Abstract
Key message
TFL1-like genes of the basal eudicot Platanus acerifolia have conserved roles in maintaining vegetative growth and inhibiting flowering, but may act through distinct regulatory mechanism.
Three TERMINAL FLOWER 1 (TFL1)-like genes were isolated and characterized from London plane tree (Platanus acerifolia). All genes have conserved genomic organization and characteristic of the phosphatidylethanolamine-binding protein (PEBP) family. Sequence alignment and phylogenetic analysis indicated that two genes belong to the TFL1 clade, designated as PlacTFL1a and PlacTFL1b, while another one was grouped in the BFT clade, named as PlacBFT. qRT-PCR analysis showed that all three genes primarily expressed in vegetative phase, but the expression of PlacTFL1a was much higher and wider than that of PlacTFL1b, with the latter only detected at relatively low expression levels in apical and lateral buds in April. PlacBFT was mainly expressed in young stems of adult trees followed by juvenile tissues. Ectopic expression of any TFL1-like gene in Arabidopsis showed phenotypes of delayed or repressed flowering. Furthermore, overexpression of PlacTFL1a gene in petunia also resulted in extremely delayed flowering. In non-flowering 35:PlacTFL1a transgenic petunia plants, the FT-like gene (PhFT) gene was significantly upregulated and AP1 homologues PFG, FBP26 and FBP29 were significantly down-regulated in leaves. Yeast two-hybrid analysis indicated that only weak interactions were detected between PlacTFL1a and PlacFDL, and PlacTFL1a showed no interaction with PhFDL1/2. These results indicated that the TFL1-like genes of Platanus have conserved roles in repressing flowering, but probably via a distinct regulatory mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
London plane tree (Platanus acerifolia Willd.) is an interspecific hybrid of P. orientalis L. and P. occidentalis L., which are basal eudicot species in the family Platanaceae belonging to the order Proteales (Byng et al. 2016). It is widely planted and applied in the cities as an excellent landscape plant, praised as ‘the king of street trees’. However, countless flowers and fruits with pollens and seed hairs, respectively, scatter everywhere in spring and early summer from the adult Platanus trees, which adversely impacts urban environment and human health and has become an increasingly serious problem in China that need to be resolved urgently (Liu et al. 2007; Lu et al. 2012). Breeding non-flowering or fruitless cultivars is desired for the species, so it is of great importance to investigate the molecular and genetic mechanisms involved in flowering regulation of Platanus.
It has been reported that genes of the phosphatidylethanolamine-binding protein (PEBP) family play crucial roles in controlling floral transition in plants (Wickland and Hanzawa 2015). In Arabidopsis, the PEBP (FT/TFL1) gene family includes six members: FLOWERING LOCUS T (FT), TWIN SISTER OF FT (TSF), TERMINAL FLOWER 1 (TFL1), BROTHER OF FT AND TFL1 (BFT), ARABIDOPSIS CENTRORADIALIS HOMOLOGUE (ATC), and MOTHER OF FT AND TFL1 (MFT) (Kobayashi et al. 1999; Jin et al. 2020). FT and TFL1 encode proteins that share highly conserved amino acid residues (~ 60% identity), but they have antagonistic functions in flowering regulation: FT is a florigen that induces flowering, while TFL1 represses flowering (Hanzawa et al. 2005; Ahn et al. 2006; Corbesier et al. 2007). TSF is the closest FT homolog and participates in flowering induction (Yamaguchi et al. 2005; Lee et al. 2019). BFT and ATC are TFL1-like genes and function as repressors of flowering (Yoo et al. 2010; Huang et al. 2012). MFT is regarded as ancestral to FT and TFL1 genes, as its orthologs exist in both basal land plants (like mosses and lycophytes) and seed plants (gymnosperms and angiosperms) (Hedman et al. 2009; Karlgren et al. 2011). In addition to promoting flowering, MFT also plays an important role in regulating seed germination (Yoo et al. 2004; Xi et al. 2010).
