Abstract
Objectives
The aim of this study was to determine whether diffusion and perfusion imaging parameters demonstrate different diagnostic values for predicting pseudoprogression between glioblastoma subgroups stratified by O6-mythylguanine-DNA methyltransferase (MGMT) promoter methylation status.
Methods
We enrolled seventy-five glioblastoma patients that had presented with enlarged contrast-enhanced lesions on magnetic resonance imaging (MRI) one month after completing concurrent chemoradiotherapy and undergoing MGMT promoter methylation testing. The imaging parameters included 10 or 90 % histogram cutoffs of apparent diffusion coefficient (ADC10), normalized cerebral blood volume (nCBV90), and initial area under the time signal-intensity curve (IAUC90). The results of the areas under the receiver operating characteristic curve (AUCs) with cross-validation were compared between MGMT methylation and unmethylation groups.
Results
MR imaging parameters demonstrated a trend toward higher accuracy in the MGMT promoter methylation group than in the unmethylation group (cross-validated AUCs = 0.70–0.95 and 0.56–0.87, respectively). The combination of MGMT methylation status with imaging parameters improved the AUCs from 0.70 to 0.75–0.90 for both readers in comparison with MGMT methylation status alone. The probability of pseudoprogression was highest (95.7 %) when nCBV90 was below 4.02 in the MGMT promoter methylation group.
Conclusions
MR imaging parameters could be stronger predictors of pseudoprogression in glioblastoma patients with the methylated MGMT promoter than in patients with the unmethylated MGMT promoter.
Key Points
• The glioblastoma subgroup was stratified according to MGMT promoter methylation status.
• Diagnostic values of diffusion and perfusion parameters for predicting pseudoprogression were compared.
• Imaging parameters showed higher diagnostic accuracy in the MGMT promoter methylation group.
• Imaging parameters were independent to MGMT promoter methylation status for predicting pseudoprogression.
• Imaging biomarkers might demonstrate different diagnostic values according to MGMT promoter methylation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The evaluation of treatment response after concurrent chemoradiotherapy (CCRT) in glioblastoma has been advanced by the development of new standardized criteria regarding changes in tumour volume after therapy by the Response Assessment in Neuro-Oncology Working Group (RANO [1]). However, radiologic images obtained in the short time interval after CCRT still contain biases for treatment response evaluation due to increasing capillary permeability or tumour cell damage by therapy [2]. Several studies using perfusion magnetic resonance imaging (MRI) showed that changes in cerebral blood volume (CBV) within tumours that occur during the early radiotherapy course can predict pseudoprogression [3] and survival [4]. Others reported that the apparent diffusion coefficient (ADC) values are useful for distinguishing between high-grade glioma and normal tissue [5] and predicting treatment responses [6, 7]. However, the exact clinical impact of advanced MR imaging techniques including diffusion-weighted imaging (DWI), dynamic susceptibility contrast-enhanced (DSC) perfusion imaging, and dynamic contrast-enhanced (DCE) perfusion imaging has not been validated by large-series prospective studies conducted exclusively for post-treatment glioblastoma.
O6-mythylguanine-DNA methyltransferase (MGMT) promoter methylation status was reported as a strong predictor of pseudoprogression and prognosis [8–10]. According to several reports about the correlation between MGMT promotor methylation status and imaging parameters, both of them should be considered when interpreting enlarged contrast-enhancing lesions on MR [3, 11]. Furthermore, the potential association between MGMT promoter methylation status and various imaging parameters may lead to the use of different diagnostic values in assessing treatment response in glioblastoma.
Our hypothesis is that diffusion and perfusion imaging parameters may demonstrate different diagnostic values for predicting pseudoprogression in glioblastoma patients according to MGMT promoter methylation status. The purpose of this study was to determine whether diffusion and perfusion imaging parameters demonstrate different diagnostic values for predicting pseudoprogression between glioblastoma subgroups stratified by MGMT promoter methylation status.
