pku.edu.cn/) (Guo and others 2007).
Analysis of Conserved Motifs and Chromosomal Location
To identify the conserved motifs within the DCL, AGO, and RDR proteins in cucumber and Arabidopsis, the online Multiple Expectation Maximization for Motif Elicitation (MEME) tool was employed to display the motifs in these proteins (http://meme.nbcr.net/meme4_1/cgi-bin/meme.cgi) (Bailey and others 2009). Parameters were set as follows: the occurrences of a single motif: zero or one per sequence; optimum motif width: ≥6 and ≤50; maximum number of motifs to identify: 10; and all other parameters were set to the default values. The SMART (http://smart.embl-heidelberg.de) program and Pfam database were used to annotate the MEME motifs (http://meme.sdsc.edu) (Bailey and others 2009). Multiple-sequence alignments of CsDCL, CsAGO, and CsRDR proteins were conducted using Clustal X (version 2.0) software with default parameters (Larkin and others 2007).
To determine the physical locations of the CsDCL, CsAGO, and CsRDR genes, the starting positions of all DCL, AGO, and RDR genes identified from the cucumber were initially determined using the tBLASTN program. MapInspect software was used to identify the map locations of cucumber DCL, AGO, and RDR genes (http://www.plantbreeding.wur.nl/uk/software_map inspect.html).
Analysis of Orthologous Relationships Between Cucumber and Other Species
To identify orthologous relationships of the CsDCL, CsAGO, and CsRDR proteins, the amino acid sequences of CsDCL, CsAGO, and CsRDR were BLASTP-searched against the Phytozome v10.1 (http://phytozome.jgi.doe.gov/pz/portal.html) of apple (Malus domestica), peach (Fragaria vesca), wild strawberry (Fragaria vesca), maize (Zea mays), and foxtail millet (Setaria italica). The unique relationship between orthologous genes was confirmed by performing reciprocal BLAST. Resultant hit with E value ≤le−4 and the score ≥400 were considered as significant orthologs. And the sequences of the RNA silencing component domain-containing proteins were aligned using Clustal X 2.0. Phylogenetic analysis was performed using the MEGA 4.0 program (Tamura and others 2007) based on the neighbor-joining (NJ) method. Moreover, the maximum parsimony method was used (with a bootstrap value of 1000 replicates) to create a phylogenetic tree and to validate the results from the NJ method. The cucumber CsDCL, CsAGO, and CsRDR genes were named based on their phylogenetic relatedness with Arabidopsis DCL, AGO, and RDR genes.
Analysis of Evolutionary Relationships
To further elucidate the evolutionary relationships of the CsDCL, CsAGO, and CsRDR proteins, PAL2NAL (Suyama and others 2006) was used to calculate the synonymous (Ks) and nonsynonymous (Ka) substitution rates for orthologous and paralogous gene pairs. Protein sequences of the gene pairs were aligned using MSA tools ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The alignment file along with the corresponding CDS sequences was imported into PAL2NAL (http://www.bork.embl.de/ pal2nal/), and the Ks and Ka were calculated by codeml in the PAML package of PAL2NAL. For each gene pair, the mean Ks values of the flanking conserved genes were calculated, and these values were then translated into divergence time in millions of years assuming a rate of 6.5 × 10−9 substitutions per site per year. The divergence time (T) was calculated as T = Ks/(2 × 6.5 × 10−9) × 10−6 Mya (Baloglu and others 2014; Lynch and Conery 2000; Yadav and others 2015).
In Silico Expression Analysis and Homology Modeling
Illumina RNA-HiSeq data of five tissues, namely root, stem, leaf, flower, and tendril, were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/sra/?term=sra046916) with the accession numbers SRA046916; SRX100325 (root); SRX100310 (stem); SRX100309 (leaf); SRX100319 (male flower); SRX100326 (tendril) (Shang and others 2014). The RNAseq data were then filtered, and the CsDCL, CsAGO, and CsRDR genes were imported into R and Bioconductor for expression analysis. Then, the pheatmap package was used to make the heatmaps (Yan and others 2014). Further, protein structure was determined as described by Yadav and others (2015), and the determination by homology modeling was performed using Phyre2 (Protein Homology/AnalogY Recognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2) under ‘intensive’ mode (Kelley and Sternberg 2009).
Plant Materials and Treatments
Cucumber (Cucumis sativus, Jinlü No.5; Horticultural Lüfeng Ltd., Tianjin City, China) seeds were germinated in flowerpots. Plants were grown in a greenhouse at 24–28 °C, and samples (roots, stems, leaves, flowers, and tendrils) were collected from seedlings at the seven main-stem node stage and stored in liquid nitrogen.