To date, TFL1-like genes have been identified and characterized in a wide variety of plant species including gymnosperms, monocots, and core eudicots (Wickland and Hanzawa 2015; Liu et al. 2016b). In Arabidopsis, TFL1 is expressed in vegetative and inflorescence meristems to maintain their vegetative character and indeterminate state, and so to control flowering time and inflorescence architecture, respectively (Bradley et al. 1997). Loss of function of TFL1 causes early flowering and determinate inflorescence by formation of a terminal flower, whereas its overexpression dramatically extends both the vegetative and reproductive phases (Ratcliffe et al. 1998). It was proposed that TFL1 regulates inflorescence indeterminacy by repressing the flower meristem identity genes LEAFY (LFY) and APETALA1 (AP1) in the center of the meristem, while LFY and AP1 repress the transcription of TFL1 in lateral floral primordia (Parcy et al. 2002). However, recent findings indicated that LFY is actually an activator of TFL1 and only indirectly represses TFL1 through AP1 (Goslin et al. 2017; Serrano-Mislata et al. 2017). The function of flowering repression has been shown to be conserved for TFL1-like genes in lots of plant species (Wickland and Hanzawa 2015); many TFL1 homologs also have conserved function in controlling inflorescence architecture, such as CENTRORADIALIS (CEN) in snapdragon (Bradley et al. 1996), RCN1 and RCN2 in rice (Nakagawa et al. 2002), VvTFL1A in grape (Fernandez et al. 2010), GhSP in cotton (McGarry et al. 2016; Si et al. 2018), and TFL1-like members in a number of legume crops (Foucher et al. 2003; Tian et al. 2010; Repinski et al. 2012; Liu et al. 2016a; Cheng et al. 2018). In addition, more diverse functions, including those involved in shoot branching and life history strategy, were characterized in TFL1-like genes (Perilleux et al. 2019). For instance, SELF-PRUNIN (SP), the CEN ortholog of tomato, regulates vegetative to reproductive switching of sympodial meristems (Pnueli et al. 1998); StTFL1 is involved in tuberization regulation in potato (Guo et al. 2010); AaTFL1 of Arabis alpina plays an important role in polycarpic development (Wang et al. 2011a, b); HvCEN promotes axillary bud initiation and tillering of barley (Bi et al. 2019a); CsCEN gene maintains the proliferative capacity of axillary meristems by antagonizing the thorn-specifying THORN IDENTITY1 (TI1) gene in Citrus (Zhang et al. 2021); TFL1 homolog KSN heterozygosity is associated with continuous flowering of Rosa rugosa Purple branch (Bai et al. 2021); and DOTFL1 may controlled pseudobulb formation in the Orchidaceae family (Li and Zhang 2021). Although the PEBP family is extensively investigated, the functions of TFL1-like genes are yet to be explored in basal eudicot species, like London plane tree.
So far, there is very limited information available about the molecular mechanisms controlling flowering in basal eudicots and perennial woody species. In this study, we isolated and characterized three TFL1-like genes from London plane tree, aiming to improve our understanding of flowering regulation in Platanus and to provide support for its genetic improvement. The gene structures, phylogenetic relationship, spatial and temporal expression patterns, and protein interaction of the three genes were investigated, and their biological functions were further characterized by overexpressing them in Arabidopsis and petunia. The results provide valuable information for understanding the evolution of TFL1-like genes in basal eudicots and for creating non-flowering and fruitless varieties of Platanus.
