Abstract
Objectives
The present study aimed to investigate the clinical prognostic significance of radiomics signature (R-signature) in patients with gastric cancer who had undergone radical resection.
Methods
A total of 181 patients with gastric cancer who had undergone radical resection were enrolled in this retrospective study. The association between the R-signature and overall survival (OS) was assessed in the primary cohort and verified in the validation cohort. Furthermore, the performance of a radiomics nomogram integrating the R-signature and significant clinicopathological risk factors was evaluated.
Results
The R-signature, which consisted of six imaging features, stratified patients with gastric cancer who had undergone radical resection into two prognostic risk groups in both cohorts. The radiomics nomogram incorporating R-signature and significant clinicopathological risk factors (T stage, N stage, and differentiation) exhibited significant prognostic superiority over clinical nomogram and R-signature alone (Harrell concordance index, 0.82 vs 0.71 and 0.82 vs 0.74, respectively, p < 0.001 in both analyses). All calibration curves showed remarkable consistency between predicted and actual survival, and decision curve analysis verified the usefulness of the radiomics nomogram for clinical practice.
Conclusions
The R-signature could be used to stratify patients with gastric cancer following radical resection into high- and low-risk groups. Furthermore, the radiomics nomogram provided better predictive accuracy than other predictive models and might aid clinicians with therapeutic decision-making and patient counseling.
Key Points
• Radiomics can stratify the gastric cancer patients following radical resection into high- and low-risk groups.
• Radiomics can improve the prognostic value of TNM staging system.
• Radiomics may facilitate personalized treatment of gastric cancer patients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Although its overall incidence has decreased, gastric cancer remains the fourth most common form of malignancy and the second leading cause of cancer-associated death worldwide [1]. Surgical resection is regarded as the main curative approach for gastric cancer; however, the 5-year survival rate following resection is still less than 33% due to high rates of recurrence and metastasis [2]. Although preoperative and postoperative adjuvant chemotherapy have been recommended for patients with resectable gastric cancer [3], not all patients benefit from these treatments [4]. For these reasons, it is crucial to stratify a patient’s risk and develop a personalized treatment plan.
In routine clinical practice, prognostic predictions for patients with gastric cancer are primarily based on the tumor-node-metastasis (TNM) staging system [5]. However, it is common for patients with gastric cancer of the same stage who have undergone complete resection to exhibit heterogeneous outcomes [6]. These suggest that the present staging system is inadequate for determining patient prognosis [7]. Although recent advances in the field of molecular biology suggest an increasingly important role for biomarkers in prognostic analysis, most biomarkers are not yet available for application in clinical settings, and their application is limited by high cost and complex protocols [8]. Therefore, it is necessary to identify a novel and reliable biomarker that can ensure more accurate prediction of outcomes in patients with resectable gastric cancer.
Non-invasive computed tomography (CT) has been widely adopted for differential diagnosis, preoperative assessment, and therapeutic evaluation in patients with gastric cancer [9, 10]. Recent studies also demonstrated that routine CT imaging could be used to predict outcomes in patients with various types of cancer, including gastric cancer [11]. However, the prognostic utility of routine medical imaging is inherently limited by interreader accuracy and reproducibility [12]. Advancements in medical imaging technology and analytical methods have led to the development of radiomics—a field of study dedicated in part to transforming these medical images into high-dimensional, mineable data that can be used to objectively and quantitatively profile tumor phenotypes in a robust and reproducible manner [13]. Radiomics hypothesizes that these models hide important biological and medical data that can provide additional information for diagnosis, prognosis, and other forms of prediction in clinical practice [14]. For example, Huang et al [15] demonstrated that a radiomics panel could be used to predict disease-free survival in patients with early-stage lung cancer when used in conjunction with the traditional prognostic model. Additional studies reported associations between radiomics features and prognosis in patients with glioma and nasopharyngeal cancer [16]. These results highlighted the potential clinical utility of radiomics features as prognostic biomarkers, which could assist with advanced clinical decision-making [17].