RNA Extraction and Quantitative Reverse-Transcription (RT)-PCR
Total RNA was extracted from the plant tissue samples using RNAiso Plus (TaKaRa) and treated with a PrimeScript™ RT Reagent Kit gDNA Eraser (TaKaRa) to remove genomic DNA contamination. RNA integrity was analyzed on a 1.2 % agarose gel, and RNA purity was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific). First-strand cDNA was synthesized with a PrimeScript™ RT Reagent Kit according to the manufacturer’s instructions. The resulting cDNA was diluted 10-fold with sterile water. Gene-specific primers for use in qRT-PCR analysis were designed using Primer 5.0 ( Table 1). The expression level of the cucumber actin 7-gene (LOC101220617) was used as an endogenous control; this gene was amplified with primers 5′-caacccaaaggctaacagag-3′ and 5′-gaatccagcacgataccagt-3′.
The qPCR was carried out in a 20 μl volume containing 1.6 μl diluted cDNA, 0.8 μl forward primer (10 μM), 0.8 μl reverse primer (10 μM), and 10 μl SYBR Premix Ex Taq II (TaKaRa). The thermal cycle conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. After 40 cycles, a melting curve was generated to analyze the specificity of the reactions. Each cDNA sample was tested with four replicates. The results from gene-specific amplification were analyzed using the comparative Cq method, which uses the formula 2−ΔΔCq for relative quantification (Livak and Schmittgen 2001); Cq represents the threshold cycle.
Results
Identification of Dicer-Like, Argonaute, and RDR Genes
To identify all Dicer-like, Argonaute, and RDR genes in the cucumber genome, we searched the annotated cucumber database with the sequences of various putative members of RNA-induced silencing complexes in cucumber, which were generated from the HMM profile in the Pfam program. Using this approach, five CsDCLs (designated CsDCL1, CsDCL2, CsDCL3, CsDCL4a, and CsDCL4b), seven CsAGOs (designated CsAGO1a to CsAGO1d, CsAGO4, CsAGO6, and CsAGO7), and eight CsRDRs (designated CsRDR1a to CsRDR1e, CsRDR2, CsRDR3, and CsRDR6) were identified. Information about these family genes including the sequence ID, aa length, MW, pI of their gene products, and the number of motifs and their physical locations on the chromosomes is listed in Table 1. The lengths of the Dicer-like, Argonaute, and RDR proteins vary, with CsDCL1 encoding a 1989 amino acid protein and most CsDCLs encoding proteins longer than 1390 amino acids, whereas CsAGO1d encodes a protein only 394 amino acid long. The pIs of all seven CsAGO gene products are above 8.5, whereas most CsDCL gene products have pIs below 7.0 (except for CsDCL2). The pIs of most CsRDR gene products are above 7.5, except for CsRDR2 (pI = 6.24; Table 1).
Diverse exon–intron structures were identified by comparing the predicted CDS with the genomic sequences of Dicer-like, Argonaute, and RDR genes in the cucumber database. Most CsAGO genes have more than six introns, whereas CsAGO7 has only two introns. Most CsRDRs (CsRDR1a, CsRDR1b, CsRDR1c, CsRDR2) have three introns, and three genes (CsRDR6, CsRDR1d, and CsRDR1e) have one intron, two introns, and four introns respectively, whereas CsRDR3 has 18 introns. By contrast, all CsDCLs have more than 20 introns (Fig. 1).
Intron–exon organization of the 20 Dicer-like, Argonaute, and RDR genes. Conserved sequences of Dicer-like, Argonaute, and RDR proteins are indicated by gray boxes. UTRs (untranslated regions) are indicated by thick black lines at both ends. Thin lines represent introns, and numbers 0, 1, and 2 indicate intron phases
Chromosomal Locations of Dicer-Like, Argonaute, and RDR Genes
Seven CsAGOs, five CsDCLs, and eight CsRDRs genes are distributed on six chromosomes, with variable distribution: there are eight genes on chromosome 1, six on chromosome 5, two each on chromosome 3 and chromosome 6, and only a single gene on chromosomes 2 and 4. Specifically, the CsAGO genes are distributed on five chromosomes, including three on chromosome 1 and four on four other chromosomes. Five CsDCL genes in cucumber are distributed on three chromosomes, including three on chromosome 1 and two (CsDCL1 and CsDCL3) on chromosomes 3 and 6, respectively. The eight CsRDRs are distributed on three chromosomes (Csa 1, Csa 2, and Csa 5), including five (CsRDR1a, CsRDR1b, CsRDR1c, CsRDR1d, and CsRDR1e) on chromosome 5. In addition, two genes (CsDCL4a and CsDCL4b) on chromosome 1 were derived from the same parent gene (Csa1M267180) and likely originated by tandem duplication (based on more than 99 % similarity at the amino acid level). Two other genes (CsRDR1b and CsRDR1c) on chromosome 5 also share the same parent gene (Csa5M239640). Moreover, the three CsRDR genes (CsRDR1a, CsRDR1b, and CsRDR1c) on chromosome 5 are adjacent to each other, as are two other CsRDR genes (CsRDR1d and CsRDR1e) on chromosome 5. Finally, two genes (CsAGO1c and CsAGO1d) on chromosome 1 are also near each other (Table 1; Fig. 2).