Materials and methods
Plant materials and sample collection
Various samples were collected from two-year-old juvenile and/or over thirty-year-old adult London plane trees grown at the campus of Huazhong Agricultural University (Wuhan, China). Juvenile trees were sampled at June, including roots (JR), stems (JS), newly growing young leaves (JYL), fully expanded mature leaves (JML) and subpetiolar buds (JSB). As described in Fig. S1, the flower development of Platanus spans two growing seasons. During the first seasons, lateral buds are formed under the petiole base (namely subpetiolar buds) on developing shoots (April–May), followed by two developmental fates. Most subpetiolar buds of adult trees (frequently located at the middle and upper part of the shoots) differentiate inflorescence and secondary shoot meristems individually in the same bud, hereinafter referred to as mixed flower buds. While, some subpetiolar buds (frequently located at the bottom part of the shoots or lower shoots of the tree) can only differentiate shoot meristems without inflorescence meristems, hereinafter referred to as vegetative subpetiolar buds. To uncover the comprehensive gene expression patterns during the whole flower and fruit development process, samples from adult trees were collected along two consecutive growing seasons (from April to April of next year), including the stems (S), newly growing young leaves (YL), fully expanded mature leaves (ML), shoot apical buds (AB), lateral subpetiolar buds (SB), vegetative subpetiolar buds (VB), mixed flower buds (MB), vegetative tissues in mixed flower buds (MB-V), inflorescences in mixed flower buds (MB-F), male inflorescences (MF), female inflorescences (FF) and fruits (F) (The corresponding descriptions of the samples are also listed in Table S1). All samples were collected from three individual trees, respectively, and immediately frozen in liquid nitrogen and stored at – 80 ℃ until they were used for RNA extraction.
Isolation of Platanus TFL1-like genes
Modified CTAB method was used to extract the total RNA of London plane tissues according to the procedures described by Li et al. (2008). Two μg of total RNA was used for first-strand cDNA synthesis using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara) following the protocol from the manufacturer and oligo (dT) primers. Degenerate primers PlacTFL1-dF and PlacTFL1-dR were used to amplify partial coding sequences of the PlacTFL1a and PlacTFL1b and designed according to nucleotide alignments of the TFL1-like genes from Vitis vinifera, Populus nigra, Malus × domestica, Citrus sinensis, and Eriobotrya japonica. The PCR was performed by denaturing cDNA at 95 ℃ for 3 min, followed by 35 cycles of 95 ℃ for 30 s, 55 ℃ for 30 s, 72 ℃ for 1 min, and a final extension at 72 ℃ for 10 min. The partial gDNA sequences were also obtained using the same pair of primers. 5′ Tail-PCR and 3′ RACE were performed to amplify the 5′ and 3′ terminal regions of PlacTFL1a, respectively; 5′/3′ Tail-PCR was carried out to obtain 5′/3′ terminal sequences of PlacTFL1b. Primers for amplifying the partial sequence of PlacBFT gene were designed according to the transcriptome sequencing data of London plane (unpublished), and 3′ RACE was used to amplify its 3′ terminal region. The RACE conditions were 95 ℃ for 3 min, followed by 36 cycles of 95 ℃ for 30 s, 60 ℃ for 30 s, 72 ℃ for 1 min, and a final extension at 72 ℃ for 10 min. TAIL-PCR was performed using a modified method (Wang et al. 2011a, b). All primers were designed using Primer 5 software and are listed in Table S2. The full-length cDNA and gDNA sequences of the three genes were amplified and cloned into a pMD18-T vector (Takara), and 3–5 positive clones were randomly selected for sequencing.
