Abstract
Objectives
To investigate the predictive value of static O-(2-18F-fluoroethyl)-L-tyrosine positron emission tomography (18F-FET PET) and cerebral blood volume (CBV) for glioma grading and determining isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion status.
Methods
Fifty-two patients with newly diagnosed gliomas who underwent simultaneous 18F-FET PET and dynamic susceptibility contrast perfusion-weighted imaging (DSC-PWI) examinations on hybrid PET/MR were retrospectively enrolled. The mean and max tumor-to-brain ratio (TBR) and normalized CBV (nCBV) were calculated based on whole tumor volume segmentations with reference to PET/MR images. The predictive efficacy of FET PET and CBV in glioma according to the 2016 World Health Organization (WHO) classification was evaluated by receiver operating characteristic curve analyses with the area under the curve (AUC).
Results
TBRmean, TBRmax, nCBVmean, and nCBVmax differed between low- and high-grade gliomas, with the highest AUC of nCBVmean (0.920). TBRmax and nCBVmean showed significant differences between gliomas with and without IDH mutation (p = 0.032 and 0.010, respectively). Furthermore, TBRmean, TBRmax, and nCBVmean discriminated between IDH-wildtype glioblastomas and IDH-mutated astrocytomas (p = 0.049, 0.034 and 0.029, respectively). The combination of TBRmax and nCBVmean showed the best predictive performance (AUC, 0.903). Only nCBVmean differentiated IDH-mutated with 1p/19q codeletion oligodendrogliomas from IDH-wildtype glioblastomas (p < 0.001) (AUC, 0.829), but none of the parameters discriminated between oligodendrogliomas and astrocytomas.
Conclusions
Both FET PET and DSC-PWI might be non-invasive predictors for glioma grades and IDH mutation status. FET PET combined with CBV could improve the differentiation of IDH-mutated astrocytomas and IDH-wildtype glioblastomas. However, FET PET and CBV might be limited for identifying oligodendrogliomas.
Key Points
• Static 18F-FET PET and DSC-PWI parameters differed between low- and high-grade gliomas, with the highest AUC of the mean value of normalized CBV.
• Static 18F-FET PET and DSC-PWI parameters based on hybrid PET/MR showed predictive value in identifying glioma IDH mutation subtypes, which have gained importance for both determining the diagnosis and prognosis of gliomas according to the 2016 WHO classification.
• Static 18F-FET PET and DSC-PWI parameters have limited potential in differentiating IDH-mutated with 1p/19q codeletion oligodendrogliomas from IDH-wildtype glioblastomas or IDH-mutated astrocytomas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The World Health Organization (WHO) classification system incorporated molecular biomarkers, especially the isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion, into the classification of brain gliomas in 2016 [1]. Retrospective and prospective studies have demonstrated a significant correlation between IDH mutation burden and prognosis of patients with glioma [2,3,4]. The majority of WHO grade II and III gliomas are of IDH mutant type, which indicates a longer survival than IDH-wildtype gliomas. Moreover, IDH-mutated with 1p/19q codeletion oligodendrogliomas are related to better outcomes and have higher chemotherapy sensitivity than non-codeleted gliomas. In contrast, IDH-wildtype glioblastomas are extremely aggressive and have the worst prognosis [5]. Accordingly, predicting IDH and 1p/19q codeleted status non-invasively would be of great value for patients with respect to treatment planning and prognostic evaluation, especially in settings where detailed molecular assay of glioma specimens is not yet routinely available.
Both clinical conventional and advanced neuroimaging techniques showed high diagnostic value for glioma grading and genotyping according to the revised 2016 WHO classification [6,7,8]. Cerebral blood volume (CBV) provided by dynamic susceptibility contrast perfusion-weighted imaging (DSC-PWI) has limited value in predicting the IDH mutation and 1p/19q codeletion status in gliomas [9, 10]. Moreover, the low resolution of CBV maps of DSC-PWI might limit the clinical practice of quantitative evaluation of glioma perfusion. Recently, amino acid positron emission tomography (PET) imaging has been recommended by the Response Assessment in Neuro-Oncology (RANO) and other working groups in gliomas for primary diagnosis, biopsy or treatment guidance, and therapeutic monitoring [11, 12]. The amino acid PET tracer O-(2-18F-fluoroethyl)-L-tyrosine (18F-FET) can identify tumor borders with superior tumor-to-background contrast, and it also can characterize molecular subtypes and be applied to non-invasive tumor grading [13,14,15,16]. Our previous work further found that the metabolically active biodistribution of gliomas delineated with 18F-FET PET significantly exceeds tumor volume on contrast-enhanced MRI, and histopathology obtained by stereotactic biopsy confirms these findings [17].