Physical locations of cucumber Dicer-like, Argonaute, and RDR genes. CsDCLs, CsAGOs, and CsRDRs represent cucumber Dicer-like, Argonaute, and RDR genes, respectively. Seven CsAGO genes are distributed on five chromosomes (Csa1, Csa3, Csa4, Csa5, and Csa6), eight CsRDR genes are distributed on three chromosomes (Csa1, Csa2, and Csa5), and five CsDCL genes are distributed on three chromosomes (Csa1, Csa3, and Csa6)
Sequence Analysis of Dicer-Like, Argonaute, and RDR Proteins
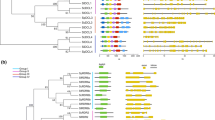
The three conserved domains, DUF, PAZ, and Piwi, are present in all CsAGO proteins except two (CsAGO1c and CsAGO1d). CsAGO1d lacks a Piwi domain, whereas CsAGO1c has only one Piwi domain. Three CsDCL proteins (CsDCL1, CsDCL4a, and CsDCL4b) have six types of conserved domains, whereas the DND1 DSRM domain is absent in the other two CsDCL proteins (CsDCL2 and CsDCL3). Finally, all CsRDR proteins contain the conserved RdRP domain (Fig. 3).
Domain distribution of cucumber Dicer-like, Argonaute, and RDR proteins. The following conserved domains are present; three (DUF, PAZ, and Piwi) in CsAGO proteins; one (RNA-dependent RNA polymerase [RdRP]) in CsRDR proteins; and six (DEAD/DEAH box helicase [DEAD], Helicase conserved C-terminal domain [Helicase C], Dicer dimerization domain [Dicer dimer], PAZ domain [PAZ], Ribonuclease III domain [Ribonuclease 3], and double-strand RNA-binding domain from DE [DND1 DSRM]) in CsDCL proteins
The online MEME server was used to identify the distribution of conserved motifs in the CsDCL, CsAGO, and CsRDR proteins in cucumber. Ten major motifs were detected in all CsDCL proteins, including a distinct copy of the newly duplicated motif 6 located between motif 2 and motif 3 (CsDCL4a and CsDCL4b). Motif 3 was duplicated in CsDCL1, and motif 5 was duplicated in CsDCL3. Only three of the seven CsAGO proteins (CsAGO1a, CsAGO1b, and CsAGO1c) contain all ten major motifs, whereas CsAGO4 lacks motif 9, CsAGO6 lacks motif 7 and motif 9, CsAGO1c lacks four motifs (motifs 4, 8, 9, and 10), and CsAGO1d has only three motifs (motifs 4, 8, and 9). Most CsRDR proteins contain all ten motifs (except for CsRDR1c, CsRDR1d, and CsRDR3), whereas three motifs (motifs 6, 8, and 9) are absent in CsRDR1c, motif 7 is absent in CsRDR1d, and five motifs (motifs 3, 6, 7, 8, and 10) are absent in CsRDR3. Finally, some proteins contain distinct copies of newly duplicated motifs, such as CsAGO4 (motif 5), CsRDR1a (motif 2), and CsRDR2 (motif 4) (Fig. 4).
