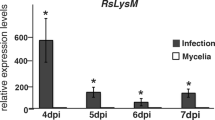
Abstract
Key message
We highlight the emerging role of the R. solani novel lipase domain effector AGLIP1 in suppressing pattern-triggered immunity and inducing plant cell death.
Abstract
The dynamic interplay between plants and Rhizoctonia solani constitutes a multifaceted struggle for survival and dominance. Within this complex dynamic, R. solani has evolved virulence mechanisms by secreting effectors that disrupt plants’ first line of defense. A newly discovered effector, AGLIP1 in R. solani, plays a pivotal role in inducing plant cell death and subverting immune responses. AGLIP1, a protein containing a signal peptide and a lipase domain, involves complex formation in the intercellular space, followed by translocation to the plant cytoplasm, where it induces cell death (CD) and suppresses defense gene regulation. This study provides valuable insights into the intricate molecular interactions between plants and necrotrophic fungi, underscoring the imperative for further exploration in this field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The interaction between plant and pathogens is a multifaceted process, facilitated by the pathogen-derived compounds essential for their pathogenicity, as well as plant-derived compounds responsible for pathogen recognition. The necrotrophic pathogen R. solani acquires nutrients from plants by secreting a wide range of cell wall-degrading enzymes (CWDEs), and various toxins (Chen et al. 2017), and subsequently colonizes plant cells following necrotic killing. However, plant immune systems are complex networks of receptors, and signaling and defensive pathways that consist of constitutive and induced defense mechanisms to counteract R. solani (Singla et al. 2024). Among these battles for survival and dominance, R. solani has evolved virulence strategies through the secretion of effectors that disrupt plants’ first line of defense (pattern recognition receptors), inhibit the production of antimicrobial compounds, modulate plant immune responses, and trigger plant cell death (CD) (Li et al. 2019; Niu et al. 2021). Effectors are predominantly proteinaceous in nature and play a crucial role in plant–pathogen interactions, facilitating pathogen colonization, nutrient acquisition and interference with various nuclear processes. Unlike bacteria, fungi lack a secretion system and deliver their effectors through direct penetration of haustoria (specialized structures) into host cells.
Fungal effectors are mainly nonenzymatic and cysteine-rich proteins with signal peptides, although some are enzymatic proteins or other molecules, such as toxins, secondary metabolites, and sRNAs that disrupt plant metabolism and immune responses (Pradhan et al. 2021). These effectors are categorized based on their localization in plant cells as extracellular (apoplastic) or intracellular (cytoplasmic) (Harris et al. 2023). Extracellular effectors are secreted into the plant cell apoplast, while intracellular effectors are directly translocated into the plant cell cytoplasm and further sub-categorized by their compartment, such as nuclear, cytoplasmic, chloroplast and mitochondrial effectors (Harris et al. 2023; Pradhan et al. 2021). Nuclear effectors disrupt various nuclear processes, including transcription machinery, host epigenetic modifications, altered gene expression, and splicing, and some target histones and immunity-associated host proteins to facilitate pathogen colonization (Harris et al. 2023).
Upon infection by biotrophic and hemibiotrophic pathogens, plants activate two lines of defense: pattern-triggered immunity (PTI), and effector-triggered immunity (ETI). PTI is the initial response, initiated when pattern recognition receptors (PRRs) recognize pathogen-associated molecular patterns (PAMPs), triggering a broad-spectrum but relatively weak immune response. In contrast, ETI is induced when plant resistance (R) proteins recognize highly variable pathogen molecules, known as avirulence (Avr) effectors, resulting in a rapid and robust hypersensitive response (HR) (Shao et al. 2021). However, the effectors in necrotrophs hijack these plant defense responses to promote pathogenesis, manipulate plant signaling pathways, suppress immune responses and induce plant CD (Shao et al. 2021). This multifunctional role of effectors has characterized them as “double-edged sword”, leading to the extraordinary adaptability of pathogens in the ever-evolving arms race (Pradhan et al. 2021). The mechanism by which effectors induce CD is unclear, but it has been suggested that signal peptides (SPs) protect effectors from being recognized by extracellular PRRs in the plasma membrane. After being secreted into the secretion in intracellular space, effectors translocate into the plant cell cytoplasm, where they are recognized by cytoplasmic receptors, triggering CD and permitting necrotrophs to acquire nutrients from dead cells. This phenomenon is termed effector-triggered susceptibility (ETS) (Shao et al. 2021). Previously, several effector proteins containing a lipase domain were identified to regulate nematotoxicity and inhibit plant defense response activity (Blümke et al. 2014; Tayyrov et al. 2021); however, a recent study reported a novel lipase domain effector AGLIP1, which promotes disease progression and induces CD in plants (Li et al. 2019). The CD in plants, an outcome of the plant R- gene response to the Avr- gene of biotrophic pathogens (gene-for-gene model), has been intensively studied. However, in necrotrophs, these effectors operate in an inverse gene-for-gene model, where the effector gene product interacts with the corresponding host susceptibility gene, resulting in host susceptibility. This hijacking of plant CD through effectors in necrotrophic pathogens is still being revealed.