Methods
Study population
Our institutional review board approved this retrospective study, and the requirement for informed consent from the study participants was waived. A retrospective review of our database identified 211 consecutive patients with pathologically proven glioblastoma between December 2010 and December 2014. Patients were included in our study if they met all of the following criteria: (a) underwent methylation-specific polymerase chain reaction (MS-PCR) of the tumour specimens to determine MGMT promoter methylation status; (b) underwent CCRT after surgery; (c) demonstrated new or progressively enlarged contrast-enhancing lesions on the follow-up MR at 4 weeks after CCRT, including DWI, DSC, and DCE perfusion MR; (d) underwent serial follow-up MR imaging with 2–3-month intervals; (e) not prescribed corticosteroid when MR imaging was performed; and (f) obtained adequate image acquisition and quality without susceptibility or motion artifacts.
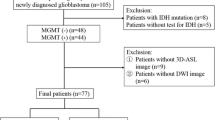
We excluded 43 patients without MS-PCR results and six patients not treated with CCRT. Additionally, 16 patients were excluded due to the absence of DWI, DSC, or DCE images, corticosteroid treatment at the time of MR, or poor quality; another 71 patients with stable contrast-enhancing lesions after CCRT were also excluded. Finally, 75 patients were enrolled in this study. The 75 patients were dichotomized into MGMT promoter methylation and unmethylation groups. Fifteen of 75 patients were reported on previously, but this prior study evaluated the diagnostic performance of multiparametric imaging for simply identifying pseudoprogression [12]. The process flowchart for this study population is shown in Fig. 1.
MGMT promoter methylation analysis
MGMT promoter methylation status was evaluated after obtaining a specimen from surgery or biopsy. Unstained tissue slides made from paraffin-embedded blocks of tumour tissues were used to extract genomic DNA. MS-PCR with modifications was used to determine the DNA methylation status of the CpG islands at the MGMT promoter [13, 14]. The annealing temperature was 59 ° C. Results from unqualified DNA that yielded uncertain PCR results were abandoned. For negative and positive controls, bisulfite-treated unmethylated and methylated control DNAs were used (Qiagen, Hilden, Germany). On 8 % polyacrylamide gels, the PCR products were separated, stained with ethidium bromide, and inspected under ultraviolet illumination. For all steps, clinical information was processed in a blind manner using a computerized barcode.
Assessment
After the second-look surgery, nine cases were confirmed as true progression and three as pseudoprogression. If the second-look operation was unavailable, the progression was decided based on MR, clinical assessment, and steroid use. An experienced neuroradiologist (H.S.K.) blinded about clinical information independently reviewed all MR images. The clinico-radiological diagnoses of 27 cases of true progression and 36 cases of pseudoprogression were established by consensus between a neuro-oncologist (J.H.K., 20 years of clinical experience in neuro-oncology) and a neuroradiologist (H.S.K.) after completely reviewing images and medical charts. Clinical and radiologic assessments were carried out at pre-CCRT; 4 weeks after completion of the CCRT; during cycles 2, 4, and 6 of the adjuvant temozolomide therapy; and every 2 or 3 months thereafter. Pseudoprogression was defined using prespecified criteria [1], and confirmatory imaging was performed after two cycles of maintenance temozolomide therapy. The diagnosis of pseudoprogression was made when there was a decrease or stabilization of the contrast-enhancing lesions on subsequent serial follow-up MR images, and combined with no required change in treatment. This diagnosis allows a slight increase in contrast-enhancing lesions as long as no change in treatment occurs during this time period. A final diagnosis of true progression was made if the enhancing lesions gradually increased on more than two serial follow-up MR images at 2-month intervals and were combined with clear neurologic deterioration attributable to enlarging contrast-enhancing lesions, causing a prompt change of treatment.
MR imaging protocol and image processing
The MR imaging studies were performed using a 3T system (Achieva; Philips Healthcare, Best, The Netherlands) with an 8-channel sensitivity-encoded head coil. The order of our brain tumour imaging protocol was as follows: T2-weighted imaging, fluid-attenuated inversion recovery imaging, DWI, non-enhanced T1-weighted imaging, DCE perfusion MR imaging, contrast-enhanced T1-weighted imaging, and DSC perfusion MR imaging. The rationale for the application of DCE imaging prior to DSC imaging is that preloading contrast agent injected during the DCE imaging can minimize T1 contrast agent leakage effects.
