Abstract
Key message
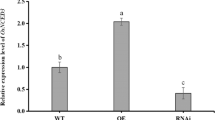
OsSPL10 is a negative regulator of rice defense against BPH, knockout of OsSPL10 enhances BPH resistance through upregulation of defense-related genes and accumulation of secondary metabolites.
Abstract
Rice (Oryza sativa L.), one of the most important staple foods worldwide, is frequently attacked by various herbivores, including brown planthopper (BPH, Nilaparvata lugens). BPH is a typical monophagous, phloem-sucking herbivore that has been a substantial threat to rice production and global food security. Understanding the regulatory mechanism of defense responses to BPH is essential for improving BPH resistance in rice. In this study, a SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 10 (OsSPL10) transcription factor was found to play a negative role in the defenses of rice against BPH. To gain insights into the molecular and biochemical mechanisms of OsSPL10, we performed combined analyses of transcriptome and metabolome, and revealed that knockout of OsSPL10 gene improved rice resistance against BPH by enhancing the direct and indirect defenses. Genes involved in plant hormone signal transduction, MAPK signaling pathway, phenylpropanoid biosynthesis, and plant–pathogen interaction pathway were significantly upregulated in spl10 mutant. Moreover, spl10 mutant exhibited increased accumulation of defense-related secondary metabolites in the phenylpropanoid and terpenoid pathways. Our findings reveal a novel role for OsSPL10 gene in regulating the rice defense responses, which can be used as a potential target for genetic improvement of BPH resistance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the most important food crops worldwide, especially in Asia and Africa. More than half of the world’s population relies on rice as the staple food (Normile 2008). There is an urgent need for increasing global rice production to keep up with the rapid population growth in the coming decades. However, rice plants in the field are often frequently attacked by a wide variety of herbivores. Hundreds of insect species are known to feed on rice, and approximately 30% of them are considered to be the main pests causing severe yield loss in rice (Lou et al. 2013). Among these rice-feeding pests, brown planthopper (BPH, Nilaparvata lugens) has become one of the most destructive pests for rice. BPH is a typical monophagous, phloem-sucking herbivore that sucks sap from the rice phloem through its stylet, and severely inhibits the growth of rice (Cheng et al. 2013). In addition to direct damage to the rice plant, it can also cause indirect damage by transmitting viruses to rice (Zhao et al. 2017). Over the past decades, pesticide application has been the main approach to control crop pests, but this approach has also brought a series of problems, including food safety risks and massive environmental detriments (Nagata 1984). Tremendous efforts have been made to screen BPH-resistance genes and develop resistant varieties to provide cost-effective and environmentally friendly alternatives (Du et al. 2009; Guo et al. 2018; Liu et al. 2015; Shi et al. 2021; Zhao et al. 2016). To promote the breeding process of BPH-resistant rice, it is important to explore endogenous genes that regulate BPH resistance in rice and characterize the underlying molecular mechanisms.
Upon perceiving the signals of feeders, plant triggers a cascade of hormone-mediated signal transduction pathways to activate the immune response of rice to insect herbivores. Phytohormone jasmonic acid (JA), ethylene (ET), and salicylic acid (SA) are generally considered as the major regulators of antiherbivore defense responses in plants (He et al. 2019; Lu et al. 2014; Ma et al. 2020; Xu et al. 2021; Zhou et al. 2009, 2014). Recent studies have shown that gibberellin (GA), brassinosteroid (BR), and cytokinin (CK) are also involved in the interactions between rice and brown planthopper (Pan et al. 2018; Zhang et al. 2017, 2022). Increasing evidence shows that crosstalk among various hormones to coordinate the expression of defense genes is essential for effective defense activation (Pan et al. 2018; Zhang et al. 2022).
Recently, transcriptomic analyses have shown that BPH feeding induces transcriptional reprogramming in rice (Tan et al. 2020). Several transcription factors (TFs) involved in the regulation of defense response, especially WRKY family genes, have been identified. For example, Bph14 interacts with OsWRKY46 and WRKY72 to initiate defense signaling (Hu et al. 2017). Another WRKY transcription factor, OsWRKY53, has been reported to function as a positive regulator of BPH resistance in rice by activating H2O2 burst and suppressing ethylene biosynthesis (Hu et al. 2016). OsWRKY70 negatively regulates gibberellin biosynthesis to enhance rice susceptibility to BPH (Li et al. 2015). In addition to WRKY TFs, two basic helix–loop–helix (bHLH) proteins, OsHLH61 and OsbHLH96, have been also identified to form a complex that mediates defense response against BPH through regulating PR genes (Wang et al. 2019).
SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPLs) genes encode plant-specific transcription factors that have been shown to regulate plant growth and development, hormone signaling pathways, and responses to stress (Jiao et al. 2010; Wang et al. 2015; Yao et al. 2022; Li et al. 2021). IDEAL PLANT ARCHITECTURE1 (IPA1) encodes SPL14 protein, which acts as a key transcription factor that promotes both yield and immunity in rice (Jiao et al. 2010). OsSPL16–GW7 module plays an important role in regulating grain quality and yield (Wang et al. 2015). A recent study has revealed that copper confers broad-spectrum virus resistance by regulating the OsSPL9–miR528–AO pathway in rice (Yao et al. 2022). In addition, OsSPL10 has been reported to positively regulate trichome development and negatively regulate salt tolerance in rice (Lan et al. 2019; Li et al. 2021). However, it remains unknown whether the SPL gene plays any role in rice defending against insect herbivores.