In this study, we applied a new method of transferring the volume of interest (VOI) of the whole solid part of the tumor on 18F-FET PET images to its CBV maps on a hybrid PET/MR scanner. We hypothesize that 18F-FET PET and DSC-PWI may provide information for glioma preoperative diagnosis. We aimed to evaluate the predictive efficacy of metabolism and perfusion amount derived from 18F-FET PET and DSC-PWI based on hybrid PET/MR for the prediction of the tumor grade, IDH mutation status, and 1p/19q codeletion status in gliomas.
Materials and methods
Patients
From February 2019 to October 2019, 72 patients with suspected supratentorial gliomas who underwent hybrid 18F-FET PET/MR imaging prior to biopsy or gross resection were retrospectively entered in this study. Among the 72 patients, the following subjects were excluded: four patients with non-neoplastic lesions or metastases, 8 patients with secondary glioblastoma, 5 patients with incomplete IDH mutation and 1p/19q codeletion diagnoses, and 3 patients with significant motion-related or other unremovable artifacts on PET/MR images. A total of 52 patients were enrolled in the final cohort. The diagnosis of glioma was supported by pathological histology according to the 2016 WHO classification system, and all included patients were divided into the following 3 groups: IDH-wildtype glioblastomas formed group 1, IDH-mutated astrocytomas formed group 2, and IDH-mutated with 1p/19q codeletion oligodendrogliomas formed group 3. There were no IDH-wildtype astrocytomas or newly diagnosed IDH-mutated glioblastomas among the 72 enrolled patients.
The Ethics Committee and Institutional Review Board of Xuanwu Hospital Capital Medical University approved this retrospective study. Written informed consent was obtained from patients before PET/MR examinations.
Hybrid 18F-FET PET/MR imaging protocol
All PET and MRI examinations were simultaneously performed on a 3-T hybrid PET/MR (GE Healthcare) with a 19-channel head-neck coil before any treatment. The duration of PET/MR scanning was approximately 35 min. All patients fasted for at least 4 h before the PET/MR examination as recommended [12]. After a daily routine emission scan for atlas-based attenuation correction with cylinder source, approximately 200 MBq of 18F-FET was injected intravenously. 18F-FET PET acquisition started 20 min after tracer injection, with a total acquisition time of 20 min. PET reconstruction was performed with a TOF-point spread function-ordered subset expectation maximization (TOF-PSF-OSEM, 6 iterations and 16 subsets) algorithm on a 128 × 128 matrix, with a 35 cm axial field-of view and a 2.78 mm slice thickness. Emission data were corrected for scattering, random effects, and dead-time coincidences.
The MRI sequences including T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), fluid-attenuated inversion recovery (FLAIR), 3-dimensional (3D) T1WI and FLAIR, DSC-PWI, and post-gadolinium 3D T1WI were acquired at the same time as PET scanning. The main MRI protocol comprised 3D T2 FLAIR (repetition time/echo time = 9000/maximum, scanning time = 5 min 21 s) and pre- and post-gadolinium 3D T1WI (repetition time/echo time = 3.2/8.5, scanning time = 5 min 21 s respectively). Post-gadolinium 3D T1WI was performed after the acquisition of DSC-PWI. The details of the scan parameters are summarized in Supplementary Table 1. After a preload of contrast agent (gadopentetate dimeglumine, 0.025 mmol/kg) to reduce T1 effects due to contrast agent leakage from the vessels [18], DSC-PWI was performed with a gradient-recalled T2*-weighted echo-planar imaging sequence 5 min later (TR/TE = 1800/15.4 ms, flip angle = 90°, thickness/space = 5/1 mm, FOV = 240 × 240 mm, NEX = 1.0, scanning time = 1 min 30 s). An 18- to 20-gauge intravenous catheter was inserted into the median cubital vein of all patients before examinations. During the first 7 phases, non-enhanced images were acquired to establish a pre-contrast baseline. When the scan came to the eighth phase of DSC-PWI, gadopentetate dimeglumine was bolus injected at a dose of 0.1 mmol/kg of body weight (flow rate, 4–5 mL/s; depending on the condition of the blood vessels) with an MRI-compatible power injector. Subsequently, a 20-mL continuous saline flush was performed at the same injection rate. Finally, a series of 50 phases and a total of 1200 images were obtained.