Sequence alignment and phylogenetic analysis of Platanus TFL1-like genes
Amino acid sequences of representative FT/TFL1-like proteins from P. acerifolia, Arabidopsis thaliana, Vitis vinifera and Nelumo nucifera were aligned using Vector NTI version 11.5 (Invitrogen). Sequences of FT/TFL1 family genes for representative species (Table S3) deposited in the National Center for Biotechnology Information (NCBI) database were retrieved for the construction of a phylogenetic tree. The alignment of amino acid sequences was made with the default settings in MUSCLE implemented in MEGA version 6.0 (Kumar et al. 2004). Phylogenetic tree was constructed using MEGA v6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Expression analysis of Platanus TFL1-like genes
Semi-quantitative RT-PCR and quantitative real-time PCR (qRT-PCR) analyses were performed to detect the expression of the Platanus TFL1-like genes. Primers were designed within the non-conservative coding region and 3′ UTR (untranslated region) using Primer 5.0 software to amplify the products between 90 and 300 bp in size (Table S2). qRT-PCR was performed in a total volume of 10 μl containing 5 μl 2 × SYBR Green Master Mix, 0.2 μl of each forward and reverse primer (10 μmol/μl), 1 μl of the RT reaction mixture as template and water to a final volume. Reactions were carried out on the ABI Prism 7500 Sequence Detection System (Applied Biosystems, USA). PCR efficiency for each primer pair was determined by a standard curve generated with serially diluted cDNA. The results from instrument onboard software Sequence Detector Version1.3.1 (PE Applied Biosystems) were further subjected to a custom-designed Microsoft Excel macro for analysis. Relative expression levels were calculated by Multiple Condition Solver REST-MCS v2 with TPI (triose phosphate isomerase) gene of P. acerifolia as a normalization (Lu et al. 2012). Calculations were based on three biological replicates and three technical replicates and data were shown as mean values ± SE (standard error).
Vector construction
To produce transgenic plants overexpressing the Platanus TFL1-like genes, we constructed 35S:PlacTFL1a, 35S:PlacTFL1b and 35S:PlacBFT on the expression vector pCAMBIA2300. The full-length coding regions of PlacTFL1a/b or PlacBFT genes were subcloned by SalI and KpnI or SalI and BamHI and ligation into pCAMBIA2300, which has cauliflower mosaic virus 35S promoter (CaMV35S) and NOS terminator. The amplified sequences were confirmed by restriction digestions and DNA sequencing. All generated constructs were introduced into Agrobacterium tumefaciens strain GV3101 or AGL0 by the electroporation method.
Plant transformation and phenotype analysis
Genetic transformation of 35S:PlacTFL1a/b and 35S:PlacBFT genes into A. thaliana was performed using the floral dip method (Clough and Bent 1998). The seeds from infected plants were cultured on agar-solidified Murashige and Skoog (MS) medium with 50 μg ml−1 kanamycin and 50 μg ml−1 cefotaxime. Kanamycin-resistant seedlings were cultured in a growth incubator with a photoperiod of 16/8 h (light/dark) at 22 ± 1 ℃ Phenotypic alternations of transgenic lines including the numbers of rosette and cauline leaves, bolting time, and anthesis time were recorded in T1 generation. For each gene constructions, homozygous transgenic lines defined by screening the kanamycin resistance of T3 generation were selected for further study, except for the lines with severe phenotype that did not produce flowers and progeny. Sixteen plants for each homozygous line were used to record phenotypic changes.
Petunia hybrida ‘W115’ was transformed with 35S:PlacTFL1a via the leaf disc method. First, 0.5% hypochlorite was used to sterilize the leaf explants for 20 min. Cut the leaves into approximately 0.5 × 0.5 cm squares and infected by Agrobacterium tumefaciens AGL0 containing 35S:PlacTFL1a expression vector for 10–15 min. Co-cultivate the explants in MS plates (MS medium supplemented with 2.0 mg/ml 6-BAP, 0.1 mg/ml NAA, 1.0 mg/ml zeatin, and 1.0 mg/ml folic acid) without antibiotics for 2–3 days at 25 ℃ under dark condition, and then transfer them into selective medium plates containing 250 μg ml−1 carbenicillin and 250 μg ml−1 kanamycin. After the shoots appeared, excise and culture them on hormone-free MS medium supplemented with 1.0 mg/ml folic acid, 250 μg ml−1 carbenicillin and 50 μg ml−1 kanamycin until rooting. Transgenic plants were identified by PCR with a primer in the 35S promoter (35SF) and a PlacTFL1a-specific primer (PlacTFL1a-vR) (Table S2).