Recently, few studies to date had focused on the application of radiomics in patients with gastric cancer. Giganti et al [18] investigated the association between CT texture and overall survival (OS) in 56 patients, which revealed that CT texture analysis could be applied in the prognostic analysis of gastric cancer. Moreover, Liu et al [19] explored the application of CT texture analysis in predicting histopathological features of gastric cancer. They found the portal venous CT texture correlated significantly with differentiation degree and Lauren classification, and the CT imaging features of standard deviation, entropy, and minimum attenuation in the arterial phase reflected the vascular invasion [19]. However, these studies were limited by their small sample sizes, lack of independent validation, and the limited clinical utility of single-feature models. Thus, the reliability and validity of radiomics biomarkers for gastric cancer require further investigation. In the present study, we aimed to develop and validate a radiomics signature (R-signature) for the prediction of OS in patients with gastric cancer following curative resection and to subsequently identify whether a novel nomogram integrating this R-signature and independent clinicopathological risk factors can provide more accurate predictions of prognosis among such patients.
Patients and methods
Patients
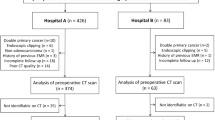
The present study was approved by the Institutional Review Board of Guizhou Provincial People’s Hospital and waived the requirement for informed consent due to the retrospective nature of the study. In total, 181 consecutive patients with gastric cancer who had undergone radical gastrectomy with D2 lymphadenectomy between September 2012 and October 2015 were enrolled in this study. The inclusion criteria were as follows: (a) underwent radical gastrectomy with D2 lymphadenectomy and R0 resection (no residual tumor), (b) pathologically confirmed as gastric cancer, (c) more than 15 examined lymph nodes, and (d) contrast-enhanced abdominal CT within 15 days before radical gastrectomy. The exclusion criteria were as follows: (a) non-tumor-related causes of death, (b) received preoperative therapy or postoperative radiotherapy, (c) with distant metastasis or peritoneal dissemination, (d) with other malignant tumors, (e) incomplete medical records, and (f) insufficient quality of CT imaging. The recruitment process is shown in Supplemental Material (Figure S1). These patients were divided into primary and validation datasets in a 2:1 ratio using an approach based on probability, with simple random sampling. Baseline demographic and clinicopathological characteristics were retrospectively extracted from the electronic medical records of each patient. Pathologic specimens were independently reviewed and restaged in accordance with the AJCC 8th edition TNM staging system (2016) by two gastrointestinal pathologists with at least 10 years of experience who were blinded to the clinical data. CT imaging data were obtained from the picture archiving and communications system of our institution. CT scanner and acquisition parameters are described in the Supplemental Material.
CT scanner and acquisition parameters
Contrast-enhanced abdominal CT was performed for all patients using a first-generation Siemens dual-source CT system (Siemens Healthcare). Oral doses of water (600–1000 mL) were administered to ensure stomach distension prior to CT examination. No anti-cholinergic agent was used. The scanning parameters were as follows: 120 reference kV with automated kilovoltage selection technology; 180 reference mAs with automated tube current modulation system; detector collimation, 64.0 mm × 0.6 mm; field of view, 350 mm × 350 mm; matrix, 512 × 512; helical pitch, 1; rotation, 0.5 s; and 5 mm reconstruction section thickness. Arterial- and portal venous-phase scans were acquired following delays of 25–30 s and 60 s, respectively, after an intraphase injection of contrast medium (3–4 mL/s, 1.5 mL/kg; Ultravist 370, Bayer Schering) via a syringe pump.
Follow-up period
The endpoint of interest was OS, which was calculated from the date of operation until the date of tumor-related death or the date of the final follow-up. All patients were followed up until October 2017. The minimum follow-up was 2 years, and the maximum was 5 years (average, 26 months). Follow-up visits were performed every 3 months for the first 2 years, every 6 months during years 2–5, and yearly thereafter. Visits were conducted on an outpatient basis or via telephone interviews.