DWI was obtained in three orthogonal directions and combined into a trace image. Using these data, ADC maps were calculated using the b values of 0 and 1000 second/mm2 on a voxel-by-voxel basis with the software incorporated into the MR imaging unit. Further information about the protocols of DWI, DSC, and DCE perfusion MR imaging are demonstrated in the supplementary material.
All MR imaging data were transferred from the MR imaging unit to an independent personal computer. A dedicated software package (Nordic ICE; Nordic Neuro Lab, Bergen, Norway) and in-house software (Matlab 2010b; Mathworks, Natick, MA) were used to obtain parametric perfusion maps. The relative CBV (rCBV) was determined with the numeric integration of the time–signal intensity (SI) curve for DSC MR imaging after correcting contrast agent leakage. On a pixel-by-pixel basis, normalized CBV (nCBV) maps were computed by dividing the individual rCBV value by an unaffected white matter rCBV value. For DCE MR imaging, the SI was normalized to the percentage change in comparison with the baseline SI value. Then, we calculated the initial area under the time SI curves (IAUCs) at 30 seconds after contrast agent administration using the trapezoidal integration of the normalized SI with time over the first 30 seconds after contrast agent arrival in the enhancing voxels of interest. We chose this model-free parameter because of our intention to maximally decrease the dependence on complex modelling of the pharmacokinetics, which can be sensitive to noise [15].
Image analysis
Intermodality coregistration and enhancing lesion segmentation
The ADC, nCBV, and IAUC maps were coregistered to each of the three-dimensional contrast-enhanced T1-weighted images. Intermodality coregistrations were performed using a non-linear transformation. For the quantitative analysis of data acquired from the DWI, DSC, and DCE MR imaging, a neuroradiology expert (H.S.K.; reader 1) and a neuroradiology fellow (J.E.P. with 5 years of experience in radiology; reader 2) segmented the entire volumes of the contrast-enhancing lesions on the coregistered contrast-enhanced T1 weighted images using a semi-automated adaptive threshold technique so that all pixels above a threshold value were taken.
DWI analysis
For the quantitative DWI analyses, the 10th percentile of the histogram cutoff for the ADC value (ADC10) was calculated. This parameter was selected because the 10th percentile parameter is analogous to the minimum value, which is commonly used with the hot-spot method.
DSC and DCE MR imaging analyses
For the cumulative nCBV and IAUC histogram parameters, the 90th percentile cutoffs were derived (nCBV90 and IAUC90, respectively). This kind of histogram parameter is more powerful than the mean and maximum values of the overall histogram for identifying areas where tumorous lesions are intermingled with treatment-associated changes and are less affected by random statistical fluctuations [16].
Statistical analyses
The imaging data were initially assessed for normality using the Kolmogorov-Smirnov test. To assess differences in demographic data between the MGMT promoter methylation and unmethylation groups, the Student’s t test for continuous variables and the chi-squared test for categorical variables were used.
In the subgroup analyses stratified by MGMT promoter methylation, differences in the mean values of the imaging parameters between the two groups were assessed using the Student’s t test. Receiver operating characteristic (ROC) curve analysis was performed on each imaging parameter to determine the optimum cutoff for differentiating between the two groups and to compare the area under the ROC curve (AUC). Bonferroni correction was used to control for multiple comparisons. Using the same population to select classifiers and estimate error rates can overfit the diagnostic performances, so imaging parameters were further evaluated using leave-one-out cross-validation. Using this method, all study patients (except for one patient) were used as the training set, and the prediction error was assessed for the excluded test set. This procedure was repeated until every case was used once as the test set.
Multivariable logistic regression modelling was used to estimate the odds ratios for the association between the combination of MGMT promoter methylation and imaging parameters with the final diagnosis. Cross-validation was also used to determine the association between the imaging parameters and final diagnosis. The ROC-based optimum cutoff values, which were calculated based on the highest Youden index [17], were used as exploratory analyses to dichotomize subgroups of each imaging parameter. The results are considered significant at P < 0.05 and corrected for the number of comparisons.