In this study, an SPL gene OsSPL10 was identified using a hybrid transcription factor resource (Zhao et al. 2015). A bioassay of OsSPL10 knockout mutants showed that OsSPL10 is a negative regulator of rice defense against BPH. Combined transcriptomic and metabolomic analyses discovered that knockout of OsSPL10 triggered a stronger defensive response through transcriptional activation of genes involved in the plant defense responses, and accumulation of defense-related secondary metabolites in phenylpropanoid and terpenoid pathway. Our findings revealed the regulatory mechanism of OsSPL10 gene in rice defense against BPH and should be useful for genetic improvement of BPH resistance in rice.
Materials and methods
Plant materials and insects
The spl10 mutants used in this study were generated from ZH11 and the HHZ as previously described (Lan et al. 2019). The seedlings were grown in a greenhouse at 28 ℃ with a photoperiod of 14 h light/10 h dark. The initial phenotype of the OsSPL10 gene was identified from a hybrid transcription factor resource (Zhao et al. 2015).
Colonies of BPH were originally obtained from rice fields at Fujian Agriculture and Forestry University in Fuzhou, Fujian, China. The colonies were reared on Taichung Native 1 (TN1) rice seedlings in a growth chamber (26 ± 1 ℃, 60% ± 10% RH, 14 h light/10 h dark).
BPH resistance evaluation
Three-week-old seedlings of transgenic rice plants were infested with second-to-third-instar BPH nymphs at fifteen insects per seedling. Once all seedlings of the susceptible control TN1 were dead, damage to each seedling was scored according to the previous study (Huang et al. 2001).
Honeydew excretion experiment
Wild-type and OsSPL10 knockout rice seedlings at the four-leaf stage were selected and each seedling was cultured in a plastic pot (10 × 15 cm) covered with a plastic cup to prevent BPH escape. A filter paper disc (12 cm in diameter) was placed on the plastic pot to collect honeydew excretion. Three gravid BPH female that had been starved for 2 h were inoculated on each plant. After feeding for 48 h, filters were collected, scanned, and analyzed using ImageJ software for determination of honeydew excretion.
BPH survival
To examine the effect of OsSPL10 knockout on the survival of BPHs, the plant was individually covered with a plastic cup into which 15 BPH nymphs were inoculated. The number of surviving BPHs on each seedling was counted daily for 7 days. The test was replicated ten times.
Number of eggs laid by BPH
At the stem base of each rice seedling, a plastic tube (5 cm in length) was used to cage BPH adults. Two BPH adults (one male and one female) were released in each tube. The number of eggs laid by BPH on each seedling was counted using a microscope after 7 days. The test was replicated ten times.
Host choice test
Two four-leaf stage rice plants (wild-type and OsSPL10 mutant) were grown in a plastic pot confined with a plastic cup into which 15 gravid BPH females were introduced. The number of BPH settled on each plant was recorded at 3, 6, 24, and 48 h after inculation. The test was replicated fifteen times for each plant pair.
Real-time qRT-PCR analysis
According to the manufacturer's recommendations, total RNA was extracted using the OminiPlant RNA kit (CWBIO). First-strand cDNA was synthesized using the FastKing RT Kit (TIANGEN BIOTECH). To determine gene expression, RT-qPCR experiments were performed with Taq SYBR Green qPCR Premix (Universal) in a StepOnePlus real-time PCR instrument (Applied Biosystems). OsActin was used as an internal control gene. All RT-qPCR reactions were performed using three independent replicate samples.
Transcriptome analysis
Wild-type ZH11 and OsSPL10 knockout rice plants at the four-leaf stage were infested by BPH for 0, 6, 24, and 48 h. The Sheaths of each plant were collected with three biological replicates at each timepoint, and total RNA was extracted using OminiPlant RNA kit (CWBIO). RNA-sequencing (RNA-seq) was performed by Novogene (Beijing, China). After adapter clipping and quality filter, clean data were remapped to the rice genome sequence (Nipponbare) using Hisat2 software and analyzed with StringTie. DESeq2 software was used to analyze the differentially expressed genes (DEGs), which were defined as those genes with significant expression changes (|log2(fold change)|> 0.5 and padj < 0.05) between ZH11 and OsSPL10 knockout plants.
Metabolome analysis
Sheath tissues were prepared as described in the transcriptome analysis. In total, 48 samples (six biological replicates per treatment group) were collected. For each sample, 100 mg of tissue was weighed and frozen in liquid nitrogen. Sample preparation for the metabonomic analysis and data analysis were performed by Novogene (Beijing, China) using standard procedures. Metabonomic analysis was performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Compound Discoverer 3.0 (CD 3.0, Thermo Fisher) was used to process the raw data files to perform peak alignment, peak picking, and quantitation for each metabolite. Differentially expressed metabolites (DEMs) were screened with |log2(fold change)|> 0.5, P value < 0.05, and VIP > 1.
Results
OsSPL10 negatively regulates rice resistance to BPH infestation
To identify transcription factors involved in the regulation of rice BPH resistance, we conducted a screen for BPH-resistant-related TFs using the hybrid transcription factor resource (Zhao et al. 2015). Two independent lines E030H-3 and E0303H-7 expressing the EAR4–SPL10 fusion protein showed stronger BPH resistance compared to the wild type (Japonica rice accession Kitaake), whereas the performance of those expressing the VP64–SPL10 fusion proteins (V030H-1 and V0303H-5) was similar to the wild-type plants (Fig. 1A).