Segmentation and feature extraction
First, image segmentation was processed before extraction of radiomics features. In order to compare the superiority between two-dimensional (2D) and three-dimensional (3D) segmentation, each 2D and 3D region of interest (ROI) was manually drawn along the margin of the tumor on the slice with the largest tumor cross section and all slices containing the tumor in portal venous phase. ROI segmentation was completed using the open-source software ITK-SNAP (www.itk-snap.org). The segmentation procedure was performed by two professional radiologists with more than 10 years of experience. For 2D segmentation, a slice with the largest tumor cross section was selected together by two radiologists. If the consensus in selecting the slice could not be reached, it would be resolved by a third experienced radiologist with 15 years. For 3D segmentation, the whole tumor volume outlining was done under the consensus of two radiologists with 5 years and 10 years, respectively.
In order to investigate the prognosis of gastric cancer, we utilized radiomics methods to extract quantitative and mineable features for ROI analysis. Two-dimensional features were extracted on the slice with the largest tumor cross section, and 3D features were extracted from the ROI of volume for each patient. The procedure of radiomics method is shown in Fig. 1. In total, four groups of radiomics features were extracted from the selected ROIs: (1) shape and size features, (2) image intensity features, (3) textural features, and (4) wavelet features. Shape and size features (shape, area, compactness, etc.) were to reflect the phenotype of each ROI. Gray intensity features were used to determine the difference between the gray histogram and gray distribution for each ROI. Textural features were used to examine voxel regulation within each ROI. Wavelet features were used to examine changes in gray intensity and textural features. A detailed explanation of the feature extraction method can be found in the Supplemental Material. All feature extraction methods were implemented using Matlab 2015a (MathWorks). In order to validate the stability of the radiomics features, we assessed the interobserver agreement of feature extraction using interclass correlation coefficients (ICCs) [20]. An ICC of greater than 0.75 was considered acceptable.
The procedure of radiomics analysis. a From four patients with the regions of interest (ROIs). b The phenotype of delineation to show the heterogeneity in the lesions. c Extraction of features from ROIs, such as semantic features, intensity, texture, and wavelet features. d Prediction and analysis for the overall survival of gastric cancer
Feature selection and development of the R-signature
Features were selected using the least absolute shrinkage and selection operator (LASSO) method and subsequently used to develop a LASSO Cox regression model. The LASSO method was an accepted algorithm for feature selection in high-dimensional variables [21]. The tuning parameter λ was essential for LASSO feature selection: larger λ values exerted no effect on the estimated regression parameters; however, as the value of λ decreased, some coefficients may decrease as well, approaching zero. Cross-validation was applied to optimize the value of λ, and the model was constructed using the selected λ. Coefficients of indistinctive covariates were reduced to zero, and the remaining non-zero coefficients were selected. We used these non-zero features to obtain Akaike information criterion (AIC) estimates of the quality of each model [22]. Finally, the non-zero coefficients of the selected features were defined as radiomics scores (R-scores). We then calculated the R-signature—the combination of R-scores for all selected features—for patients in both the primary and validation cohorts.
Prognostic model and individualized nomogram analysis
The potential association between R-signatures and OS was first assessed in the primary cohort and validated in the validation cohort. We performed the Kaplan-Meier survival analysis for both cohorts. All patients would be divided into high-risk and low-risk groups based on the median R-score: patients with scores higher than the median were placed in high-risk groups. We validated the R-signature in the clinical subgroups by stratified analysis to determine the OS. We used Cox proportional hazards models to calculate the Harrell concordance index (C-index) of the R-signature. Multivariate Cox regression analyses were applied to develop a diagnostic model for gastric cancer survival using the primary cohort. Backward stepwise selection was performed using the likelihood ratio test, with the AIC as the stopping rule. To provide the clinician with a quantitative tool for the prediction of OS for each patient, we developed a clinical nomogram and a radiomics nomogram using data from the primary cohort. The clinical nomogram was developed using independent clinical risk factors. Multivariate Cox regression was used to develop the radiomics nomogram based on the R-signature and these clinical risk factors. Furthermore, we validated these two nomograms by plotting calibration curves demonstrating the performance of OS estimates, based on the significance of the C-index. The C-index, which ranges from 0.5 to 1.0, is commonly used to evaluate the performance of prognostic models in survival analysis. Its higher values revealed that it had greater ability to group patients into different disease progressions. We performed bootstrap analysis for C-index statistics that had been corrected for potential overfitting. We plotted calibration curves to assess the true survival probability against the predicted probability-based nomogram. Moreover, the G-rho rank test was used to calculate the relative hazard ratio (HR) [23]. A decision curve analysis was also performed to demonstrate the net clinical benefit of the radiomics nomogram and the clinical nomogram at certain threshold probabilities.