Data postprocessing
CBV maps were generated automatically from DSC-PWI data by a single-compartment model and an arterial input function using AW 4.7 postprocessing workstation (GE Healthcare). 18F-FET PET images, 3D T1 images, T2 FLAIR images, and CBV maps were postprocessed and analyzed in PMOD version 3.505 (PMOD). Different modalities were co-registered using non-affine deformations and manually adjusted by referring to anatomic landmarks. The static PET images and CBV maps were resliced to the same voxel size as 3D T1 CE MRI with 1 × 1 × 1 mm.
For the quantitative analysis of 18F-FET PET and CBV data, two volumes of interest (VOIs) were drawn for each case: (1) A large crescent-shaped VOI was set at the level of semi-oval center of the unaffected hemisphere including white and gray matter as a reference to calculate relative parameters [19]. (2) For patients with increased FET uptake, the second VOI was delineated on 18F-FET PET images by an auto-contouring process with a tumor-to-brain ratio (TBR) threshold of 1.6 or more based on a previous biopsy-controlled study [20]. For patients without increased FET uptake, the second VOI was delineated on 3D FLAIR images using a 3D auto-contouring segmentation method with an individually determined visual threshold judged by two experienced neuroradiologists in consensus. The included blood vessels and other non-tumorous tissues with the standardized uptake value (SUV) or signal intensity exceeding the threshold were manually removed from the VOI. Necrotic areas of the lesions without FET uptake were also excluded from the final volume [21]. Then, the second VOI was transferred to the co-registered 18F-FET PET images and CBV maps to obtain the maximum and mean SUV and CBV values of the lesions (Fig. 1). The mean and maximum TBR (TBRmean and TBRmax, respectively) and normalized CBV (nCBVmean and nCBVmax, respectively) were calculated by dividing the SUVmean/max and CBVmean/max of the lesions by the reference value.
18F-FET PET images, the 3D tumor mask, and CBV maps of a 53-year-old male with IDH-wildtype glioblastoma. The 8F-FET PET images (a) in the axial, coronal, and sagittal planes from left to right show a lesion delineated with a red border with a TBR threshold of 1.6. The 3D tumor mask (b) extracted from 18F-FET PET images is exactly transpired and co-registered to the corresponding CBV maps (c). 18F-FET PET, O-(2-18F-fluoroethyl)-L-tyrosine positron emission tomography; 3D, 3-dimensional; CBV, cerebral blood volume; IDH, isocitrate dehydrogenase; TBR, tumor-to-brain ratio
Neuropathological evaluation
The enrolled gliomas were histologically classified according to the 2016 WHO classification of gliomas. The IDH mutation status was detected by an immunohistochemical assay to first evaluate the IDH1 gene mutation status at the most common codon R132H. If the result was negative, further genomic sequencing analysis was performed. The 1p/19q codeletion status was analyzed by fluorescence in situ hybridization. Pathology was determined by two neuropathologists separately. If there was any discrepancy in the diagnosis, a consensus was reached.
Statistical analysis
Statistical analysis was performed using SPSS version 22 (IBM), MedCalc Statistical Software version 18.2.1 and Graph Pad Prism version 8.0.2. Descriptive statistics are presented as the mean and standard deviation. The Shapiro-Wilk test was used to determine the normality of quantitative data. For comparison of differences between two groups, the normally distributed continuous variable was compared using the independent t test for uniform variance data, with adjustment by the non-parametric Mann-Whitney U test in cases of unequal variance or non-normally distributed data. The Kruskal-Wallis test was performed to compare the differences among three groups. Receiver operating characteristic (ROC) curve analysis was performed in SPSS version 22 (IBM) and MedCalc Statistical Software version 18.2.1, to screen the optimal parameter and its best cutoff value for the discrimination of different grades and glioma genotypes through the assessment of the area under the ROC curve (AUC).
Results
Patient characteristics
Fifty-two patients (31 men, 21 women; mean age 51.23 ± 15.59 years) with a primary diagnosis of WHO grade II, III, and IV gliomas were included in this study (Table 1). Histopathological and molecular analyses revealed 30 IDH-wildtype glioblastomas (group 1) and 22 IDH-mutated gliomas (group 2, 9/22 IDH-mutated astrocytomas; group 3, 13/22 IDH-mutated with 1p/19q codeletion oligodendrogliomas).