SPL gene involved in the regulation of BPH tolerance is identified using a hybrid transcription factor resource. A BPH resistance score of Kitaake and OsSPL10-transgenic rice plants. Bars indicate SE (n = 10). Asterisks indicate significant differences compared to WT (*, P < 0.05; **, P < 0.01). WT, Japonica rice accession Kitaake. E0303H, a transgenic line expressing EAR4 (tetrameric repeats of EAR)–SPL10 fusion protein. V0303H, a transgenic line expressing VP64 (tetrameric repeats of VP16)–SPL10 fusion protein. B SPL10 expression in response to BPH infestation in the sheaths of rice seedlings. Bars indicate SE (n = 3)
OsSPL10 is a member of the SPL protein and has been reported to play a dual role in trichome development and salt tolerance in rice (Lan et al. 2019; Li et al. 2021). To examine the expression of OsSPL10, ZH11 seedlings at the four-leaf stage were infested with 15 BPH nymphs. Real-time quantitative PCR (RT-qPCR) showed that OsSPL10 expression was highly induced in the sheaths after BPH infestation (Fig. 1B), suggesting that it may function in response to BPH feeding.
Two spl10 mutants (ZH11-KO and HHZ-KO) in the ZH11 and HHZ background obtained via the CRISPR/Cas9 editing system were used to further determine the function of OsSPL10 in rice resistance to BPH (Lan et al. 2019). The BPH survival rates were tested to investigate the effect of OsSPL10 gene knockout on the growth and development of BPH nymphs. The survival rates of BPH on both ZH11-KO and HHZ-KO plants were significantly lower than those fed on wild-type plants (Fig. 2A, B). In the honeydew excretion test, BPHs fed on both ZH11-KO and HHZ-KO plants excreted a lower volume of excretion than those fed on wild-type plants (Fig. 2C, D), indicating that BPH feeding activity was significantly suppressed in the mutants. In addition, the female adults on two spl10 mutants laid fewer eggs than those on wild-type plants (Fig. 2E, F). These results indicate that knockout of OsSPL10 gene enhances rice resistance to BPH.
SPL10 negatively regulates BPH tolerance in rice. A BPH survival rates on ZH11 and spl10 knockout mutant plants after a 7-day infestation. Bars indicate SE (n = 8). B BPH survival rates on Huanghuazhan (HHZ) and spl10 knockout mutant plants after a 7-day infestation. Bars indicate SE (n = 8). C Areas of honeydew excretion by female BPH adults feeding on ZH11 and spl10 knockout plants. Bars indicate SE (n = 30). D Areas of honeydew excretion by female BPH adults feeding on HHZ and spl10 knockout plants. Bars indicate SE (n = 30). E Numbers of eggs laid by female BPH adults on ZH11 and spl10 knockout plants. Bars indicate SE (n = 12). F Numbers of eggs laid by female BPH adults on HHZ and spl10 knockout plants. Bars indicate SE (n = 12). Asterisks indicate significant differences compared to WT (*, P < 0.05; **, P < 0.01)
Transcriptome analysis reveals OsSPL10-regulated genes
To better understand the molecular mechanism by which OsSPL10 gene mutation enhances BPH tolerance in rice, we performed transcriptome analysis of ZH11-KO mutant plants and wild-type plants before (0 h) and after BPH feeding (6 h, 24 h, and 48 h). Differentially expressed genes (DEGs) were defined as genes that had significant expression change between ZH11-KO and ZH11 plants at different timepoints after BPH feeding (Supplemental Table S1). Among the DEGs, 2069, 2955, 779, and 2242 genes were upregulated, and 1971, 2531, 301, and 1771 genes were downregulated at 0, 6, 24, and 48 h of BPH infestation, respectively (Fig. 3A). DEGs commonly regulated by OsSPL10 at all four-timepoints were identified by Venn analysis, including 23 upregulated and 19 downregulated genes in ZH11-KO plants in relative to ZH11 plants (Fig. 3B).
Transcriptomic analysis of spl10 mutant and ZH11 plants before and after BPH feeding. A Number of differentially expressed genes (DEGs) in the sheaths between spl10 and ZH11 seedlings at different timepoints after BPH infestation. B, C, Venn analysis of DEGs identified in K0 vs W0, K6 vs W6, K24 vs W24, and K48 vs W48. DEGs are shown as up- B and downregulated C genes in sheaths. W0, uninfested wild-type plants; W6, W24, and W48, wild-type plants infested by BPH at 6, 24, 48 h; K0, uninfested ZH11-KO; K6, K24, and K48, ZH11-KO plants infested by BPH at 6, 24, 48 h
Nine DEGs related to ribosome biogenesis, plant hormone transduction, plant–pathogen interaction, and secondary metabolism were selected to confirm the results of transcriptome analysis by RT-qPCR. The expression profiles of these genes were consistent with transcriptome analysis, indicating the validation of the results from our transcriptome experiment (Supplemental Fig. S1).
Knockout of OsSPL10 induces defensive response to BPH infestation
To determine the putative functions of DEGs, we performed KEGG and GO enrichment analyses to explore the pathways and biological processes in which OsSPL10 participates. When comparing K0 with W0, KEGG analysis indicated that the genes involved in DNA replication, mismatch repair, homologous recombination, nucleotide excision repair, and ribosome biogenesis were upregulated, while the genes linked to phenylpropanoid biosynthesis and plant hormone signal transduction were downregulated. GO analysis revealed that enriched GO terms in DEGs were related to cell cycle, chromosome organization, DNA metabolic process, cell cycle process, cell wall organization, or biogenesis (Fig. 4A, Supplemental Table S2, and Supplemental Table S3).