Statistical analysis
Statistical analyses were performed using R software (version 3.4.2; http://www.R-project.org) and Matlab 2015b (MathWorks). The R packages used included “Survival,” “glmnet,” “rms,” “Hmisc,” and “Glmnet,” which were utilized for feature selection using the LASSO algorithm. To assess the risk of model overfitting, we calculated the complexity of the estimated model and the optimal combination of the selected radiomics features based on the AIC. Based on the results of the multivariate analysis, a nomogram was developed using the rms package, and the performance of the nomogram was evaluated based on the C-index, which was calculated using the Hmisc package. We used SPM12 in Matlab 2015b to analyze the CT data. Features and clinical variables were compared between the two cohorts, and clinical variables were compared between high-risk and low-risk groups in primary and validation cohorts independently. The two-sample t test or the Wilcoxon rank-sum test was used to compare continuous variables, and Pearson’s chi-square test or Fisher’s exact test was used to compare categorical variables. A weighted log-rank test was used to compare the survival curves of the high-risk and low-risk groups. The level of statistical significance was set at p < 0.05. We used Spearman’s correlation test to examine the potential correlation between the selected radiomics features and pathological tumor characteristics.
Results
Analysis of clinical data
The clinical characteristics of the primary and validation cohorts are presented in Table S1, and the clinical characteristics between the high-risk and low-risk groups in primary and validation cohorts are shown in Table 1. The result revealed no significant differences in clinical variables or R-scores between the primary and validation cohorts (the p range of 0.23–0.56). As of the final follow-up, the number of cancer-related deaths was 57 (49.5%) and 36 (54.5%) for the primary and validation cohorts. The mean OS was 26 months and 25 months, respectively. Our feature stability analysis showed that the ICC for most of radiomics feature was high. Therefore, all outcomes were based on the measurements of the first radiologist.
Construction of the regression model based on R-signature
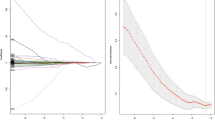
For 2D analysis, we extracted 273 radiomics features from the selected ROIs for the prediction of OS. First, we randomly selected 30 patients to determine the ROIs from which features would be extracted. We observed no significant differences between measurements reported by two independent readers for any of the selected features, with p values ranging from 0.134 to 0.915. The interobserver ICCs of all metrics were calculated based on the measurements of the two readers, which revealed that five radiomics features did not align with the inclusion criteria. The ICCs of all other features ranged from 0.75 to 1, indicative of good stability. A total of 268 radiomics features were reduced to 12 potential features (Fig. 2). We then refined the model based on the AIC, following which six features were selected for the development of the R-signature (Fig. 2c). The R-score was then calculated according to the following formula: R-score = (SSF_Area_to_circumference_ratio × 0.887) − (WLH_IIF_skewness × 0.163) − (WHH_IIF_mean × 3.13) − (WHH_GLCM_IMC2 × 7.02) − (WHL_GLRLM_LRHGLE × 0.0124) + (WHH_GLRLM_energy × 0.000001). The p values for proportional hazard (PH) assumptions ranged from 0.233 to 0.994. Therefore, we regarded the subsequent estimates of OS as meaningful.