Predictive efficacy of 18F-FET PET and CBV among genotypes
The distribution of TBRmean, TBRmax, and nCBVmean was significantly different among the 3 groups (p = 0.045, 0.037, and 0.003, respectively), while there was no significant difference in nCBVmax among 3 groups (p = 0.111). All 3 parameters (TBRmean, TBRmax, and nCBVmean) discriminated between IDH-wildtype glioblastomas (group 1) and IDH-mutated astrocytomas (group 2) (p = 0.049, 0.034, and 0.029, respectively), while only nCBVmean differentiated between IDH-wildtype glioblastomas (group 1) and IDH-mutated with 1p/19q codeletion oligodendrogliomas (group 3) (p < 0.001). Furthermore, no parameters discriminated between IDH-mutated astrocytomas (group 2) and IDH-mutated with 1p/19q codeletion oligodendrogliomas (group 3) (p = 0.806, 0.501, and 1.000 respectively, Fig. 2). Imaging examples of gliomas from each genotype are provided in Fig. 3.
Box plots comparing TBRmean, TBRmax, nCBVmean, and nCBVmax of 18F-FET PET and DSC-PWI in IDH-wildtype glioblastomas, IDH-mutated without 1p/19q codeletion astrocytomas, and IDH-mutated with 1p/19q codeletion oligodendrogliomas. The significant p values for intergroup comparison are indicated in the figures. NS indicates no significance. TBR, tumor-to-brain ratio; nCBV, normalized cerebral blood volume (the ratio of the CBV of lesion to the CBV of the unaffected hemisphere); 18F-FET PET, O-(2-18F-fluoroethyl)-L-tyrosine positron emission tomography; DSC-PWI, dynamic susceptibility contrast perfusion-weighted imaging; IDH, isocitrate dehydrogenase
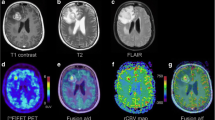
Axial slices of T2WI (left column), FLAIR (second column), T1WI (third column), contrast-enhanced T1WI (forth column), 18F-FET PET images (fifth column), and CBV maps (right column) of 3 patients with different genotypes. The top row (a) shows a 53-year-old male with a WHO grade IV IDH-wildtype glioblastoma in right parietal lobe. The TBRmean, TBRmax, nCBVmean, and nCBVmax values of the lesion are 2.48, 4.83, 2.78, and 6.63, respectively, which are higher than those of IDH-mutated without 1p/19q codeletion astrocytoma. The middle row (b) shows a 34-year-old female with a WHO grade III IDH-mutated with 1p/19q codeletion oligodendroglioma in left frontal lobe. The TBRmean, TBRmax, nCBVmean, and nCBVmax values of the lesion are 3.98, 7.35, 1.08, and 3.33 respectively. The bottom row (c) shows a 31-year-old female with a WHO grade II IDH-mutated without 1p/19q codeletion astrocytoma in left frontal-parietal lobe. The TBRmean, TBRmax, nCBVmean, and nCBVmax values of the lesion are 2.35, 3.74, 0.83, and 3.24 respectively. FLAIR, fluid-attenuated inversion recovery; T1WI, T1-weighted imaging; 18F-FET PET, O-(2-18F-fluoroethyl)-L-tyrosine positron emission tomography; CBV, cerebral blood volume; IDH, isocitrate dehydrogenase; WHO, World Health Organization; TBR, tumor-to-brain ratio; nCBV, normalized cerebral blood volume (the ratio of the CBV of lesion to the CBV of the unaffected hemisphere)
Since that TBRmean, TBRmax, and nCBVmean could differentiate IDH-wildtype glioblastomas (group 1) from IDH-mutated astrocytomas (group 2), we further evaluated the AUC of the combination of TBRmean, TBRmax, and nCBVmean (pairwise combination and combination of three parameters) in discriminating groups 1 and 2. We found that the AUC of combined parameters based on logistic regression was higher than that of a single parameter alone. The AUC, cutoff values, sensitivity, and specificity for parameters with significant differences are summarized in Table 2.
Predictive efficacy of 18F-FET PET and CBV for IDH mutation status
TBRmax and nCBVmean showed significant differences between gliomas with and without IDH mutation (t = 2.208, p = 0.032; Z = − 2.564, p = 0.010, respectively). ROC curve analysis indicated that the cutoff value of TBRmax and nCBVmean of 3.806 and 1.078, respectively, were able to discriminate IDH subtypes (AUC value, 0.678 vs. 0.815; sensitivity, 93.10% vs. 88.46%; specificity, 59.09% vs. 75.00%). However, the combined analysis of TBRmax and nCBVmean did not significantly improve the predictive efficacy for IDH mutation status, with AUC, sensitivity, and specificity values of 0.685, 92.00%, and 56.25%, respectively. The details of the results are presented in Table 3.