In K6 vs W6 comparison, we identified 10 upregulated, and 15 downregulated KEGG categories. Interestingly, plant hormone signal transduction, fatty acid elongation, MAPK signaling pathway, and plant–pathogen interaction were identified as the most enriched pathways of upregulated DEGs (Fig. 4B and Supplemental Table S2). GO terms enriched in upregulated DEGs were also related to response to wounding, cell wall organization, response to acid chemical, and jasmonic acid signaling pathway, which are closely associated with herbivore-induced defense (Supplemental Table S3). Similarly, KEGG analysis showed that phenylpropanoid biosynthesis, plant–pathogen interaction, plant hormone signal transduction, and MAPK signaling pathway were the most enriched pathways of upregulated DEGs in K24 vs W24. These pathways presumably play important roles in underlying resistance to BPH infestation (Fig. 4C and Supplemental Table S2). GO analysis showed that the upregulated DEGs were mainly linked to the hydrogen peroxide catabolic process, cellular oxidant detoxification, hemicellulose metabolic process, xylan catabolic process, extracellular region, and chitinase activity (Supplemental Table S3).
A further comparison between K48 and W48 revealed that upregulated KEGG categories mainly included genes involved in the ribosome, citrate cycle (TCA cycle), purine metabolism, alanine, aspartate, and glutamate metabolism, while downregulated KEGG categories contained genes linked to plant hormone signal transduction, MAPK signaling pathway, basal transcription factors, cysteine and methionine metabolism, circadian rhythm (Fig. 4D and Supplemental Table S2). By GO enrichment analysis, we identified significantly enriched GO terms as ribosome biogenesis, ncRNA metabolic process, regulation of transcription, and regulation of RNA biosynthetic process (Supplemental Table S3).
Defense phytohormone-related genes induced by OsSPL10 knockout
Phytohormones, such as ethylene, jasmonic acid, and salicylic acid, are central regulators of plant defense. Plant hormone signaling network is usually activated after insect attack, enabling plants to activate effective defense response against herbivore infestation. The OsSPL10 downstream hormone-responsive genes were identified by comparing the DEGs with the previously reported genes induced by ethylene precursor ACC, JA, and SA (Garg et al. 2012). A total of 46 ACC-, 107 JA-, and 113 SA-responsive genes were identified in the DEGs (Fig. 5, Supplemental Table S4). Most hormone-responsive DEGs were induced by BPH feeding. For ACC-responsive genes, two genes involved in ethylene perception and signaling, OsETR2 (Os04g0169100) and OsCTR2 (Os02g0527600) were upregulated in ZH11-KO mutant after BPH infestation. Induced expression of genes related to the JA response, including OsJAZ6 (Os03g0402800), OsWRKY71 (Os02g0181300), and OsMYB110 (Os10g0478300), were also identified. These results indicated the involvement of ethylene and JA signaling in the regulation of OsSPL10-mediated defense response against BPH infestation.
Heatmap showing the expression pattern of defensive phytohormone-responsive genes in spl10 mutant and ZH11 plants at different timepoints after BPH infestation. A ACC- B JA- C SA-responsive genes in response to BPH infestation. Red represents upregulation, and blue represents downregulation (colour figure online)
Major transcription factor families regulated by OsSPL10 in response to BPH
Since transcription factors are the key players in the regulatory networks underlying plant responses to biotic and abiotic stress, differentially expressed TFs among different treatment groups were identified by searching the Plant Transcription Factor Database (PlnTFDB, V3.0) (http://plntfdb.bio.uni-potsdam.de/v3.0/). 145 upregulated and 97 downregulated, 183 upregulated and 92 downregulated, 73 upregulated and 12 downregulated, 94 upregulated and 121 downregulated differentially expressed TFs were identified in K0 vs W0, K6 vs W6, K24 vs W24, and K48 vs W48, respectively (Supplemental Table S5). The most members of identified TFs were AP2/ERF (61 genes), MYB (62 genes), bHLH (56 genes), C2H2 (33 genes), bZIP (35 genes), NAC (31 genes), and WRKY (27 genes), HD-ZIP (28 genes). After BPH feeding, most of these TFs were upregulated in ZH11-KO mutant plants compared to the ZH11 plants, except for AP2/ERF, HD-ZIP, and bZIP, which were downregulated at one or two timepoints of BPH infestation.
Metabolomic changes in spl10 mutant
To investigate metabolomic changes caused by OsSPL10 mutation after BPH inoculation, we conducted a metabolomic analysis of samples from spl10 and ZH11 plants before and after BPH infestation using an ultra‐performance liquid chromatography‐tandem mass spectrometry (UPLC‐MS/MS) detection platform. The large data sets obtained were subjected to Principal component analysis (PCA), which showed some degree of segregation between the spl10 and ZH11 samples at four timepoints (Supplemental Fig. S2). Partial Least Squares Discriminant Analysis (PLS-DA) was subsequently performed, and the score showed that different genotypes displayed significant segregation in the PLS-DA results (Supplemental Fig. S3).
Differentially expressed metabolites (DEMs) between ZH11-KO and ZH11 plants were defined at different timepoints after BPH feeding (Supplemental Table S6). The abundance of 577 metabolites was significantly changed in the four comparison groups. In the absence of pest damage, compared with ZH11 plants, 134 metabolites were increased and 76 were decreased in spl10 plants. In K6 vs W6, K24 vs W24, and K48 vs W48 comparison groups, 128, 42, and 149 increased metabolites, and 36, 67, and 129 decreased metabolites were detected, respectively (Supplemental Fig. S4A). Venn analysis showed that 115, 88, 60, and 179 specific metabolites were detected in each comparison. All comparison groups shared 6 common metabolites (Supplemental Fig. S4B). Based on HMDB database, metabolites were divided into 16 classes, among which lipids and lipid-like molecules were the most abundant metabolite (32% of the metabolites), followed by organic acids and derivatives (17% of the metabolites). Phenylpropanoids and polyketides accounted for 12% (Supplemental Fig. S5) (Supplemental Table S7).