Multiple alignments of Dicer-like, Argonaute, and RDR proteins. Ten motifs were identified using MEME software. Different motifs are indicated by different colors. Ten conserved motifs are located in most members of the Dicer-like, Argonaute, and RDR families. The order of motifs corresponds to the positions of motifs in the individual protein sequences. The names of members from different subfamilies and combined P values are shown on the left, and the scale at the bottom indicates the relative size of each motif
Analysis of Orthologous Relationships Between Cucumber and other Species
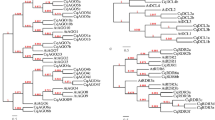
To investigate the phylogenetic relationships among the DCL, AGO, and RDR proteins and to assess the evolutionary history of these gene families, full-length protein sequences from cucumber and other species (apple, peach, wild strawberry, and so on) were used to construct a neighbor-joining phylogenetic tree. A monophyletic family comprises 7 CsAGO, 16 MdAGO, 12 PpAGO, 12 FvAGO, 19 SiAGO, 17 ZmAGO, and 11 AtAGO proteins exhibiting high sequence conservation, whereas 94 AGO proteins from cucumber and other species exclusively belong to seven subfamilies (AGO1, AGO2, AGO4, AGO5, AGO6, AGO7, and AGO10). Three CsAGOs (CsAGO1a, CsAGO1b, and CsAGO1c) are included in the same cluster, AGO10, whereas four others are grouped into four other subfamilies, respectively, AGO1, AGO4, AGO6, and AGO7. Based on the domain compositions and phylogenetic relationships of the 38 (five Cs [Cucumis sativus] DCLs, two Md [Malus domestica], eight Pp [Prunus persica], eight Si [Setaria italica], seven Fv [Fragaria vesca], four Zm [Zea mays], and four At [Arabidopsis thaliana] DCL) protein sequences, 38 DCLs exhibited high sequence conservation with their counterparts, and five CsDCL proteins (CsDCL1, CsDCL2, CsDCL3, CsDCL4a, and CsDCL4b) were divided into four subfamilies (DCL1, DCL2, DCL3, and DCL4); when more than one ortholog is present, a lower-case letter following the protein name is used based on sequence similarity. Eight CsRDR proteins, sixteen MdRDR proteins, nine PpRDR proteins, fifteen SiRDR proteins, six FvRDR proteins, and six AtRDR proteins were divided into four subfamilies; CsRDR6 is included in cluster RDR6, whereas CsRDR3 and CsRDR2 are grouped into RDR3 and RDR2, respectively, and five CsRDRs (CsRDR1a, CsRDR1b, CsRDR1c, CsRDR1d, and CsRDR1e) share high sequence conservation with AtRDR1 (Fig. 5a–c).
Phylogenetic relationships of cucumber and other species Dicer-like, Argonaute, and RDR genes. At, Arabidopsis thaliana; Cs, Cucumis sativus; MDP, Malus domestica; ppa, Prunus persica; Si, Setaria italica; mrna, Fragaria vesca; GRMZM, and AC, Zea mays. a Sequence-based clustering of AGO proteins; b Sequence-based clustering of DCL proteins, c Sequence-based clustering RDR proteins. Among those, the Arabidopsis accession numbers and abbreviations are as follows: AtAGO2 (AT1G31280.1), AtAGO3 (AT1G31290.1), AtAGO1a (AT1G48410.1), AtAGO1b (AT1G48410.2), AtAGO7 (AT1G69440.1), AtAGO4 (AT2G27040.1), AtAGO5 (AT2G27880.1), AtAGO6 (AT2G32940.1), AtAGO8 (AT5G21030.1), AtAGO9 (AT5G21150.1), AtAGO10 (AT5G43810.1), AtDCL1 (AT1G01040.1), AtDCL2 (AT3G03300.1), AtDCL3 (AT3G43920.1), AtDCL4 (AT5G20320.1), AtRDR1 (AT1G14790.1), AtRDR2 (AT4G11130.1), AtRDR3 (AT2G19910.1), AtRDR4 (AT2G19920.1), AtRDR5 (AT2G19930.1), AtRDR6 (AT3G49500.1)
Analysis of Evolutionary Relationships
Orthologs of CsAGO, CsDCL, and CsRDR proteins were identified in apple, peach, wild strawberry, maize, and foxtail millet. Among seven CsAGO genes, the collinearity pattern of one (~47 %) AGO gene with apple, four (~47 %) AGO genes with peach, five (~26 %) with wild strawberry, one with maize, and three (~21 %) with foxtail millet (Fig. 6; Supplementary Table S1). Meanwhile, of the five CsDCL genes, two (40 %) are present in peach, four (80 %) in maize, and four (80 %) in foxtail millet, whereas there is no gene found in apple and wild strawberry (Fig. 6; Supplementary Table S2). Similarly, CsRDR genes showed the syntenic relationship with one (~47 %) RDR gene with apple, four (~47 %) RDR genes with peach, two (~26 %) with wild strawberry, and one with maize and two (~21 %) with foxtail millet (Fig. 6; Supplementary Table S3).