Radiomics feature selection and weight of each coefficient. Feature selection for the prediction of OS using the LASSO Cox regression model [1]. a Tuning parameter (λ) selection in the LASSO model involved the use of tenfold cross-validation with minimum criteria. b We examined the coefficients of the 273 radiomics features to identify 12 potential predictors. c We used the AIC to optimize the radiomics model developed with these 12 features. Finally, six features with p values less than 0.296 were used to build the LASSO Cox regression model. The y-axis represents the contribution of the feature to the development of the R-signature. The x-axis represents the features that contribute to the R-signature in the LASSO Cox regression model. Features with negative coefficients are labeled with a minus sign (-)
For 3D analysis, we extracted 485 radiomics features from the selected ROIs for the prediction of OS. We calculated the R-signature—the combination of nine selected 3D features—for patients in both the primary and validation cohorts. The R-score was then calculated according to the following formula: R-score = − (SSF_Spherical_disproportion × 0.242) + (WHHH_IIF_median × 0.106) + (GLCM_maximum_probability × 0.002) + (WLHH_GLCM_energy × 0.128) − (WLHL_GLCM_maximum_probability × 0.031) − (WHHL_GLCM_maximum_probability × 0.009) − (WHHH_GLCM_correlation × 0.166) + (WLLH_GLRLM_LRHGLE × 0.207) + (WHHH_GLRLM_RLN × 0.204).
Validation of the predictive utility of the R-signature
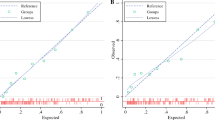
The LASSO Cox regression model revealed that the 2D R-signature combined by 2D radiomics features was associated with OS in the primary cohort (p = 1.1e−12; HR = 1.37; 95% confidence interval (CI) = 1.25, 1.50). Moreover, this result was replicated in the validation cohort (p = 1.3e−4; HR = 4.0; 95% CI = 1.97, 8.15) (Fig. 3a, b). We also observed that lower R-scores were generally associated with better OS. When patients were stratified based on clinical risk factors, R-signature was significant to classify the patients into high- and low-risk groups (Supplementary Figure S2). Especially, the 2D R-signature can classify the patient in TNM stage of II and III (Fig. 3c, d).
Results of the Kaplan-Meier survival analyses according to the R-signature. a We observed a significant association between the R-signature and OS in the primary cohort. b The association was confirmed in the validation cohort a. Dashed line = two-sided CI of the survival curves (solid line). Results of the Kaplan-Meier survival analyses according to the R-signature for patients in the TNM stage II (c) and TNM stage III (d) in the validation cohort
For 3D radiomics features, we performed the Kaplan-Meier survival analysis for both cohorts. The LASSO Cox regression model revealed that the 3D R-signature was associated with OS in the primary cohort (p = 1.1e−9; HR = 6.99; 95% CI = 4.21, 11.63). However, the performance of LASSO Cox model was validated in the validation cohort (p = 9.8e−1; HR = 1.72; 95% CI = 0.83, 3.55), which showed no significant association for prediction of OS.
Therefore, the R-signature combined by 2D radiomics features was included in the subsequent analysis.
Performance of R-signature in individualized nomogram with OS prediction
A Cox regression analysis identified R-signature, T stage, N stage, and differentiation as independent risk factors. The individualized prediction model for OS was visualized as a nomogram (Fig. 4). The utility of the R-signature is demonstrated in Fig. 4c, which includes the clinical nomogram and the R-signature. Figure 4b, d depicts the calibration curves of the nomograms for the probability of OS at 1 year, 2 years, or 3 years after surgery. Our findings indicated that inclusion of the R-signature in the radiomics nomogram improved performance relative to observation alone. We also calculated the C-index, calibrated C-index, HR, and AIC estimates for each of the models (R-signature, radiomics nomogram, and clinical nomogram) (Table 2). The radiomics nomogram demonstrated that the predictive capability of the model was improved when the R-signature was integrated with independent clinicopathological risk factors (C-index for the radiomics nomogram = 0.82; 95% CI = 0.77, 0.87). The lowest AIC (408.3), highest C-index, and highest HR were observed for the radiomics nomogram, which exhibited better predictive performance than the clinical nomogram (C-index = 0.71; 95% CI = 0.64, 0.79). To demonstrate the contribution of the R-signature, we plotted decision curves for the clinical nomogram and radiomics nomogram. As shown in Fig. 4e, the net benefit of the radiomics nomogram was higher than that of the clinical nomogram at a certain threshold probability.