SPL10 regulates antibiosis of rice to BPH through phenylpropanoid pathway
We integrated transcriptomic and metabolomic data to further understand the mechanism underlying the response of spl10 mutant to BPH infestation. The KEGG enrichment analysis revealed a positive correlation between many genes and metabolites in four comparisons, including phenylpropanoid biosynthesis and terpenoid backbone biosynthesis (Supplemental Fig. S7). In the phenylpropanoid pathway, genes encoding enzymes such as phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), o-methyltransferase (COMT), and beta-glucosidase (BGLU) were upregulated in OsSPL10 mutant. As expected, our metabolomic analysis showed that the contents of phenylalanine, ferulic acid, cinnamic acid, cinnamaldehyde, coniferaldehyde, and coumarin in this pathway were also increased in spl10 mutant, especially after BPH feeding (Fig. 6A, B, and C). Next, we investigated the effects of artificial diet feeding of three metabolites, phenylalanine, ferulic acid, and cinnamic acid, on the mortality of BPH female adults. Our results indicated that the mortalities of BPH female adults increased with increasing concentrations of ferulic acid and cinnamic acid (Fig. 6D).
OsSPL10 participates in regulating the genes and metabolites involved in phenylpropanoid biosynthesis. A Phenylpropanoid metabolism pathway schematic. Uppercase letters are genes that encode enzymes. Metabolites shaded in orange are differential abundant metabolites. BGLU, beta-glucosidase; PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl-alcohol dehydrogenase; POD, peroxidase; COMT, o-methyltransferase; CCoAOMT, Caffeoyl-CoA-O-methyltransferase. B Heatmap showing relative expression of DEGs involved in the phenylpropanoid metabolism pathway. C Relative abundance of differential abundant metabolites in spl10 mutant and ZH11 plants after BPH infestation. D Survival rate of BPH under the artificial feeding of metabolites. Bars indicate SE (n = 8). Asterisks indicate significant differences compared to WT (*, P < 0.05; **, P < 0.01) (colour figure online)
SPL10 regulates antixenosis of rice to BPH through terpene synthesis pathway
Next, we focused on the genes and metabolites involved in terpenoid metabolism according to KEGG database (Fig. 7A and Supplemental Fig. S6). The genes encoding crucial enzymes in the methylerythritol phosphate (MEP) pathway and the mevalonic acid (MVA) pathway were upregulated in the spl10 mutant plant. In addition, the expression of several genes encoding enzymes in the mono/diterpenoid biosynthesis and carotenoid biosynthesis pathways were also altered by BPH feeding (Fig. 7B). Consistently, metabolomic analysis showed that OsSPL10 mutation led to increased levels of glyceraldehyde-3-phosphate, isopentenyl pyrophosphate, farnesyl diphosphate, and mevalonate in spl10 mutant after BPH feeding (Fig. 7C). Interestingly, the rice volatiles analysis indicated that OsSPL10 mutation could also affect the volatiles released from rice plants (Table 1). Compared with WT plants, the level of limonene was significantly enhanced in the OsSPL10 mutant before and after BPH feeding (Table 1). Y-tube assays showed that limonene was repellent to BPH (Supplemental Fig. S8). Consistently, in the choice test between WT plants and spl10 mutants (ZH11-KO and HHZ-KO), a lower number of BPHs settled on spl10 mutants than on WT plants at 1, 3, 6, 9, 12, 24, and 48 h after BPH inoculaiton (Fig. 7D).
OsSPL10 participates in regulating the genes and metabolites involved in terpenoid biosynthesis. A Terpenoid metabolism pathway schematic. Uppercase letters are genes that encode enzymes. Metabolites shaded in orange are differential abundant metabolites. G3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME2P, 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate; HMG-CoA, hydroxymethylglutaryl-CoA; DMAPP, dimethylallyl pyrophosphate; IPP, isopentenyl pyrophosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; CPP, copalyl diphosphate; TPS, terpenoid synthase; VON, 9-cis-Violaxanthin; NON, 9′-cis-Neoxanthin; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXS, DXP synthase; CMK, 4-diphosphocytidyl-2-Cmethyl-D-erythritol kinase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HMGS, HMG-CoA synthase; HMGR, HMG-CoA reductase; GPS, GPP synthase; FPS, FPP synthase; GGPS, GGPP synthase; PS, phytoene synthase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase. B Heatmap showing relative expression of the genes involved in the terpenoid biosynthesis pathway. C Relative abundance of differential abundant metabolites in spl10 mutant and ZH11 plants after BPH infestation. D Comparison of settlement preference of BPH between spl10 mutants and ZH11 plants. Bars indicate SE (n = 22). Asterisks indicate significant differences compared to WT (*, P < 0.05; **, P < 0.01) (colour figure online)
Discussion
SPL proteins, as plant-specific transcription factors, play diverse functions in a wide range of processes, including plant growth and development, and response to environmental cues. In this study, we discovered that an SPL protein OsSPL10 plays a negative role in the regulation of resistance to BPH in rice. Integration of transcriptomic and metabolomic analyses revealed that OsSPL10 mutation could trigger the defense response of rice against BPH, including induced gene expression in plant hormone signal transduction, MAPK signaling pathway, phenylpropanoid biosynthesis, and plant–pathogen interaction pathway, and enhanced production of metabolites in phenylpropanoid pathway and terpenoid metabolism. Our study reveals a novel role of OsSPL10 in rice response to biotic stress in addition to its role in plant development and abiotic stress response.