Further, the ratios of nonsynonymous (Ka) versus synonymous (Ks) substitution rate (Ka/Ks) for the orthologous gene pairs of DCL, AGO, and RDR highlighted the evolutionary relationships of these genes (Fig. 6; Supplementary Table S1–S3). The analysis revealed the recent divergence of cucumber from peach, apple, and wild strawberry around 100–240 Mya, whereas there was a much earlier divergence of cucumber to maize and foxtail millet (~560–2740 Mya) (Fig. 6; Supplementary Table S1–S3).
In Silico Expression Profiles and Homology Modeling of CsDCL, CsAGO, and CsRDR Genes
The expression pattern of CsAGO, CsDCL, and CsRDR genes in six tissues, namely, root, stem, leaf, male flower, female flower, and tendril, was analyzed using the RNA-sequence data. The heat map showed a differential expression pattern of all the genes. Among CsAGOs, CsAGO1a and CsAGO4 were found to be highly expressed in all six tissues. Tissue-specific higher expression of CsAGO1b, CsAGO1c, and CsAGO7 was observed in leaf. In particular, all CsDCLs were predominantly expressed in tendrils at lower levels, whereas all the CsDCLs showed moderate expression in other tissues. Among CsRDRs, higher expression of CsRDR1a and CsRDR1b was observed in most of the tissues especially in root. CsRDR2 and CsRDR6 showed moderate expression in all tissues, whereas CsRDR1d and CsRDR1e showed a relatively lower expression in most tissues (Fig. 7). Using BLASTP algorithm, three-dimensional protein structures were predicted for 7 CsAGO, 5 CsDCL, and 8 CsRDR proteins on the basis of homology searching in the PDB database and Phyre2 in intensive mode. The protein structures are modeled at greater than 90 % confidence (Supplementary Figs. 1–3).
Real-Time Quantitative RT-PCR Analysis of the Expression Levels of the Dicer-Like, Argonaute, and RDR Genes
Seven CsAGOs, five CsDCLs, and eight CsRDRs were chosen for expression analysis based on representing the subfamilies of respective gene families. Among 7 CsAGOs, CsAGO1c, CsAGO1d, and CsAGO7 were highly expressed in the leaves, among which, CsAGO1c and CsAGO1d were highly upregulated (13.6-fold change and 9.6-fold change, respectively) in leaves, CsAGO7 was greatly upregulated (19.2-fold change) in leaves; whereas CsAGO1c (3.92-fold change), CsAGO1d (3.0-fold change), CsAGO6 (3.5-fold change), and CsAGO7 (3.4-fold change) had relatively higher upregulation in tendrils. Meanwhile, all CsAGOs had low expression levels in stems. Among CsDCLs, a relatively higher upregulation of all CsDCLs was observed in tendrils (4.2-fold change of CsDCL1, 3.0-fold change of CsDCL2, 2.0-fold change of CsDCL3, 1.7-fold change of CsDCL4a, and 1.9-fold change of CsDCL4b, respectively) than in other organs, while except for tendrils, almost no expression was detected for CsDCL1, CsDCL4a or CsDCL4b. CsRDR1a, CsRDR2, CsRDR3, and CsRDR6 had relatively higher upregulation (1.3-fold change, 3.8-fold change, 2.8-fold change, and 1.3-fold change) in tendrils than in other organs. And almost no expression was detected in stems or flowers (Fig. 8).
Expression of CsAGO, CsDCL, and CsRDR genes in cucumber roots (Rt), stems (St), leaves (Le), flowers (Fl), and tendrils (Te). Fold changes of CsAGO, CsDCL, and CsRDR genes are shown. Expression levels were quantified by qRT-PCR. Cucumis sativus actin-7-like (LOC101220617) was used as a reference gene. The expression levels in roots were arbitrarily set to 1. Error bars represent the standard deviations of four technical PCR replicates
Discussion
Identification of CsAGOs, CsDCLs, and CsRDRs in Cucumber
The AGO, DCL, and RDR gene families play important roles in small RNA-mediated gene silencing, and many AGOs, DCLs, and RDRs genes have been identified in numerous plants, such as Arabidopsis (Fang and Spector 2007), rice (Yang and others 2013), maize (Qian and others 2011), soybean (Liu and others 2014), Salvia miltiorrhiza (Shao and Lu 2013), and tomato (Bai and others 2012), many of which were identified through computational prediction based on sequence similarity. In Arabidopsis, whereas three AtAGOs (AtAGO3, AtAGO5, and AtAGO8) were predicted computationally (http://www.arabidopsis.org/), the remaining seven were experimentally tested (Shao and Lu 2013). Moreover, six rice OsAGO genes (OsAGO1a, OsAGO1b, OsAGO1c, OsAGO1d, OsAGO7, and OsPNH1) have been cloned (Shao and Lu 2013). In this study, we performed genome-wide prediction of 20 CsAGOs, CsDCLs, and CsRDRs using computational approaches; the number of identified cucumber CsAGOs, CsDCLs, and CsRDRs genes is comparable to that of Arabidopsis. The results of this study are useful for further elucidating the functions of CsAGOs, CsDCLs, and CsRDRs in cucumber as well as gene model prediction.