Inter-reader agreement was assessed using the intraclass correlation coefficient (ICC) with 95 % confidence intervals and by applying a two-way ICC with a random rater’s assumption. Instances of P < 0.05 were considered to indicate significant statistical differences. Statistical analyses were performed using SPSS (version 21.0; SPSS, Chicago, IL) and R version 2.15.3 (R Project for Statistical Computing; http://www.r-project.org).
Results
Clinical parameters of subgroups
Among 75 patients, 34 (45.3 %) were classified to the MGMT promoter methylation group and 41 (54.7 %) to the MGMT promoter unmethylation group. No clinical parameters were significantly different between the two groups (Table 1). Contrast-enhancing lesion-related clinical symptoms were presented in 17 (50.0 %) of 34 MGMT promoter methylation patients and 28 (68.3 %) of 41 MGMT promoter methylation patients (P = 0.170). In total, 25 cases (73.5 %) of pseudoprogression and nine cases (26.5 %) of true progression were in the MGMT promoter methylation group, while the MGMT promoter unmethylation group included 14 cases (34.1 %) of pseudoprogression and 27 cases (65.9 %) of true progression. This difference was statistically significant (P = 0.002) according to the chi-squared test. Representative cases are shown in Figs. 2 and 3.
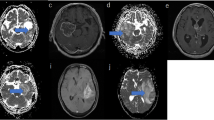
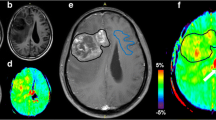
Images obtained from a 38-year-old female glioblastoma patient with MGMT promoter methylation and true tumour progression. a Contrast-enhanced T1-weighted image obtained 7 weeks after CCRT. A necrotic contrast-enhancing lesion can be observed in the right temporal lobe. b, c Contrast-enhanced T1-weighted image obtained 15 weeks after CCRT and 2 days after surgery for a recurrent tumour; b in this panel, the contrast-enhancing lesion is enlarged. The ADC map (d) shows restricted diffusion in the corresponding contrast-enhancing lesion shown in (b). Both the nCBV (e) and IAUC (f) maps show increased perfusion pixel values in the corresponding contrast-enhancing lesion shown in (b), suggesting true tumour progression (i.e., true positive)
Images obtained from a 59-year-old male glioblastoma patient with MGMT promoter unmethylation and pseudoprogression. a, b Contrast-enhanced T1-weighted images obtained before and 5 weeks after CCRT. A newly developed contrast-enhancing lesion is seen around the surgical cavity. c Contrast-enhanced T1-weighted images obtained 10 weeks after CCRT. The size of the contrast-enhancing lesion increased. The ADC map (d) shows equivocal finding of restricted diffusion in the corresponding contrast-enhancing lesion shown in (c). Both the nCBV (e) and IAUC (f) maps show no increase of perfusion pixel values in the corresponding contrast-enhancing lesion shown in (c), suggesting pseudoprogression. g Contrast-enhanced T1-weighted images obtained 20 weeks after CCRT. The contrast-enhancing lesion is stabilized, confirming pseudoprogression
Diagnostic accuracy of the imaging parameters in the subgroups
In the MGMT promoter methylation group, the mean values of the DWI, DSC, and DCE MR imaging parameters were significantly different between the true progression and pseudoprogression groups for both readers (corrected P = .001–.006 for reader 1; corrected P = .001–.039 for reader 2). In the MGMT promoter unmethylated group, the mean values of the nCBV90 and IAUC90 values were significantly different between the two groups for both readers (corrected P = .001–.003 for both readers). However, the mean ADC10 values were not significantly different between groups for both readers (Table 2 and Fig. 4).
On the ROC curve analysis, the AUCs were highest for nCBV90 derived from DSC MR imaging and lowest for the ADC10 derived from DWI in both groups for both readers (Table 3). For comparing two perfusion parameters in terms of differentiating true progression from pseudoprogression, the nCBV90 derived from DSC MR imaging showed a higher AUC than the IAUC90 derived from DCE perfusion MR imaging in both MGMT methylation and unmethylation groups for both readers. However, these results were not statistically significant (Table 3). All of the three imaging parameters demonstrated a trend towards higher AUCs in the MGMT promoter methylation group than in the MGMT promoter unmethylation group for differentiating true progression from pseudoprogression. On the leave-one-out cross-validation, the cross-validated AUCs and the accuracies of the imaging parameters demonstrated similar trends, as did the ROC curve analyses using the same training and validation sets (Table 4 and Fig. 5).