Phytohormones, including JA, SA, and ET, play important roles in the defense against BPHs in rice. Several studies reported that JA pathways genes, including biosynthetic and signaling genes, are significantly upregulated by BPH attack. Overexpression of OsRCI-1, a BPH-induced lipoxygenase gene, increased the levels of JA and JA-Ile, conferring enhanced BPH resistance in rice (Liao et al. 2022). Knockout of biosynthetic and signaling genes AOC and MYC2 increased the susceptibility of rice plants to BPH, implying that JA pathway is involved in BPH resistance (Xu et al. 2021). Consistently, our transcriptomic data showed that knockout of OsSPL10 activated the expression of genes associated with JA-mediated signaling pathway. Expression levels of JAZ genes, including OsJAZ4, OsJAZ6, OsJAZ7, OsJAZ9, OsJAZ10, and OsJAZ13, were upregulated in spl10 mutant upon BPH feeding (Figs. 4, 5, and Supplemental Table S2). In addition, increased transcript levels were also observed for JA-responsive genes, such as OsWRKY71 and OsMYB110 (Fig. 5, and Supplemental Table S4). OsWRKY71 expression is induced by SA, MeJA, ACC, and pathogen infection. Overexpression of OsWRKY71 constitutively increased the expression of two defense marker genes, OsNPR1 and OsPR1b, contributing to enhanced resistance to virulent bacterial pathogens Xanthomonas oryzae pv. oryzae (Xoo) in rice (Liu et al. 2007). MYB110 is a cinnamate/monolignol pathway gene regulator and functions downstream of the MAMP-responsive MAPK cascade (MKK4–MPK3/MPK6) (Kishi-Kaboshi et al. 2018). OsWRKY53 has been reported to be phosphorylated by MAPK3 and MPK6 and enhances rice defenses against BPH by regulating the levels of H2O2 and ethylene (Hu et al. 2016). Interestingly, the expression of both OsMKK4 and OsWRKY53 was significantly elevated in spl10 mutant (Fig. 4 and Supplemental Table S2). Recently, increasing evidence demonstrates the crucial role of ethylene in the regulation of BPH resistance in rice. Silencing of the 1-aminocylopropane-1-carboxylic acid synthase gene OsACS2 increases resistance to BPH and decreases resistance to the striped stem borer (SSB) in rice (Lu et al. 2014). Two ET signaling components, ETHYLENE INSENSITIVE3-BINDING F-BOX PROTEIN1 (OsEBF1), and ETHYLENE INSENSITIVE3-LIKE1 (OsEIL1), positively and negatively regulated BPH resistance, revealing a negative regulation of ET pathway in response to BPH attack (Ma et al. 2020). Our results showed that the expression of OsETR2 and OsCTR2, which are involved in ethylene perception and signaling, was significantly higher in spl10 mutant than that in WT plant upon BPH feeding (Fig. 5 and Supplemental Table S4). We speculate that knockout of OsSPL10 increases the JA signaling, and suppresses the ET signaling, resulting in enhanced rice resistance to BPH.
The phenylpropanoid pathway is a major pathway in plants, which is required for the biosynthesis of lignin and serves as a starting point for the production of many other important defensive compounds, such as SA, flavonoids, and coumarins (Barros et al. 2016; Lefevere et al. 2020; Wu et al. 2022). In this study, a combination of the transcriptomic and metabolic analyses revealed that transcriptional upregulation of PAL, C4H, 4CL, CCR, and COMT genes, was accompanied by the elevation of the main metabolites in the phenylpropanoid pathways (Fig. 6). PAL catalyzes the first step in the phenylpropanoid pathway, a recent study demonstrated that PAL is a key enzyme involved in rice defense against BPH (He et al. 2019). Flavanone 3‐hydroxylase gene (OsF3H), which is also involved in the phenylpropanoid pathway, has been identified to play a dual role in resistance against brown planthopper and rice blast fungus Magnaporthe oryzae (Chen et al. 2022). We also found that both ferulic acid and cinnamic acid showed mortal effect on BPH females (Fig. 6D). These data suggest that increased genes and metabolites in the phenylpropanoid pathway play an important role in OsSPL10-mediated BPH resistance in rice.
Volatile compounds released by rice plants can affect the behavior of both herbivores and natural enemies. For example, BPH female adults preferentially oviposit on SSB-infested rice plants, due to the repellent role of SSB-induced volatiles for egg parasitoid A. nilaparvatae (Hu et al. 2020). Terpenoids, which are the most common group of secondary metabolites and volatile compounds, are important for multitrophic interactions and insect community composition (Munawar et al. 2022; Xiao et al. 2012). Our results showed that knockout of OsSPL10 increased the expression of the genes involved in terpenoid metabolism (Fig. 7B). As a result, the contents of isopentenyl pyrophosphate and farnesyl diphosphate, the main precursors in the biosynthesis of monoterpenes, sesquiterpenes, and triterpenes, and diterpenes were elevated in spl10 mutant (Fig. 7C). It should be noted that the induction of the OsTPS19 gene, which encodes limonene synthase, was observed (Fig. 7B) (Chen et al. 2018). As expected, we also found that knockout of OsSPL10 increased the level of a volatile compound, limonene, which has the repellent role for BPH feeding and oviposition (Table 1 and Supplemental Fig. S8).
Taken together, we demonstrate that a member of SPL transcription factor OsSPL10 is a negative regulator of BPH resistance in rice. Activation of JA signaling and suppression of ET signaling may contribute to OsSPL10-mediated BPH resistance. Enhanced expression of defense associated genes and increased accumulation of defensive metabolites in phenylpropanoid and terpenoid pathway also play a role in OsSPL10-mediated BPH resistance. Our study identifies a novel role of the transcription factor OsSPL10 in rice defense against BPH. Manipulation of OsSPL10 may have potential benefits for the development of insect-resistant rice cultivars.
Data availability
All data supporting the conclusions of this article are provided within the article (and its Additional files).