Expression analysis of exogenous and endogenous genes in transgenic plants
To understand the functional conservation of Platanus TFL1-like proteins and whether key regulatory genes in Arabidopsis were affected by the transgene of PlacTFL1-like genes, we performed qRT-PCR experiment to investigate the expression levels of AtAP1, AtFUL, AtLFY and AtSOC1. Seedlings 21 d after sowing were sampled to isolate total RNA in wild type and transgenic Arabidopsis. Expression of petunia FT-like gene (PhFT; GenBank accession no. GU939627) and AP1 homologues (PFG, FBP26, and FBP29) was investigated using qRT-PCR in apical buds and leaves of 35S:PlacTFL1a transgenic petunia plants to understand the underlying mechanism of the repressed flowering phenotypes.
RNA extraction of wild-type and transgenic plants were conducted by the Trizol reagent (Takara, Japan) and reverse transcription were carried out using the method described above. AtEF1α and PhEF1α were used as the endogenous reference genes to normalize the data (Mallona et al. 2010). Primers are listed in Table S2.
Yeast two-hybrid assays
The full-length coding sequences of London plane TFL1-like and FD-like (PlacFDL, GenBank accession no. MH845055.1) genes, as well as petunia FD-like genes (PhFDL1 and PhFDL2, Appendix S1), were cloned and introduced to the bait plasmid pGBKT7 and prey plasmid pGADT7, respectively. All constructions were confirmed by sequencing. Yeast cells were transformed using the Frozen-EZ Yeast Transformation II Kit (Zymo Research Corp, USA). All baits were tested for autoactivation capacity prior to the screening for potential protein–protein interactions, and none of them showed autoactivation. Co-transformed yeast cells were selected on SD plates lacking Leu and Trp. Interactions were determined by spotting assay on selective SD media lacking Leu, Trp, His and Ade, supplemented with X-α-Gal.
Results
Isolation and phylogenetic analysis of Platanus TFL1-like genes
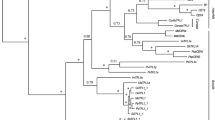
Using 3’ RACE combined with Tail-PCR methods, three TFL1-like genes were isolated from London plane tree. Sequence alignment and phylogenetic analysis indicated that two genes belong to the TFL1 clade, designated as PlacTFL1a and PlacTFL1b (GenBank accession no. MG344736 and MG344737), while the other one belongs to the BFT clade, designated as PlacBFT (GenBank accession no. MG344738) (Figs. 1, 2). Comparison of their cDNA and genomic sequences revealed that PlacTFL1a/b and PlacBFT genes have identical gene structure (including four exons and three introns) and ORF (open reading frame) length (519 bp encoding 172 aa), and that the four exons of the three genes demonstrated consistent length of 198 bp, 62 bp, 41 bp and 218 bp, respectively (Fig. 1a). However, the length of introns in PlacTFL1a, PlacTFL1b and PlacBFT are different, and their gDNA are 1,632 bp, 1,355 bp, and 1,324 bp in length, respectively. PlacTFL1a and PlacTFL1b genes share high similarity, with the identity of 91.7% and 90.1% at nucleotide and amino acid levels, respectively, whereas PlacBFT was less closely related to PlacTFL1a/b, showing only 72.4% and 71.3% identity of nucleotide and 75.1% and 73.4% identity of amino acid with PlacTFL1a and PlacTFL1b, respectively (Fig. 1).
Gene structure and sequence alignment of FT/TFL1 family genes. a Comparison of structure of the three TFL1-like gene sequences from London plane. Numbers indicate the base pairs in the exons (black boxes) and introns (thin lines). b Alignment of the deduced amino acid sequences of the products of FT/TFL1 family in Platanus acerifolia, Arabidopsis thaliana, Petunia, Vitis vinifera and Nelumo nucifera. Intron positions are indicated by black arrowheads. Asterisks indicate amino acids that are critical to the definition of proteins in the FT/TFL1 family
Multiple sequence alignment of the deduced amino acid sequences of the FT/TFL1-like proteins in P. acerifolia and several other species showed that PlacTFL1a/b and PlacBFT proteins had characteristic features of the TFL1-like proteins. The conserved key amino acid residues His88 and Asp144 of TFL1 (Ahn et al. 2006) were found at corresponding positions (His84 and Asp140) of PlacTFL1a/b and PlacBFT proteins, in addition, the amino acids Ser142, Ala173, Arg175, and Arg176 that were shown to be differently selected in TFL1-like proteins after the duplication resulting in FT and TFL1 clades (Wang et al. 2015) were also conserved in PlacTFL1a/b and PlacBFT (Fig. 1b). Phylogenetic analysis using the amino acid sequences of FT/TFL1 family proteins from London plane and other 12 representative species showed that the gene family consists of TFL1-like, FT-like, and MFT-like clades. TFL1-like clade is further divided into ATC, TFL1 and BFT subclades, and ATC subclade only contains genes from core eudicots (Fig. 2). PlacTFL1a/b and PlacBFT showed the closest relationship with their corresponding orthologs of Nelumbo nucifera, NnuFTL7/8 and NnuFTL9/10, respectively.