Clinical and radiomics nomograms for the prediction of OS, presented alongside the assessments used for model calibration, and decision curve analysis for the clinical and radiomics nomograms. a Clinical nomogram developed based on several representative clinical risk variables. Results for the primary cohort are shown. b Calibration curves for the clinical nomogram. Calibration of the clinical nomogram was based on the correspondence between the predicted and observed outcomes at 1 year, 2 years, and 3 years. c Radiomics nomogram showing the weight of each risk factor when the R-score is included in the clinical nomogram. d Calibration curve for the radiomics nomogram. The green line represents the radiomics nomogram, while the pink line represents the clinical nomogram. e Decision curve analysis to show the comparison of performance. The horizontal black line represents the assumption that no patients should take the necessary measures, while the blue line represents the assumption that all patients should. The y-axis represents the net benefit, which was calculated by adding points associated with benefits and subtracting those associated with harms. Based on the threshold probabilities obtained, our findings indicated that the radiomics nomogram provided a greater net benefit than the clinical nomogram
Potential association between radiomics features and pathological tumor characteristics
We found that five radiomics features associated with prognosis were respectively correlated with at least one clinicopathological characteristics, including differentiation, tumor size, N stage, TNM stage, and neural invasion (Spearman’s rho coefficient = 0.26 to 0.38; p < 0.05). More details are shown in supplementary material and Figure S3.
Discussion
In the present study, we developed and validated a novel, CT-based, prognostic R-signature to improve the prediction of OS among patients with gastric cancer following curative resection. Our results demonstrated that this signature could be used to stratify these patients into high- and low-risk groups based on OS, even those within the same clinical stage (TNM stages II–III). We further developed a radiomics nomogram based on the R-signature and independent clinicopathological risk factors (T stage, N stage, and differentiation), which provided better predictive accuracy than the use of the clinicopathological nomogram, and R-signature alone. Thus, the R-signature enhanced the prognostic utility of the TNM stage.
Intratumor heterogeneity is correlated with poor outcomes among patients with gastric cancer [24]. Indeed, the TNM staging system has continuously improved in accordance with the evolution of methods for diagnosis and treatment. However, it has been recognized that the TNM staging system—which is entirely based on the anatomical characteristics of the tumor—fails to reflect the innate biological heterogeneity of the tumor and is, thus, insufficient for providing complete and precise prognostic information. In contrast to traditional clinical staging, the radiomics approach extracts features based on the characteristics of the entire tumor as identified via medical imaging, providing a robust yet non-invasive method for characterizing intratumor heterogeneity. Therefore, radiomics features can provide more accurate and comprehensive information for use in conjunction with the TNM staging system [25]. To test this hypothesis, we performed a stratification analysis in the validation cohort of the present study. Due to the small sample size, we were unable to perform this analysis in patients with stage I tumors. However, the R-signature stratified patients with stage II and III tumors into high-risk and low-risk subgroups, further suggesting that this signature could be used to provide additional prognostic information and refine the traditional risk stratification system for patients with gastric cancer. Tumor heterogeneity is also dependent on initial variations in genetic and molecular characteristics, which are generally implicated in tumor recurrence and metastasis, and thus may be a crucial biomarker for prognosis [26]. Previous studies reported that proteogenomic and phenotypic information could be inferred from radiologic images of the tumor [27]. Therefore, radiomics approaches can permit non-invasive assessment of both genetic and molecular heterogeneity and have the potential to advance clinical decision-making.