References
Barros J, Serrani-Yarce JC, Chen F, Baxter D, Venables BJ, Dixon RA (2016) Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nature Plants 2:16050
Chen X, Chen H, Yuan JS, Kollner TG, Chen Y, Guo Y, Zhuang X, Chen X, Zhang YJ, Fu J, Nebenfuhr A, Guo Z, Chen F (2018) The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol J 16:1778–1787
Chen S, Sun B, Shi Z, Miao X, Li H (2022) Identification of the rice genes and metabolites involved in dual resistance against brown planthopper and rice blast fungus. Plant Cell Environ 45:1914–1929
Cheng X, Zhu L, He G (2013) Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant 6:621–634
Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, He G (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106:22163–22168
Garg R, Tyagi AK, Jain M (2012) Microarray analysis reveals overlapping and specific transcriptional responses to different plant hormones in rice. Plant Signal Behav 7:951–956
Guo J, Xu C, Wu D, Zhao Y, Qiu Y, Wang X, Ouyang Y, Cai B, Liu X, Jing S, Shangguan X, Wang H, Ma Y, Hu L, Wu Y, Shi S, Wang W, Zhu L, Xu X, Chen R, Feng Y, Du B, He G (2018) Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat Genet 50:297–306
He J, Liu Y, Yuan D, Duan M, Liu Y, Shen Z, Yang C, Qiu Z, Liu D, Wen P, Huang J, Fan D, Xiao S, Xin Y, Chen X, Jiang L, Wang H, Yuan L, Wan J (2019) An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci USA 117:271–277
Hu L, Ye M, Li R, Lou Y (2016) OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant Signal Behav 11:e1169357
Hu L, Wu Y, Wu D, Rao W, Guo J, Ma Y, Wang Z, Shangguan X, Wang H, Xu C, Huang J, Shi S, Chen R, Du B, Zhu L, He G (2017) The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell 29:3157–3185
Hu X, Su S, Liu Q, Jiao Y, Peng Y, Li Y, Turlings TC (2020) Caterpillar-induced rice volatiles provide enemy-free space for the offspring of the brown planthopper. Elife. https://doi.org/10.7554/eLife.55421
Huang Z, He G, Shu L, Li X, Zhang Q (2001) Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet 102:929–934
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genet 42:541–544
Kishi-Kaboshi M, Seo S, Takahashi A, Hirochika H (2018) The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol 59:903–915
Lan T, Zheng Y, Su Z, Yu S, Song H, Zheng X, Lin G, Wu W (2019) OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L). G3 (bethesda) 9:4107–4114
Lefevere H, Bauters L, Gheysen G (2020) Salicylic acid biosynthesis in plants. Front Plant Sci 11:338
Li R, Zhang J, Li J, Zhou G, Wang Q, Bian W, Erb M, Lou Y (2015) Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. Elife 4:e04805
Li J, Tang B, Li Y, Li C, Guo M, Chen H, Han S, Li J, Lou Q, Sun W, Wang P, Guo H, Ye W, Zhang Z, Zhang H, Yu S, Zhang L, Li Z (2021) Rice SPL10 positively regulates trichome development through expression of HL6 and auxin-related genes. J Integr Plant Biol 63:1521–1537
Liao Z, Wang L, Li C, Cao M, Wang J, Yao Z, Zhou S, Zhou G, Zhang D, Lou Y (2022) The lipoxygenase gene OsRCI-1 is involved in the biosynthesis of herbivore-induced JAs and regulates plant defense and growth in rice. Plant Cell Environ 45:2827–2840
Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164:969–979
Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, Zhou F, Zhou K, Zheng X, Ren Y, Chen L, Wang Y, Zhao Z, Lin Q, Wu F, Zhang X, Guo X, Cheng X, Jiang L, Wu C, Wang H, Wan J (2015) A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol 33:301–305
Lou Y, Zhang G, Zhang W, Hu Y, Zhang J (2013) Biological control of rice insect pests in China. Biol Control 67:8–20
Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y (2014) Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant 7:1670–1682
Ma F, Yang X, Shi Z, Miao X (2020) Novel crosstalk between ethylene- and jasmonic acid-pathway responses to a piercing-sucking insect in rice. New Phyto 225:474–487
Munawar A, Zhang Y, Zhong J, Ge Y, Abou El-Ela AS, Mao Z, Ntiri ES, Mao LJ, Zhu Z, Zhou W (2022) Heat stress affects potato’s volatile emissions that mediate agronomically important trophic interactions. Plant Cell Environ 45:3036–3051
Nagata T (1984) Insecticide resistance in the brown planthopper. Chin J Entomol 4:117–124
Normile D (2008) Reinventing rice to feed the world. Science 321:330–333
Pan G, Liu Y, Ji L, Zhang X, He J, Huang J, Qiu Z, Liu D, Sun Z, Xu T, Liu L, Wang C, Jiang L, Cheng X, Wan J (2018) Brassinosteroids mediate susceptibility to brown planthopper by integrating with the salicylic acid and jasmonic acid pathways in rice. J Exp Bot 69:4433–4442
Shi S, Wang H, Nie L, Tan D, Zhou C, Zhang Q, Li Y, Du B, Guo J, Huang J, Wu D, Zheng X, Guan W, Shan J, Zhu L, Chen R, Xue L, Walling LL, He G (2021) Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol Plant 14:1714–1732
Tan J, Wu Y, Guo J, Li H, Zhu L, Chen R, He G, Du B (2020) A combined microRNA and transcriptome analyses illuminates the resistance response of rice against brown planthopper. BMC Genomics 21:144
Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X (2015) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Wang M, Yang D, Ma F, Zhu M, Shi Z, Miao X (2019) OsHLH61-OsbHLH96 influences rice defense to brown planthopper through regulating the pathogen-related genes. Rice 12:9