Diagnostic efficacy of 18F-FET PET and CBV for glioma grading
According to the previous WHO grading system classification, the distribution of TBRmean, TBRmax, and nCBVmean was significantly different among WHO grade II, III, and IV gliomas (p = 0.002, 0.001, and < 0.001, respectively), while there was no significant difference in nCBVmax among the 3 different grades (p = 0.057). Only nCBVmean differentiated between WHO grade II and III gliomas (p < 0.001). All 3 parameters (TBRmean, TBRmax, and nCBVmean) discriminated between WHO grade II and IV gliomas (p = 0.002, 0.002, and < 0.001, respectively), while no parameters could discriminate between WHO grade III and IV gliomas (p all > 0.05). ROC curve analysis of PET and CBV parameters indicated that the nCBVmean reached a higher AUC value (AUC, 0.912; cutoff value, 1.12; sensitivity, 87.50%; specificity, 89.66%) than TBRmean and TBRmax in discriminating between WHO grade II and IV gliomas (AUC = 0.783 and 0.781 for TBRmean and TBRmax, respectively). The AUC of nCBVmean to differentiate between WHO grade II and III gliomas was 0.857, with a cutoff value 1.06, a sensitivity of 91.67%, and a specificity of 80.00%.
Furthermore, 18F-FET PET could help discriminate high-grade gliomas (WHO grades III and IV) from low-grade gliomas (WHO grade II), with p values of 0.009 for TBRmean and 0.004 for TBRmax. All perfusion values (nCBVmean and nCBVmax) were significantly different between high- and low-grade gliomas (p < 0.001 and = 0.0038, respectively). ROC curve analysis of PET and CBV parameters indicated that the nCBVmean reached the highest AUC value (AUC, 0.920; cutoff value, 1.06; sensitivity, 91.67%; specificity, 90.00%). The details of the results are presented in Fig. 4 and Table 4.
Discussion
Non-invasive measures to predict grades and molecular subtypes, such as IDH and 1p/19q codeletion status, play pivotal roles in determining the diagnosis and prognosis of gliomas. Our study demonstrated that 18F-FET PET and DSC-PWI parameters based on hybrid PET/MR can help predict WHO grades and IDH mutation status of gliomas with high diagnostic efficacy. In addition, they can also differentiate IDH-mutated astrocytomas from IDH-wildtype glioblastomas. However, IDH-mutated with 1p/19q codeletion oligodendrogliomas remains indistinguishable from IDH-mutated astrocytomas. Surprisingly, nCBVmean is a potential imaging marker to discriminate between IDH-mutated with 1p/19q codeletion oligodendrogliomas and IDH-wildtype glioblastomas.
Recently, it has been proven that amino acid PET tracer 18F-FET can provide information for glioma grading [12,13,14, 22, 23]. Pöpperl et al [24] found a statistically significant difference in TBRmax between low- and high-grade gliomas, with an AUC of 0.798, which is in agreement with our results. A similar result was also reported by Rapp et al [25], who indicated that TBRmax less than 2.5 may differentiate low-grade gliomas from high-grade gliomas, with an AUC of 0.77. However, the optimal cutoff values in the previous studies are obviously lower than those in our study (2.58, 2.5, and 3.81). We assume that the main reason for this discrepancy is the inconsistent method of background activity assessment. Unterrainer et al [26] evaluated the variability of different approaches for background reference delineation and found that a crescent-shaped VOI resulted in minimal intra- and inter-reader variability. Accordingly, our study may provide update reference for the use of 18F-FET PET in glioma preoperative grading.
Our study suggested that IDH-wildtype gliomas such as glioblastomas have a significantly higher tracer uptake than IDH-mutated gliomas. The TBR of 18F-FET PET may be informative to non-invasively predict IDH mutation status in gliomas. This result is in accordance with other studies [15, 27]. Non-invasive prediction of IDH mutation status in gliomas supplies an improved assessment of prognosis and response to therapies. To date, the diagnostic efficacy of amino acid PET in glioma genotypes is still inadequate, as demonstrated in the literature. Verger et al [15] found that static and dynamic 18F-FET PET parameters could not differentiate IDH-mutated with 1p/19q codeletion oligodendrogliomas from IDH-wildtype glioblastomas and IDH-mutated astrocytomas, which is in consistent with our results. One reason for this observation is the relatively high FET uptake in oligodendrogliomas [24]. Oligodendrogliomas may exhibit higher FET uptake than astrocytomas and show an overlap with glioblastoma. Kebir et al [27] used 11C-methionine, another amino acid PET tracer, to analyze its potential in the diagnosis of glioma subtyping and achieved similar results.