TFL1-like genes display distinct expression patterns in London plane
To get insight into the potential roles of the three TFL1-like genes in Platanus, we investigate their spatiotemporal expression patterns in juvenile (two-year-old) and adult (30-year-old) London plane trees. RT-PCR was performed to pretest the expression patterns of the three genes in various tissues and ontogenetic stages. The results showed that all three Platanus TFL1-like genes were primarily expressed in vegetative tissues, and no expression of them was detected in different development stages of inflorescences and fruits (Fig. S2).
The expression levels were further detected in vegetative tissues and several development stages of inflorescences and fruits from juvenile and/or adult trees by qRT-PCR, which showed that PlacTFL1a was expressed in all sampled tissues of juvenile plants, with high expression in young leaves and stems, low expression in mature leaves and SBs, and weak expression in roots (Fig. 3a). In adult trees, PlacTFL1a was expressed mainly in stems, young leaves, shoot apical buds, VBs, SBs before inflorescence initiation (April and May), and vegetative tissues in MBs, but was rarely detected in mature leaves, inflorescences, and fruits (Fig. 3b). The expression levels of PlacTFL1a in stems, shoot apical buds, and SBs increased at May compared to April, but its expression in SBs suddenly dropped at June when the inflorescences began to differentiate. In the VBs that did not form inflorescences, PlacTFL1a maintains high expression levels at July, and declines during later months, till very weak or no expression being detected during dormant period (Dec to Jan); after the dormancy release, expression of PlacTFL1a resumed gradually again at Feb to March (Fig. 4). In the vegetative tissues of MBs, PlacTFL1a displayed the same expression pattern as in the VBs (Fig. 4). Compared to PlacTFL1a, PlacTFL1b showed much lower levels as well as narrower regions of expression. For instance, it was only weakly expressed in SBs and not detected in roots, stems, and young or mature leaves of juvenile trees (Fig. 3a); in adult trees, PlacTFL1b was mainly expressed in apical and subpetiolar buds at April (Fig. 3b), with less transcripts in VBs and vegetative tissues of MBs during the growing period (Fig. 4). PlacBFT was also expressed in all detected tissues of juvenile plants, but predominantly in stems and roots followed by SBs and mature leaves (Fig. 3a). In adult trees, PlacBFT was predominantly expressed in stems (April and May), and lower expression was detected in SBs at April and VBs at March, with almost no expression in other tissues (Figs. 3b; 4).