Wu F, Duan Z, Xu P, Yan Q, Meng M, Cao M, Jones CS, Zong X, Zhou P, Wang Y, Luo K, Wang S, Yan Z, Wang P, Di H, Ouyang Z, Wang Y, Zhang J (2022) Genome and systems biology of Melilotus albus provides insights into coumarins biosynthesis. Plant Biotechnol J 20:592–609
Xiao Y, Wang Q, Erb M, Turlings TC, Ge L, Hu L, Li J, Han X, Zhang T, Lu J, Zhang G, Lou Y (2012) Specific herbivore-induced volatiles defend plants and determine insect community composition in the field. Ecol Lett 15:1130–1139
Xu J, Wang X, Zu H, Zeng X, Baldwin IT, Lou Y, Li R (2021) Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore. New Phyto 230:1639–1652
Yao S, Kang J, Guo G, Yang Z, Huang Y, Lan Y, Zhou T, Wang L, Wei C, Xu Z, Li Y (2022) The key micronutrient copper orchestrates broad-spectrum virus resistance in rice. Sci Adv 8:eabm0660
Zhang J, Luo T, Wang W, Cao T, Li R, Lou Y (2017) Silencing OsSLR1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Plant Cell Environ 40:2147–2159
Zhang X, Liu D, Gao D, Zhao W, Du H, Qiu Z, Huang J, Wen P, Wang Y, Li Q, Wang W, Xu H, He J, Liu Y, Wan J (2022) Cytokinin confers brown planthopper resistance by elevating jasmonic acid pathway in rice. Int J Mol Sci. https://doi.org/10.3390/ijms23115946
Zhao T, Liu J, Li HY, Lin JZ, Bian MD, Zhang CY, Zhang YX, Peng YC, Liu B, Lin C (2015) Using hybrid transcription factors to study gene function in rice. Sci China Life Sci 58:1160–1162
Zhao Y, Huang J, Wang Z, Jing S, Wang Y, Ouyang Y, Cai B, Xin X-F, Liu X, Zhang C, Pan Y, Ma R, Li Q, Jiang W, Zeng Y, Shangguan X, Wang H, Du B, Zhu L, Xu X, Feng Y-Q, He SY, Chen R, Zhang Q, He G (2016) Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci USA 113:12850–12855
Zhao S, Hong W, Wu J, Wang Y, Ji S, Zhu S, Wei C, Zhang J, Li Y (2017) A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife. https://doi.org/10.7554/eLife.27529
Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, Lou Y (2009) Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J 60:638–648
Zhou G, Ren N, Qi J, Lu J, Xiang C, Ju H, Cheng J, Lou Y (2014) The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol Plant 152:59–69
Funding
This work is supported by the National Natural Science Foundation of China (32101676, U2005208, 31971833), the Natural Science Foundation of Fujian Province, China (2021J05019, 2020J02030, 2021J01075), the Fujian Agriculture and Forestry University Natural Science Funds for Distinguished Young Scholar (xjq21001), Young Talents Special Project of Yunnan Xingdian Talent Support Plan (Grant No. XDYC-QNRC-2022–0717).
Author information
Authors and Affiliations
Contributions
LL and ZS performed most of the work and initiated the draft. RW, YD, and ZZ helped with some experiments and data analysis. RZ, YS and TL conceived the study, obtained funding, and revised the final version of the manuscript. All authors read and approved the final article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Li Tian.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1:
Figure S1. Validation of nine DEGs using qRT-PCR. Figure S2. Principal component analysis (PCA) of all detected metabolites in spl10 and ZH11 seedlings at 0 (A), 6 (B), 24 (C), and 48h (D) after BPH infestation (Six biological replicates per treatment). Figure S3. Partial least squares-discriminant analysis (PLS-DA) of all detected metabolites in spl10 and ZH11 seedlings at 0 (A), 6 (B), 24 (C), and 48h (D) after BPH infestation (Six biological replicates per treatment). Figure S4. Metabolomic analysis of spl10 and ZH11 seedlings before and after BPH feeding. Figure S5. HMDB annotation for all DEMs between spl10 and ZH11 seedlings at different timepoints after BPH infestation. Figure S6. KEGG annotation for all DEMs between spl10 and ZH11 seedlings at different timepoints after BPH infestation. Figure S7. Correlation analysis of transcriptomic and metabolomic data from BPH-infested spl10 and ZH11 plants. Figure S8. Limonene showing the repellent role for BPH.
Supplementary file 2:
Table S1. List of DEGs between spl10 and ZH11 plants after BPH infestation.
Supplementary file 3:
Table S2. KEGG enrichment analysis of DEGs between spl10 and ZH11 plants at four timepoints after BPH infestation.
Supplementary file 4:
Table S3. GO enrichment analysis of DEGs between spl10 and ZH11 plants at four timepoints after BPH infestation.
Supplementary file 5:
Table S4. Expression changes of defensive phytohormone-responsive genes in spl10 mutant and ZH11 plants at different timepoints after BPH infestation.
Supplementary file 6:
Table S5. List of differentially expressed transcription factors (TFs) regulated by SPL10.
Supplementary file 7:
Table S6. List of differentially expressed metabolites between spl10 and ZH11 plants after BPH infestation.
Supplementary file 8:
Table S7. List of primers used in this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, L., Sun, Z., Wang, R. et al. Integration of transcriptome and metabolome analyses reveals the role of OsSPL10 in rice defense against brown planthopper. Plant Cell Rep 42, 2023–2038 (2023). https://doi.org/10.1007/s00299-023-03080-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-023-03080-z