In our study, we used a new method that defined the entire tumor volume on 18F-FET PET as the VOI and then co-registered it to CBV maps for parameter measurement. To our knowledge, although there have been a few studies on the clinical value of CBV in the management of gliomas, the regions of interest (ROIs) were drawn with visual definition of the highest signal areas [28, 29]. CBV values obtained by this method cannot distinguish IDH-mutated and wildtype gliomas [30]. Some studies performed biopsy in gliomas under the guidance of 18F-FET PET/MR, finding that a TBR of 1.6 was the best threshold for the detection of tumor tissue and that tumor volumes on 18F-FET PET are larger than those on contrast-enhanced MRI [20]. Accordingly, we extracted a whole tumor mask including the solid portion of the tumor on 18F-FET PET and co-registered it to CBV maps to reflect the overall characteristics of the lesion. CBV values acquired by this method could provide more information for diagnosis and molecular subtype prediction in gliomas than previously used method. Brendle et al [29] compared the glioma grading performance of nCBV from DSC-PWI by using different ROI delineation methods. They found that the ROI definition based on a T2 hyperintense tumor on T2 FLAIR sequences yielded the best performance of glioma grading in DSC-PWI, with the AUC, cutoff value, specificity, and sensitivity values of 0.74, 1.4, 59%, and 86% respectively. Compared with the diagnostic performance of DSC-PWI in glioma grading of this study (AUC, specificity, and sensitivity values of 0.92, 91.67%, and 90% respectively), we found that an ROI definition based on 18F-FET PET was superior to other ROI definition methods for glioma grading.
In addition to providing grade information with higher accuracy than previous studies [31], CBV is able to supply information for molecular subtyping prediction. Among the parameters derived from 18F-FET PET and DSC-PWI, only nCBVmean could distinguish IDH-mutated with 1p/19q codeletion oligodendrogliomas from IDH-wildtype glioblastomas, but all parameters failed to discriminate between IDH-mutated with 1p/19q codeletion oligodendrogliomas and IDH-mutated astrocytomas. Lee et al [32] found that nCBVmean of grade II–III IDH-mutated with 1p/19q codeletion oligodendrogliomas was larger than that of IDH-mutated astrocytomas. A similar result was found by Latysheva et al using histogram-derived CBV [9]. In our study, the nCBVmean of IDH-mutated with 1p/19q codeletion oligodendrogliomas tended to be higher than that of IDH-mutated astrocytomas. The main reason for the inconsistent results may be the small sample sizes of both groups. The predictive value of DSC-PWI in determining the 1p/19q codeletion status remains to be validated in a larger sample size.
Moreover, our study suggests that 18F-FET PET parameters and nCBVmean could discriminate between IDH-mutated astrocytomas and IDH-wildtype glioblastomas. The combination of TBRmax and nCBVmean significantly increased the predictive efficacy of hybrid 18F-FET PET/MR in the differentiation of IDH-mutated astrocytomas and IDH-wildtype glioblastomas, with an AUC of 0.903. These findings highlight the application value of the combination of 18F-FET PET and DSC-PWI in the most common groups of gliomas.
Some limitations to our study need to be considered. First, the predictive value of dynamic 18F-FET PET parameters in different molecular subtypes has not been analyzed because some patients with gliomas could not endure the 40- to 60-min examinations without moving. Second, the small sample size may have influenced the results, which need to be validated in a larger cohort. Furthermore, only the most common genotypes of gliomas were analyzed in this study, while other genotypes, such as IDH-wildtype astrocytoma and newly diagnosed IDH-mutated glioblastoma, were not identified in this study due to their rarity.
In conclusion, this pilot study investigated the predictive performance of simultaneous 18F-FET PET and DSC-PWI for determining the molecular subtypes of gliomas according to the revised 2016 WHO classification. 18F-FET PET and DSC-PWI are potential imaging applications that can be used to predict the grade and IDH mutation status of gliomas. The combination of parameters derived from 18F-FET PET and DSC-PWI based on hybrid PET/MR could improve the prediction efficacy of IDH-mutated astrocytomas and IDH-wildtype glioblastomas, two of the most common subtypes of gliomas. However, 18F-FET PET and DSC-PWI demonstrated limited predictive potential in differentiating IDH-mutated with 1p/19q codeletion oligodendrogliomas from IDH-mutated astrocytomas and IDH-wildtype glioblastomas.