Prognosis among patients with gastric cancer following curative resection is dependent on complex and multifactorial interactions. A single yet strong independent risk factor is insufficient for accurate prediction of gastric cancer prognosis. Furthermore, single factor is suitable for application at the population/group level only, rather than the individual level. Among the available predictive models, nomograms are advantageous in that they incorporate tumor-, patient-, and treatment-related factors for a more accurate and quantified prognostic evaluation at the individual level [28]. Previous studies indicated that nomograms exhibit better performance for predicting survival than the traditional TNM staging system in patients with gastric cancer [29]. In the present study, we integrated the R-signature and independent clinicopathological risk factors (T stage, N stage, and differentiation) to develop a radiomics nomogram, which enabled significantly better discrimination, calibration, and net reclassification than the clinical nomogram and the R-signature alone when predicting 1-year, 2-year, and 3-year OS. These results demonstrated that the R-signature provided complementary prognostic information, thereby improving the prognostic performance of the TNM staging system.
Taken together, our findings indicated that the radiomics nomogram would be a potential tool to improve the screening of patients for individualized treatment. Patients whose predicted OS is poor are more likely to experience relapse and metastasis [2]. In such cases, aggressive postoperative adjuvant therapy or intensive follow-up is indicated, while unnecessary or inadequate treatment will be avoided in patients with favorable prognoses [30]. Furthermore, the simple and visual representation provided by the nomogram will help patients understand their survival expectations, enhance clinical decision-making, and promote communication between doctors and patients.
The present study possesses some limitations of note. First, our nomogram only included traditional clinical and pathologic factors. The inclusion of additional variables such as molecular biomarkers may have provided more accurate estimates of survival. Second, this study was a single-center, retrospective analysis and subjected to the inherent limitations of such investigation. Third, the median follow-up periods were relatively short (26 months), which could reduce the incidence of endpoint events, limiting our ability to identify potentially important variables. Fourth, R-signature possessed prognostic significance for patients with gastric cancer following radical resection, but the features of which merely possessed weak correlations with pathological tumor characteristics. We considered that, in addition to the small sample size, lacking the other clinical, pathological, and molecular characteristics of tumor might be the important reason. Lastly, ROI was segmented in the largest cross section instead of whole-tumor volume. The 3D radiomics features were worse than the 2D radiomics features in the prognostic analysis in this study. However, previous studies indicated that the whole tumor volume analysis could provide more comprehensive tumor biological characteristics.
In conclusion, our findings demonstrated that R-signature could be used to stratify patients with gastric cancer following radical resection into high- and low-risk groups. In addition, the R-signature could be implemented to improve the prognostic value of the TNM staging system. As our radiomics nomogram exhibited improved discrimination, calibration, and net reclassification ability relative to other models for prognostic prediction, this nomogram might aid clinicians with therapeutic decision-making and individualized patient counseling.
Abbreviations
- AIC:
-
Akaike information criterion
- CT:
-
Computed tomography
- HR:
-
Hazard ratio
- ICCs:
-
Interclass correlation coefficients
- LASSO:
-
Least absolute shrinkage and selection operator
- OS:
-
Overall survival
- R-scores:
-
Radiomics scores
- R-signature:
-
Radiomics signature
- ROI:
-
Region of interest
- TNM:
-
Tumor-node-metastasis
References
Fitzmaurice C, Dicker D, Pain A et al (2015) The global burden of cancer 2013. JAMA Oncol 1:505–527
Tegels JJ, De Maat MF, Hulsewé KW, Hoofwijk AG, Stoot JH (2014) Improving the outcomes in gastric cancer surgery. World J Gastroenterol 20:13692–13704
Bang YJ, Kim YW, Yang HK et al (2012) Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet 379:315–321
Jiang Y, Zhang Q, Hu Y et al (2016) ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. https://doi.org/10.1097/SLA.0000000000002116