Leave-one-out cross-validation used to determine the diagnostic accuracy of DWI, DSC, and DCE MR imaging for differentiating true tumour progression from pseudoprogression in subgroups stratified by MGMT promoter methylation for both readers a MGMT promoter methylated group; b MGMT promoter unmethylated group
Independence and added value of the imaging parameters to MGMT promoter methylation status
Exploratory analyses using the optimum cutoff values based on the highest Youden index were performed to test the independence and added value of the imaging parameters to the MGMT promoter methylation status for predicting pseudoprogression. On multivariate logistic regression analyses using MGMT promoter methylation status, all of the three imaging parameters independently influenced the incidence of pseudoprogression (P = .004–.001 for reader 1; P = .041–.001 for reader 2, respectively). The combination of MGMT promoter methylation status with each imaging parameters improved the AUCs from 0.70 to 0.75–0.90 for both readers in comparison with MGMT promoter methylation status alone (Table 5). The cross-validated AUCs and accuracies of the imaging parameters demonstrated similar trends, as did the ROC curve analyses using the same training and validation sets (Table 6).
Probability of pseudoprogression using the decision tree
A diagnostic decision tree was constructed based on the exploratory analyses using our own optimum cutoff values (Fig. 6). The probability of pseudoprogression was highest (95.7 %) when nCBV90 was below 4.02 in the MGMT promoter methylation group and lowest (0 %) when nCBV90 was above 4.02 in the MGMT promoter unmethylation group. The probability of pseudoprogression was 90.0 % when the ADC10 was above 0.94 in the MGMT promoter methylation group.
Inter-reader agreement for measuring imaging parameters
The ICCs were 0.71, 0.86, and 0.74 for the calculations of ADC10, nCBV90, and IAUC90, respectively.
Discussion
In our study, we found that the DSC and DCE perfusion parameters demonstrated significant differences between true progression and pseudoprogression in glioblastoma demonstrating either MGMT promoter methylation or unmethylation, although the ADC value demonstrated a significant difference between these two entities in the MGMT promoter methylation group only. Moreover, diffusion and perfusion MR imaging parameters demonstrated better diagnostic performance to predict pseudoprogression in the MGMT promoter methylation group compared to the unmethylation group. When comparing perfusion parameters obtained with both DCE and DSC techniques, we found that the best overall diagnostic accuracy to predict pseudoprogression was achieved with the combination of DSC perfusion MR imaging and MGMT promoter methylation. These results suggest that the MR imaging protocol that includes DSC perfusion MR imaging is most efficient in the prediction of pseudoprogression in the MGMT promoter methylation patient, as compared with that which includes DCE perfusion MR imaging. On the multivariate analysis, diffusion and perfusion MR imaging parameters were independent to MGMT promoter methylation status for predicting pseudoprogression, and provided added value in terms of diagnostic accuracy. Among the three imaging parameters, nCBV90 showed the highest diagnostic accuracy for predicting pseudoprogression in both MGMT promoter methylation and unmethylation groups. Lastly, the probability of pseudoprogression was highest (22 of 23 patients; 95.7 %) when the nCBV90 was below 4.02 in the MGMT promoter methylation group on our exploratory analyses. These results imply that the role of the imaging parameters for assessing treatment response could be limited by MGMT promoter unmethylation, which leads to predetermined chemoresistance of the tumour, regardless of other tumour microenvironments estimated using various imaging parameters. However, imaging parameters (especially perfusion parameters) could provide added value to MGMT promoter methylation status for predicting pseudoprogression in the MGMT promoter methylation group, which reflects the early treatment response to CCRT. This suggests that tumour vascularity and permeability might also be important for the treatment response in glioblastoma demonstrating MGMT promoter methylation.