Abbreviations
- 18F-FET PET:
-
O-(2-18F-Fluoroethyl)-L-tyrosine positron emission tomography
- AUC:
-
Area under the curve
- DSC-PWI:
-
Dynamic susceptibility contrast perfusion-weighted imaging
- IDH:
-
Isocitrate dehydrogenase
- nCBV:
-
Normalized cerebral blood volume
- ROC:
-
Receiver operating characteristic
- TBR:
-
Tumor-to-brain ratio
- VOI:
-
Volume of interest
- WHO:
-
World Health Organization
References
Rogers TW, Toor G, Drummond K et al (2018) The 2016 revision of the WHO Classification of Central Nervous System Tumours: retrospective application to a cohort of diffuse gliomas. J Neurooncol 137:181–189. https://doi.org/10.1007/s11060-017-2710-7
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508. https://doi.org/10.1056/NEJMoa1407279
Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ (2011) Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol 12:83–91. https://doi.org/10.1016/S1470-2045(10)70053-X
Buckner J, Giannini C, Eckel-Passow J et al (2017) Management of diffuse low-grade gliomas in adults - use of molecular diagnostics. Nat Rev Neurol 13:340–351. https://doi.org/10.1038/nrneurol.2017.54
Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS (2018) Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol 4:1254–1262. https://doi.org/10.1001/jamaoncol.2018.1789
Broen MPG, Smits M, Wijnenga MMJ et al (2018) The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol 20:1393–1399. https://doi.org/10.1093/neuonc/noy048
Smits M, van den Bent MJ (2017) Imaging correlates of adult glioma genotypes. Radiology 284:316–331. https://doi.org/10.1148/radiol.2017151930
Englander ZK, Horenstein CI, Bowden SG et al (2018) Extent of bold vascular dysregulation is greater in diffuse gliomas without isocitrate dehydrogenase 1 R132H mutation. Radiology 287:965–972. https://doi.org/10.1148/radiol.2017170790
Latysheva A, Emblem KE, Brandal P et al (2019) Dynamic susceptibility contrast and diffusion MR imaging identify oligodendroglioma as defined by the 2016 WHO classification for brain tumors: histogram analysis approach. Neuroradiology 61:545–555. https://doi.org/10.1007/s00234-019-02173-5
Hyun SC, Sung KH, Chai JS et al (2019) Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: a systemic review and meta-analysis. Eur Radiol 29:745–758. https://doi.org/10.1007/s00330-018-5608-7
Albert NL, Weller M, Suchorska B et al (2016) Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol 18:1199–1208. https://doi.org/10.1093/neuonc/now058
Law I, Albert NL, Arbizu J et al (2018) Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [18F]FDG: version 1.0. Eur J Nucl Med Mol Imaging 46:540–557. https://doi.org/10.1007/s00259-018-4207-9
Galldiks N, Stoffels G, Ruge MI et al (2013) Role of O-(2-18F-fluoroethyl)-L-tyrosine PET as a diagnostic tool for detection of malignant progression in patients with low-grade glioma. J Nucl Med 54:2046–2054. https://doi.org/10.2967/jnumed.113.123836
Kunz M, Thon N, Eigenbrod S et al (2011) Hot spots in dynamic (FET)-F-18-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol 13:307–316. https://doi.org/10.1093/neuonc/noq196
Verger A, Stoffels G, Bauer EK et al (2018) Static and dynamic 18F–FET PET for the characterization of gliomas defined by IDH and 1p/19q status. Eur J Nucl Med Mol Imaging 45:443–451. https://doi.org/10.1007/s00259-017-3846-6
Vettermann F, Suchorska B, Unterrainer M et al (2019) Non-invasive prediction of IDH-wildtype genotype in gliomas using dynamic 18F-FET PET. Eur J Nucl Med Mol Imaging 46:2581–2589. https://doi.org/10.1007/s00259-019-04477-3
Song S, Cheng Y, Ma J et al (2020) Simultaneous FET-PET and contrast-enhanced MRI based on hybrid PET/MR improves delineation of tumor spatial biodistribution in gliomas: a biopsy validation study. Eur J Nucl Med Mol Imaging 47:1458–1467. https://doi.org/10.1007/s00259-019-04656-2
Welker K, Boxerman J, Kalnin A et al (2015) ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. AJNR Am J Neuroradiol 36:E41–E51. https://doi.org/10.3174/ajnr.A4341
Munck Af Rosenschold P, Costa J, Engelholm SA et al (2015) Impact of [18F]-fluoro-ethyl-tyrosine PET imaging on target definition for radiation therapy of high-grade glioma. Neuro Oncol 17:757–763. https://doi.org/10.1093/neuonc/nou316