Phylogenetic Analysis and Conservation of CsAGOs, CsDCLs, and CsRDRs
Plants AGO, DCL, and RDR proteins share some highly conserved domains. MEME analysis showed that the majority of the motifs were well conserved in the CsAGO, CsDCL, and CsRDR proteins. The phylogeny and domain analysis revealed the occurrence of significant domain variations and conservations in all three proteins. For example, AGOs share three conserved domains including DUF1785, PAZ, and PIWI. PAZ functions in binding sRNA duplexes, and PIWI is involved in RNA cleavage (Song and Joshua-Tor 2006; Wang and others 2008), whereas the function of DUF1785 remains to be elucidated. RDRs share one conserved RdRP domain, whereas DCLs share six conserved domains including DEAD, Helicase C, DUF283, PAZ, Ribonuclease III, and dsRB. In the current study, we found that two of seven CsAGOs (CsAGO1c and CsAGO1d) lack one or two conserved domains, whereas all CsRDR proteins contain the conserved RdRP domain. Finally, although three CsDCLs contain all of the conserved domains, two of five CsDCLs (CsDCL2 and CsDCL3) have lost the dsRB domain. It remains to be determined whether the proteins that have lost conserved domains still function in small RNA-mediated silencing.
AGO is essential for siRNA biogenesis; plants encode multiple AGOs to meet the diversified functions of small RNA silencing (Bartel 2004). The cucumber genome encodes seven AGOs, three of which (CsAGO1a, CsAGO1b, and CsAGO1c) share high similarity to each other and AtAGO10, together with CsAGO1d, which is highly similar to AtAGO1, belongs to the first subfamily. These proteins might associate with miRNA and ta-siRNAs to cleave target mRNA, thereby silencing specific genes (Yu and Wang 2010). CsAGO4 and CsAGO6, which are highly similar to AtAGO4 and AtAGO6, respectively, belong to the second subfamily. These proteins might bind to 24 nt ra-siRNAs to direct DNA methylation (Havecker and others 2010). CsAGO7, which is highly similar to AtAGO7, comprises the third clades, together with AtAGO2 and AtAGO3.
DCL plays an important role in small RNA-mediated silencing in plants. Plants contain four groups of DCLs, which function in the generation of both miRNAs and siRNAs; these DCLs have overlapping and diversified functions in miRNA and siRNA biogenesis (Margis and others 2006). Arabidopsis, tomato, sorghum, and soybean each possess four DCL families (Baulcombe 2004; Bai and others 2012; Liu and others 2014; Curtin and others 2012). In this study, we determined that cucumber possess four DCL subfamilies. Among these, two DCL4 paralogs, DCL4a and DCL4b, share high sequence similarity. DCL4a and DCL4b might have arisen from gene replication and may have evolved new functions related to those of the original gene. These genes belong to the same clade as Arabidopsis AtDCL4, which produces 21 nt siRNA or some miRNAs (Xie and others 2005). CsDCL1, CsDCL2, and CsDCL3, which have high similarity to Arabidopsis AtDCL1, AtDCL2, and AtDCL3, respectively, might have similar functions. In Arabidopsis, AtDCL1 cleaves pri-miRNA to release 21 nt miRNAs (Song and others 2007), whereas AtDCL2 produces 22 nt viral-derived siRNAs in infected plants (Bouche and others 2006) and AtDCL3 generates 24 nt ra-siRNAs (Henderson and others 2006).
RDR is an essential player in siRNA biogenesis as well. Arabidopsis, apple, peach, wild strawberry, foxtail millet, and maize plants possess four groups of RDRs: RDR1, RDR2, RDR3, and RDR6. In Arabidopsis, RDR2 converts ssRNAs to precursor dsRNAs of ra-siRNAs (Xie and others 2004), whereas RDR6 produces ta-siRNA precursors (Yoshikawa and others 2005). RDR1 acts redundantly with RDR6 in viral-derived siRNA biogenesis (Wang and others 2010). The function of the RDR3 family is currently unknown. In this study, we identified cucumber homologs corresponding to RDR1, RDR2, RDR3, and RDR6 in Arabidopsis. Five CsRDR1 paralogs (CsRDR1a to CsRDR1e), which are highly similar to each other, are also similar to AtRDR1, suggesting that the RDR1 gene family in plants is derived from a common ancestor. CsRDR2, CsRDR3, and CsRDR6 have high similarity to AtRDR2, AtRDR3, and AtRDR6, respectively, and might have similar functions.