In contrast to our results, a previous study reported that the rCBV demonstrates more predictability in the MGMT promoter unmethylation group than in the methylation group [3]. However, this previous study demonstrated a remarkable incidence of true progression in their MGMT promoter unmethylation group, whereas we observed a remarkable incidence of pseudoprogression in our MGMT promoter methylation group. Besides, we used the histogram parameters of nCBV from the entire corresponding contrast-enhancing lesion instead of the “hot-spot” method. Regarding the DCE MR, to the best of our knowledge, no previous studies reported the diagnostic performance of DCE MR imaging parameters for predicting pseudoprogression in glioblastoma stratified by MGMT promoter methylation. We found a similar trend with the results of DSC MR imaging, which demonstrated a higher diagnostic performance for IAUC90 in the MGMT methylation group. Therefore, we suggest that perfusion imaging parameters could provide added diagnostic value for predicting pseudoprogression in the MGMT promoter methylation group.
Although it remains unclear as to why different diagnostic values of the imaging parameters were observed between MGMT promoter methylation and unmethylation groups, one possible explanation is that the higher diagnostic performance of the imaging parameters in the MGMT promoter methylation group might be associated with a higher proportion of pseudoprogression in the MGMT methylation group, whereas a relatively even distribution of true progression and pseudoprogression is seen in the unmethylation group. However, even if the correlation between the MGMT promoter methylation status and incidence of pseudoprogression may lead to a decrease in the power of the relationship between imaging parameters and pseudoprogression, there was a significant relationship between the three imaging parameters and the incidence of pseudoprogression in the MGMT promoter methylation group.
Interestingly, the combination of MGMT methylation and perfusion parameters provided additional diagnostic value for predicting pseudoprogression compared to the combination of MGMT promoter methylation and diffusion parameter. A previous study proposed that tumours with low ADC values can be enriched in MGMT promoter-methylated tumour cells [18], in which elevated minimum ADC values and/or lower mean ADC values were associated with MGMT promoter methylation status. Therefore, the lowest diagnostic accuracy of ADC for predicting pseudoprogression in subgroups might be attributed to the stronger association between MGMT promoter methylation status and ADC values than those of perfusion parameters. Regarding the perfusion MR, one previous study reported that the MGMT promoter methylation status in glioblastoma is not associated with the rCBV ratio [19]. In contrast, a recent study suggested that the MGMT protein modulates angiogenesis of glioblastoma by changing the levels of vascular endothelial growth factor receptors [20], and accordingly, different rCBV ratios could be expected with respect to the MGMT methylation status of the tumour. Thereby, it may explain why the diagnostic values of the perfusion parameters differed between the MGMT promoter methylation and unmethylation groups in our study.
On our exploratory analyses, we found that the combination of MGMT promoter methylation and nCBV90 below 4.02 can predict pseudoprogression with a probability of 95.7 %. A previous study also reported that there is a 91.3 % probability of pseudoprogression in patients with the methylated MGMT promoter if the first MR image after CCRT shows enlargement of the enhancing lesion [8]. Therefore, these results also imply that the setting of the molecular profile and imaging parameters may reflect treatment-induced blood–brain barrier disruption, which is related to the efficacy of CCRT.
Our study has several limitations. First, it was a retrospective study with relatively small number of populations which causes uneven categorization of patient groups. Therefore, a further prospective study is needed to validate our results. Second, the pathophysiological mechanisms that underlie the different diagnostic values of the imaging parameters between the two subgroups remain unclear, and future investigation with an experimental approach for tumorigenesis and genomics is warranted.
In conclusion, diffusion and perfusion imaging parameters may demonstrate different diagnostic values for predicting pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation status. Our results suggest that MR imaging parameters could be stronger predictors of pseudoprogression in glioblastoma patients with the methylated MGMT promoter than in patients with the unmethylated MGMT promotor. Among the three imaging parameters, DSC perfusion MR imaging shows the highest diagnostic accuracy for predicting pseudoprogression in both MGMT promoter methylation and unmethylation groups. Therefore, the addition of DSC perfusion MR imaging may provide increased accuracy in glioblastoma demonstrating MGMT promoter methylation for predicting pseudoprogression.