Bando E, Makuuchi R, Tokunaga M, Tanizawa Y, Kawamura T, Terashima M (2017) Impact of clinical tumor-node-metastasis staging on survival in gastric carcinoma patients receiving surgery. Gastric Cancer 20:448–456
Kim YI (2014) Is retrieval of at least 15 lymph nodes sufficient recommendation in early gastric cancer? Ann Surg Treat Res 87:180–184
Fridman WH, Mlecnik B, Bindea G, Pagès F, Galon J (2011) Immunosurveillance in human non-viral cancers. Curr Opin Immunol 23:272–278
Sohn BH, Hwang JE, Jang HJ et al (2017) Clinical significance of four molecular subtypes of gastric cancer identified by The Cancer Genome Atlas Project. Clin Cancer Res 23:4441–4449
Liang P, Ren XC, Gao JB, Chen KS, Xu X (2017) Iodine concentration in spectral CT: assessment of prognostic determinants in patients with gastric adenocarcinoma. AJR Am J Roentgenol 209:1033–1038
Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H (2016) Gastric cancer. Lancet 388:2654–2664
Komori M, Asayama Y, Fujita N et al (2013) Extent of arterial tumor enhancement measured with preoperative MDCT gastrography is a prognostic factor in advanced gastric cancer after curative resection. AJR Am J Roentgenol 201:W253–W261
Kumar V, Gu Y, Basu S et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248
Verma V, Simone CB 2nd, Krishnan S, Lin SH, Yang J, Hahn SM (2017) The rise of radiomics and implications for oncologic management. J Natl Cancer Inst 109:djx055
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 281:947–957
Grossmann P, Narayan V, Chang K et al (2017) Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. https://doi.org/10.1093/neuonc/nox092
O’Connor JP (2017) Radiomics: rethinking the role of clinical imaging. Elife 6:e30563
Giganti F, Antunes S, Salerno A et al (2017) Gastric cancer: texture analysis from multidetector computed tomography as a potential preoperative prognostic biomarker. Eur Radiol 27:1831–1839
Liu S, Liu S, Ji C et al (2017) Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. Eur Radiol 27:4951–4959
Weir JP (2005) Quantifying test-retest reliability using the intraclass correlation coefficient and the SEM. J Strength Cond Res 19:231–240
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Methodol 58:267–288
Bozdogan H (1987) Model selection and Akaike’s information criterion (AIC): the general theory and its analytical extensions. Psychometrika 52:345–370
Hernán MA (2010) The hazards of hazard ratios. Epidemiology 21:13–15
Morris LG, Riaz N, Desrichard A et al (2016) Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7:10051
Sala E, Mema E, Himoto Y et al (2017) Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol 72:3–10
Zhang J, Fujimoto J, Zhang J et al (2014) Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346:256–259
Rutman AM, Kuo MD (2009) Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 70:232–241
Harrell F (2001) Regression modeling strategies. Springer, New York
Cao Y, Liu H, Zhang H et al (2017) CXC chemokine receptor 1 predicts postoperative prognosis and chemotherapeutic benefits for TNM II and III resectable gastric cancer patients. Oncotarget 8:20328
Nishida T (2012) Adjuvant therapy for gastric cancer after D2 gastrectomy. Lancet 379:291–292
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81360565, 61661010, 81227901, 81771924, 81501616, 61231004, 81671851, and 81527805), the National Key R&D Program of China (2017YFA0205200, 2017YFC1308700, 2017YFC1309100), the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-SW-STS-160), the Beijing Municipal Science and Technology Commission (Z161100002616022), the Guizhou Provincial Department of Science and Technology and Guizhou Provincial People’s Hospital United Foundation (QKHLHZ[2015]7115), the Guizhou Provincial People’s Hospital Doctoral Foundation (GZSYBS[2015]02), the Science and Technology Foundation of Guizhou Province (QKHJC[2016]1096), the Technology and Innovation Foundation for the Returned Overseas Chinese Scholars (QRXMZZ(2016)03), and the Guizhou Science and Technology Department Key Project (QKF[2017]25).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jie Tian.
Conflict of interest
The authors declare that they have no conflict of interest.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Electronic supplementary material
ESM 1
(DOCX 7872 kb)
Rights and permissions
About this article
Cite this article
Li, W., Zhang, L., Tian, C. et al. Prognostic value of computed tomography radiomics features in patients with gastric cancer following curative resection. Eur Radiol 29, 3079–3089 (2019). https://doi.org/10.1007/s00330-018-5861-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5861-9