Analysis of Orthologous and Evolutionary Relationships with Other Species
Orthologs of CsAGO, CsDCL, and CsRDR proteins were identified between cucumber and C3 plants (apple, peach, and wild strawberry) and C4 plants (maize and foxtail millet). Among seven CsAGO genes, the collinearity pattern of one AGO genes with apple, four with peach, five with wild strawberry, one with maize, and three with foxtail millet. Meanwhile, of the five CsDCL genes, two are present in peach, four in maize, and four in foxtail millet, whereas there is no gene found in apple and wild strawberry. Similarly, CsRDR genes showed the syntenic relationship with one RDR gene with apple, four with peach, two with wild strawberry, one with maize, and two with foxtail millet. Further, the ratios of nonsynonymous (Ka) versus synonymous (Ks) substitution rate (Ka/Ks) for the orthologous gene pairs of DCL, AGO, and RDR highlighted the evolutionary relationships of these genes. The analysis revealed the recent divergence of cucumber from peach, apple, and wild strawberry around 100–240 Mya, whereas there was a much earlier divergence of cucumber to maize and foxtail millet (~560 to 2740 Mya). The synteny analysis revealed the close evolutionary relationship between cucumber and C3 plants, whereas there was a much earlier divergence of cucumber to maize and foxtail millet. This ortholog information of AGO, DCL, and RDR gene families between cucumber and other species could assist in gene identification, selection of candidate genes for further characterization, regulatory motif discovery, gene functional annotation, and revealing gene clusters.
Expression Pattern of CsAGOs, CsDCLs, and CsRDRs Gene Families in Cucumber
AGO, DCL, and RDR proteins are reported to control the small RNA-mediated gene silencing pathways and epigenetic regulation of the genome (Sahu and others 2013). Hence, the in silico expression pattern of CsAGO, CsDCL, and CsRDR genes in six tissues (root, stem, leaf, male flower, female flower, and tendril) was analyzed using the RNA-sequence data. The heat map showed a differential expression pattern of all the genes. The in silico expression data would be useful in studying functional response patterns of the genes, genotyping analysis, parsing pathways, and performing case versus control studies.
Meanwhile, we compared the expression levels of CsAGOs, CsDCLs, and CsRDRs in stems, leaves, flowers, and tendrils with those in roots (control). Among CsAGOs, CsAGO1c, CsAGO1d, and CsAGO7 were highly upregulated. Although CsAGO1a, CsAGO1b, CsAGO1c, and CsAGO1d share a close evolutionary relationship, their expression patterns were somewhat different. Interestingly, all CsAGOs were significantly upregulated in leaves and tendrils compared with other tissues, whereas nearly all CsAGOs were significantly downregulated in stems and flowers. These results suggest that all CsAGOs function in tendrils and leaves during plant vegetative and reproductive development. Moreover, all CsDCLs were upregulated in tendrils than in other tissues, suggesting that these genes function in tendril development. In addition, a relatively higher upregulation of all CsDCLs was observed in tendrils, which meant that these genes may function in tendril development, whereas all CsDCLs (except CsDCL3) were downregulated in stems, leaves, and flowers, and the evolutionarily related CsDCL4a and CsDCL4b shared the same expression pattern. Finally, the expression of nearly half of the CsRDRs (CsRDR1a, CsRDR2, CsRDR3, and CsRDR6) was upregulated in tendrils, and downregulated in all other organs, which meant that these genes may function in tendril development. Meanwhile, the expression patterns of CsRDR1a, CsRDR1b, CsRDR1c, CsRDR1d, and CsRDR1e were somewhat different despite their close evolutionary relationship. These variations in the gene expression pattern suggest the role of these genes in the complex molecular network of the RNA silencing process. These data would provide a preliminary knowledge to expedite further functional characterization of CsAGO, CsDCL, and CsRDR genes.