Abbreviations
- CCRT:
-
Concurrent chemoradiotherapy
- MR:
-
Magnetic resonance
- CBV:
-
Cerebral blood volume
- ADC:
-
Apparent diffusion coefficient
- MGMT:
-
O6-mythylguanine-DNA methyltransferase
- DWI:
-
Diffusion-weighted imaging
- DSC:
-
Dynamic susceptibility contrast-enhanced
- DCE:
-
Dynamic contrast-enhanced
- IAUC:
-
Initial area under the time–signal intensity curve
- ROC:
-
Receiver operating characteristic
References
Wen PY, Macdonald DR, Reardon DA et al (2010) Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 28:1963–1972
Brandes AA, Rigon A, Zampieri P et al (1998) Carboplatin and teniposide concurrent with radiotherapy in patients with glioblastoma multiforme: a phase II study. Cancer 82:355–361
Kong DS, Kim ST, Kim EH et al (2011) Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: the role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. AJNR Am J Neuroradiol 32:382–387
Catalaa I, Henry R, Dillon WP et al (2006) Perfusion, diffusion and spectroscopy values in newly diagnosed cerebral gliomas. NMR Biomed 19:463–475
Castillo M, Smith JK, Kwock L, Wilber K (2001) Apparent diffusion coefficients in the evaluation of high-grade cerebral gliomas. AJNR Am J Neuroradiol 22:60–64
Pope WB, Kim HJ, Huo J et al (2009) Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology 252:182–189
Pope WB, Lai A, Mehta R et al (2011) Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol 32:882–889
Brandes AA, Franceschi E, Tosoni A et al (2008) MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol 26:2192–2197
Brandes AA, Tosoni A, Franceschi E et al (2009) Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J Clin Oncol 27:1275–1279
Weller M, Tabatabai G, Kastner B et al (2015) MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR trial. Clin Cancer Res 21:2057–2064
Drabycz S, Roldan G, de Robles P et al (2010) An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage 49:1398–1405
Park JE, Kim HS, Goh MJ, Kim SJ, Kim JH (2015) Pseudoprogression in patients with glioblastoma: assessment by using volume-weighted voxel-based multiparametric clustering of MR imaging data in an independent test set. Radiology 275:792–802
Esteller M, Garcia-Foncillas J, Andion E et al (2000) Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343:1350–1354
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG (1999) Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59:793–797
Evelhoch JL, LoRusso PM, He Z et al (2004) Magnetic resonance imaging measurements of the response of murine and human tumors to the vascular-targeting agent ZD6126. Clin Cancer Res 10:3650–3657
Kim HS, Goh MJ, Kim N, Choi CG, Kim SJ, Kim JH (2014) Which combination of MR imaging modalities is best for predicting recurrent glioblastoma? Study of diagnostic accuracy and reproducibility. Radiology 273:831–843
Youden WJ (1950) Index for rating diagnostic tests. Cancer 3:32–35
Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B (1987) Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A 84:6899–6903
Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC (2012) Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 54:555–563
Chahal M, Xu Y, Lesniak D et al (2010) MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro-Oncology 12:822–833
Acknowledgments
The scientific guarantor of this publication is Prof. Sang Joon Kim. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant number: NRF-2014R1A2A2A01004937). The Institutional Review Board approved our human study (The Institutional Review Board of Asan Medical Center [http://eirb.amc.seoul.kr]: S2014-2090-0001). Written informed consent was waived by the Institutional Review Board. Methodology: retrospective, diagnostic or prognostic study, performed at one institution. The authors thank the Biomedical Imaging Infrastructure, Department of Radiology, Asan Medical Center for the technical support of image processing. Some subjects or cohorts have been previously reported in Park JE, Kim HS, Goh MJ, Kim SJ, Kim JH. Pseudoprogression in patients with glioblastoma: assessment by using volume-weighted voxel-based multiparametric clustering of MR imaging data in an independent test set. Radiology 2015 Jun;275(3):792–802).
Author information
Authors and Affiliations
Corresponding author
Additional information
Ra Gyoung Yoon and Wooyul Paik contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yoon, R.G., Kim, H.S., Paik, W. et al. Different diagnostic values of imaging parameters to predict pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation. Eur Radiol 27, 255–266 (2017). https://doi.org/10.1007/s00330-016-4346-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-016-4346-y