Interestingly, a recent study by Li et al. (2019) revealed a novel R. solani effector that promotes cell death in plants and suppresses PTI. There are four key points of this study: (1) AGLIP1 is a putative effector in R. solani and has the ability to trigger cell death in Nicotiana benthamiana and rice; (2) the signal peptide–AGLIP1–lipase domain complex (SP-AGLIP1-LP) regulates CD-elicitation; (3) AGLIP1 acts upstream in the endoplasmic reticulum (ER) of N. benthamiana and rice to promote plant CD; and (iv) ectopic expression of AGLIP1 promotes virulence and suppresses PTI in transgenic Arabidopsis thaliana.
The role of SP-AGLIP1-LP in triggering plant cell death
Using heterologous transient expression analysis, Li et al. (2019) identified a novel putative effector, AGLIP1, as an inducer of plant CD both in N. benthamiana and rice. To classify the AGLIP1 effector, the authors used Pfa and BLAST searches against NCBI databases. AGLIP1 encodes a 302 amino acid protein containing an N-terminal SP and a C-terminal lipase domain with enzymatic active sites at S174 and D230, which are essential for inducing cell death in both N. benthamiana and rice. To further investigate whether CD is induced by HR or by cellular toxicity, the authors analyzed the expression of the ETI marker genes NbPR1 and NbHsr203J and found no significant differences in the DEX-induced expression of AGLIP1. These findings indicate that AGLIP1-induced plant CD may be due to cellular toxicity rather than HR.
To predict the role of the lipase active sites and SP of AGLIP1 in inducing plant cell death, the authors performed site-directed mutagenesis by splicing overlap extension (SOE) of the conserved lipase domain active sites. Expression analysis revealed that the mutants AGLIP1S174A, AGLIP1D230A and AGLIP1NSP (lacking SP) were unable to induce cell death in N. benthamiana. However, when recombinant plasmids containing the AGLIP1 sequence and its variant with a luciferase protein driven by the cauliflower mosaic virus 35S promoter were tested in rice protoplasts, no inhibitory effect was detected for AGLIP1S174A, AGLIP1D230A or AGLIP1NSP, while other variants exhibited low luciferase activity (Li et al. 2019). These findings demonstrate that the lipase domain active sites S174 and D230 and the SP protein produce a complex of SP-AGLIP1-LP, which regulates CD- elicitation in plants and functions in the intercellular space to prevent recognition of the effector by PRRs in the plasma membrane. However, Li et al. (2019) reported that AGLIP1 localized to the endoplasmic reticulum (ER) in both N. benthamiana and rice cells. Similarly, two other novel effectors, RsSCR10 (a small cysteine-rich protein) and RsIA_NP8 from R. solani AG1 IA, are SP dependently induced cell death and oxidative bursts in tobacco and rice (Niu et al. 2021; Wei et al. 2020). In plant cells, the effector complex SP-AGLIP1-LP translocates into the plant cytoplasm and strongly recognized by plant cytoplasmic receptors, which triggers CD (Fig. 1). These necrotic killing benefits the pathogen by facilitating the colonization and acquisition of nutrients from dead plant tissues (Li et al. 2019). These findings suggest that the necrotrophic fungus R. solani has evolved a mechanism of effector-mediated hijacking of the plant CD for its own benefit.
Conceptual model of the effector AGLIP1 in Rhizoctonia solani, illustrating its role in inducing necrotic cell death (CD) and suppressing pattern-triggered immunity (PTI) in plants. AGLIP1, a lipase domain effector, contains two active sites: S174 and D230 at the C-terminus and SP proteins at the N-terminus, which function in the intercellular space to prevent recognition by plasma membrane PRRs and suppress plant PTI. Subsequently, AGLIP1 translocates into the plant cytoplasm and localizes to the endoplasmic reticulum (ER), where it performs multiple functions. In the cytoplasm, AGLIP1 is intensely recognized by plant cytoplasmic receptors, leading to the initiation of CD, which benefits the pathogen by facilitating colonization and enabling the acquisition of nutrients from dead plant tissues. Figure created with BioRender.com
The role of the lipase domain effector AGLIP1 in suppressing PTI
Several effector proteins containing a lipase domain including, CLT1 and CLT2 in the model mushroom Coprinopsis cinerea, and FGL1 in Fusarium graminearum to function as nematotoxic agents (Tayyrov et al. 2021) and suppress plant defense responses (Blümke et al. 2014). Recently, Li et al. (2019) reported a novel lipase domain effector protein, AGLIP1, which encodes a 302 amino acid protein containing an N-terminal SP and a C-terminal lipase domain with enzymatic active sites at S174 and D230. More importantly, the plant immunity-suppressing activity of AGLIP1 was confirmed by Agrobacterium-mediated transformation of AGLIP1-expressing transgenic Arabidopsis lines using a DEX-inducible promoter, which expressed four PAMPs-induced defense genes, At1g51890 (leucine-rich repeat), At2g17740 (cysteine/histidine-rich C1 domain family protein), At5g57220 (a member of CYP81F) and FRK1 (Flg22-induced receptor like kinase 1). Surprisingly, the expression of the PR genes induced by flg22 and chitin was suppressed in all the transgenic lines. These findings indicate that AGLIP1 enhances the virulence of R. solani by suppressing PTI. Furthermore, transgenic Arabidopsis lines ectopically expressing AGLIP1 and inoculated with Pseudomonas syringae pv. tomato (Pst) DC3000 hrcC mutant completely inhibited defense gene regulation and facilitated bacterial multiplication and the development of disease symptoms. However, AGLIP1 was cytotoxic to rice and N. benthamiana and was found to be ineffective in Arabidopsis. These findings suggest that the effector AGLIP1 performs multiple functions in plant cells.