Pauleit D, Floeth F, Hamacher K et al (2005) O-(2-[18F]fluoroethyl)-L-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain 128:678–687. https://doi.org/10.1093/brain/awh399
Lohmann P, Stavrinou P, Lipke K et al (2019) FET PET reveals considerable spatial differences in tumour burden compared to conventional MRI in newly diagnosed glioblastoma. Eur J Nucl Med Mol Imaging 46:591–602. https://doi.org/10.1007/s00259-018-4188-8
Filss CP, Galldiks N, Stoffels G et al (2014) Comparison of 18F-FET PET and perfusion-weighted MR imaging: a PET/MR imaging hybrid study in patients with brain tumors. J Nucl Med 55:540–545. https://doi.org/10.2967/jnumed.113.129007
Langen KJ, Watts C (2016) Neuro-oncology: amino acid PET for brain tumours - ready for the clinic? Nat Rev Neurol 12:375–376. https://doi.org/10.1038/nrneurol.2016.80
Pöpperl G, Kreth FW, Mehrkens JH et al (2007) FET PET for the evaluation of untreated gliomas: correlation of FET uptake and uptake kinetics with tumour grading. Eur J Nucl Med Mol Imaging 34:1933–1942. https://doi.org/10.1007/s00259-007-0534-y
Rapp M, Heinzel A, Galldiks N et al (2013) Diagnostic performance of F-18-FET PET in newly diagnosed cerebral lesions suggestive of glioma. J Nucl Med 54:229–235. https://doi.org/10.2967/jnumed.112.109603
Unterrainer M, Vettermann F, Brendel M et al (2017) Towards standardization of 18F-FET PET imaging: do we need a consistent method of background activity assessment? EJNMMI Res 7:48. https://doi.org/10.1186/s13550-017-0295-y
Kebir S, Weber M, Lazaridis L et al (2019) Hybrid C-11-MET PET/MRI combined with “machine learning” in glioma diagnosis according to the revised glioma WHO Classification 2016. Clin Nucl Med 44:214–220. https://doi.org/10.1097/RLU.0000000000002398
Liang J, Liu D, Gao P et al (2017) Diagnostic values of DCE-MRI and DSC-MRI for differentiation between high-grade and low-grade gliomas: a comprehensive meta-analysis. Acad Radiol 25:338–348. https://doi.org/10.1016/j.acra.2017.10.001
Brendle C, Hempel J, Schittenhelm J et al (2018) Glioma grading by dynamic susceptibility contrast perfusion and (11)C-methionine positron emission tomography using different regions of interest. Neuroradiology 60:381–389. https://doi.org/10.1007/s00234-018-1993-5
Anzalone N, Castellano A, Cadioli M et al (2018) Brain gliomas: multicenter standardized assessment of dynamic contrast-enhanced and dynamic susceptibility contrast MR images. Radiology 287:933–943. https://doi.org/10.1148/radiol.2017170362
Verger A, Filss CP, Lohmann P et al (2017) Comparison of 18F-FET PET and perfusion-weighted MRI for glioma grading: a hybrid PET/MR study. Eur J Nucl Med Mol Imaging 44:2257–2265. https://doi.org/10.1007/s00259-017-3812-3
Lee JY, Ahn KJ, Lee YS, Jang JH, Jung SL, Kim BS (2018) Differentiation of grade II and III oligodendrogliomas from grade II and III astrocytomas: a histogram analysis of perfusion parameters derived from dynamic contrast-enhanced (DCE) and dynamic susceptibility contrast (DSC) MRI. Acta Radiol 59:723–731. https://doi.org/10.1177/0284185117728981
Acknowledgments
The authors thank Jie Ma, Yu Yang, Dongmei Shuai, and Qingtang Lin for assistance with the patient studies, and Cheng Peng and Zhigang Liang for the radiosynthesis of 18F-FET.
Funding
This study has received funding by Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20180802) and Beijing Higher Education Young Elite Teacher Project (CN) (CIT&TCD201904091).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jie Lu.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
The Institutional Review Board of Xuanwu Hospital Capital Medical University approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuangshuang Song and Leiming Wang contribute equally to this work as co-first author.
Supplementary Information
ESM 1
(DOC 33 kb)
Rights and permissions
About this article
Cite this article
Song, S., Wang, L., Yang, H. et al. Static 18F-FET PET and DSC-PWI based on hybrid PET/MR for the prediction of gliomas defined by IDH and 1p/19q status. Eur Radiol 31, 4087–4096 (2021). https://doi.org/10.1007/s00330-020-07470-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07470-9