References
Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, Yang YH, Xie BY (2012) Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 501(1):52–62
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–W208
Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014) Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 9(4):e96014
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Baulcombe D (2004) RNA silencing in plants. Nature 431(7006):356–363
Bernstein E, Caudy AA, Hammond SM, Harmon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818):363–366
Bouche N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25(14):3347–3356
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896
Curaba J, Chen X (2008) Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem 6:3059
Curtin SJ, Kantar MB, Yoon HW, Whaley AM, Schlueter JA, Stupar RM (2012) Co-expression of soybean Dicer-like genes in response to stress and development. Funct Integr Genomics 12:671–682
Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–553
Ding SW (2010) RNA-based antiviral immunity. Nat Rev Immunol 9:632–644
Djupedal I, Ekwall K (2009) Epigenetics: heterochromatin meets RNAi. Cell Res 19:282–295
Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, Llave C (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol 11:5167–5177
Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17(9):818–823
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) The Pfam protein families database. Nucleic Acids Res 42(Database Issue):D222–D230
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22(2):321–334
Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38(6):721–725
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9:22–32
Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, Kapoor S (2008) Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom 9:451
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:142–144
Liu X, Lu T, Dou YC, Yu B, Zhang C (2014) Identification of RNA silencing components in soybean and sorghum. BMC Bioinform 15:4
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C (T)) method. Methods 25(4):402–408
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580(10):2442–2450
Moazed D (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420
Pattanayak D, Solanke AU, Kumar PA (2013) Plant RNA interference pathways: diversity in function, similarity in action. Plant Mol Biol Rep 31:493–506
Qian YX, Cheng Y, Cheng X, Jiang HY, Zhu SW, Cheng BJ (2011) Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep 30:1347–1363
Qu F, Ye XH, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. PNAS 105(38):14732–14737
Sahu PP, Pandey G, Sharma N, Puranik S, Muthamilarasan M, Prasad M (2013) Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep 32:1151–1159
Shang Y, Ma YS, Zhou Y, Zhang HM, Duan LX, Chen HM, Zeng JG, Zhou Q, Wang SH, Gu WJ, Liu M, Ren JW, Gu XF, Zhang SP, Wang Y, Yasukawa K, Bouwmeester HJ, Qi XQ, Zhang ZH, Lucas WJ, Huang SW (2014) Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346:1084
Shao FJ, Lu SF (2013) Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genom 14:512
Song JJ, Joshua-Tor L (2006) Argonaute and RNA-getting into the groove. Curr Opin Struct Biol 16(1):5–11
Song L, Han MH, Lesicka J, Fedoroff N (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA 104(13):5437–5442
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Vaistij FE, Jones L (2009) Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol 3:1399–1407
Vaucheret H (2008) Plant ARGONAUTES. Trends Plant Sci 13(7):350–358
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687
Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ (2008) Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456(7224):921–926
Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW (2010) RNA mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107(1):484–489
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2(5):E104
Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102(36):12984–12989
Yadav CB, Muthamilarasan M, Pandey G, Prasad M (2015) Identification, characterization and expression profiling of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in foxtail millet. Plant Mol Biol Rep 33(1):43–55
Yan HW, Zhang W, Lin YX, Dong Q, Peng XJ, Jiang HY, Zhu SW, Cheng BJ (2014) Different evolutionary patterns among intronless genes in maize genome. Biochem Biophys Res Commun 449:146–150
Yang Y, Zhong J, Ouyang YD, Yao JL (2013) The integrative expression and co-expression analysis of the AGO gene family in rice. Gene 528(2):221–235
Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19(18):2164–2175
Yu B, Wang H (2010) Translational inhibition by microRNAs in plants. Prog Mol Subcell Biol 50:41–57
Zhao HL, Zhao K, Wang J, Chen X, Chen Z, Cai YH, Xiang Y (2014) Comprehensive analysis of Dicer-like, Argonaute, and RNA-dependent RNA polymerase gene families in grapevine (Vitis Vinifera). J Plant Growth Regul. doi:10.1007/s00344-014-9448-7
Zilberman D, Cao X, Jacobsen SE (2003) Argonaute 4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719
Acknowledgments
This work was supported by Grants from the Higher Education Revitalization Project of Anhui Province (2013zdjy057), the Academic Backbone Cultivation Project of Anhui Agricultural University (2014XKPY-12), the Scientific Research Foundation for the Stability and Introduction of the Talents (wd2011-14), and the Academician Innovative Project of Anhui Province (AH201310364017). We thank members of the Key Laboratory of Crop Biology of Anhui Province for their assistance in this study.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Defang Gan and Dandi Liang have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gan, D., Liang, D., Wu, J. et al. Genome-Wide Identification of the Dicer-Like, Argonaute, and RNA-Dependent RNA Polymerase Gene Families in Cucumber (Cucumis sativus L.). J Plant Growth Regul 35, 135–150 (2016). https://doi.org/10.1007/s00344-015-9514-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-015-9514-9