Concluding remarks and future perspectives
A groundbreaking study by Li et al. (2019) revealed a novel lipase domain, AGLIP1, which enhances the virulence of R. solani by promoting successful infection and colonization and inducing plant cell death. AGLIP1 encodes a 302 amino acid protein with an N-terminal SP and a C-terminal lipase domain, both of which are required for inducing plant cell death. These findings challenge previous assumptions about the simplicity of necrotrophic pathogenesis and underscore the importance of proteinaceous effectors in fungal pathogenicity. This study also represents a significant advancement in our understanding of the molecular mechanisms underlying the interaction between plants and necrotrophic fungi such as R. solani. Recent advances in molecular biology, plant pathology and bioinformatics have revealed a more complex picture of necrotrophic pathogenesis, with the discovery of various virulence factors in pathogens. Despite the significant crop losses caused by necrotrophic fungi worldwide, our understanding of the arms race between necrotrophic fungi and plants is improving. This finding offers potential insights for disease control strategies using CRISPR/Cas, RNAi, and small RNA technologies, highlighting the need for further investigation into effector-mediated plant–fungal interactions. These findings are expected to make significant contributions to the field of plant and necrotrophic fungal interactions, advancing our knowledge of fungal biology and enhancing our ability to develop effective disease management strategies.
Data availability
The authors declare that no new data were generated for this article.
References
Blümke A, Falter C, Herrfurth C et al (2014) Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol 165(1):346–358. https://doi.org/10.1104/pp.114.236737
Chen X, Lili L, Zhang Y et al (2017) Functional analysis of polygalacturonase gene RsPG2 from Rhizoctonia solani, the pathogen of rice sheath blight. Eur J Plant Pathol 149(2):491–502. https://doi.org/10.1007/s10658-017-1198-5
Harris W, Kim S, Vӧlz R, Lee YH (2023) Nuclear effectors of plant pathogens: distinct strategies to be one step ahead. Mol Plant Pathol 24(6):637–650. https://doi.org/10.1111/mpp.13315
Li S, Peng X, Wang Y et al (2019) The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front Microbiol. https://doi.org/10.3389/fmicb.2021.684923
Niu X, Yang G, Lin H, Liu Y, Li P, Zheng A (2021) A novel, small cysteine-rich effector, RsSCR10 in Rhizoctonia solani is sufficient to trigger plant cell death. Front Microbiol. https://doi.org/10.3389/fmicb.2021.684923
Pradhan A, Ghosh S, Sahoo D, Jha G (2021) Fungal effectors, the double edge sword of phytopathogens. Curr Genet 67(1):27–40. https://doi.org/10.3389/fmicb.2019.02228
Shao D, Smith DL, Kabbage M, Roth MG (2021) Effectors of plant necrotrophic fungi. Front Plant Sci 12:687713. https://doi.org/10.3389/fpls.2021.687713
Singla P, Bhardwaj RD, Sharma S, Sunidhi (2024) Plant–fungus interaction: a stimulus–response theory. J Plant Growth Regul 43(2):369–381. https://doi.org/10.1007/s00344-023-11100-1
Tayyrov A, Wei C, Fetz C et al (2021) Cytoplasmic lipases—a novel class of fungal defense proteins against nematodes. Front Fungal Biol. https://doi.org/10.3389/ffunb.2021.696972
Wei M, Wang A, Liu Y, Ma L, Niu X, Zheng A (2020) Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01115
Acknowledgements
Kumar P is a Research Associate researcher at the ICAR-Indian Institute of Maize Research (ICAR-IIMR), Ludhiana, India.
Funding
Pk is a recipient of Research Associate funding from the ICAR- Indian Institute of Maize Research (ICAR-IIMR), Ludhiana, India.
Author information
Authors and Affiliations
Contributions
Kumar P conceived the idea of this Focus Article. Kumar P wrote the original draft and prepared the illustrations. Kumar P and Kumari P read and edited the manuscript. The final draft was read and approved by all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, P., Kumari, P. Lipase domain effector AGLIP1 in Rhizoctonia solani triggers necrotic killing in plants. Plant Cell Rep 43, 145 (2024). https://doi.org/10.1007/s00299-024-03235-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00299-024-03235-6