Expression profiling of TFL1-like genes in London plane. a Relative expression of TFL1-like genes in juvenile plants. b Relative expression of TFL1-like genes in adult trees. JR roots of juvenile, JS stems of juvenile, JYL young leaves of juvenile, JML mature leaves of juvenile, JSB subpetiolar buds of juvenile, S stems, YL young leaves, ML mature leaves, AB shoot apical buds, SB subpetiolar buds, MB mixed flower buds, MB-F inflorescences in mixed flower buds, MF male inflorescences, MF-P fleshy peduncles of male inflorescences, FF female inflorescences. The numbers indicate the sampling month of the tissues. The level of expression was normalized to London plane TPI gene. Error bars represent SE for three replicates
Expression of PlacTFL1a/b and PlacBFT genes in different stages of VB and MB-V. VB, vegetative subpetiolar buds; MB-V, vegetative tissues in mixed flower buds. The numbers indicate the sampling month of the tissues. The level of expression was normalized to London plane TPI gene. Error bars represent SE for three replicates
Ectopic expression of Platanus TFL1-like genes in Arabidopsis inhibits flowering
To figure out the potential functions of Platanus TFL1-like genes in flowering regulation, CaMV35S was used to ectopically express PlacTFL1a/b and PlacBFT genes in Arabidopsis. Thirty-eight, fifty-one and thirty-three independent transgenic lines were achieved for 35S:PlacTFL1a, 35S:PlacTFL1b and 35S:PlacBFT, respectively. Overexpression of the three Platanus TFL1-like genes in Arabidopsis resulted in similar phenotypic alterations in delaying or repressing flowering (Fig. 5a–d, k; Table S4). Among the transgenic lines, five from 35S:PlacTFL1a, eight from 35S:PlacTFL1b, and three from 35S:PlacBFT displayed the strongest phenotypes, which produced an average of 50–54 rosette leaves before bolting under long day conditions (16/8 h, day/night), with the maximum number of rosette leaves up to 75 (Fig. 5e). After bolting, the inflorescence meristems of these plants maintained the status to form secondary and tertiary inflorescences reiteratively rather than to produce flowers until death (Fig. 5f), so we cannot obtain seeds and progenies from these strong phenotypic lines. For the transgenic lines with moderate or weak phenotypes, homozygous plants were chosen for further observation. We found most transgenic lines with moderate or weak phenotypes enhanced late flowering in their homozygous generation. For instance, homozygous lines #20 and #24 of 35S:PlacTFL1a and #19 of 35S:PlacTFL1b bolted with twenty to forty rosette leaves, but they always maintained at the inflorescence-producing status without flowers (Fig. 5g, k; Table S4). Transgenic lines with relatively weak phenotypes ultimately converted to flowering when inflorescences developed to a certain degree (Fig. 5h). In addition, overexpression of the three PlacTFL1-like genes in Arabidopsis also showed higher plant stature, larger leaves, thicker inflorescence stems, and more cauline leaves compared with the wild-type plants (Fig. 5d–f, i, k; Table S4).qRT-PCR analysis of transgenes in wild-type Arabidopsis (Col-0) and transgenic lines indicated that the phenotypic variations of the three PlacTFL1-like transgenic plants are related in a certain extent to the expression levels of the transgenes, namely higher expression levels tend to result in more severe phenotypic changes such as later bolting and flowering time or more rosette leaves (Fig. 5j, k; Table S4). Expression analysis of the flowering time related genes in 35S:PlacTFL1-like transgenic plants showed that AP1, FUL, LFY and SOC1 were intensively down-regulated in 21-day-old seedlings of 35S:PlacTFL1a line 20 and 35S:PlacTFL1b line 19, especially AP1 and FUL genes showed the strongest downregulating (Fig. 6). It is worth noting that only AP1 and FUL genes were significantly declined in 35S:PlacBFT line 26 (Fig. 6).
Phenotype analysis of transgenic Arabidopsis plants overexpressing PlacTFL1a/b or PlacBFT genes. a–c Wild-type (left) and 35S:PlacTFL1a transgenic line #10, 35S:PlacTFL1b transgenic line #48, 35S:PlacBFT transgenic line #26 (right), respectively. d Wild-type (left) and 35S:PlacTFL1a transgenic plants with moderate (line #15, middle) and strong (line #10, right) phenotype. e Rosette leaves of 35S:PlacTFL1a transgenic line #10. f Strong phenotypic lines of 35S:PlacTFL1a (line #10). g, h The top of inflorescences in 35S:PlacTFL1a transgenic plants with strong (g, line #20) and moderate (h, line #15) phenotype. i Inflorescence stem of 35S:PlacTFL1a transgenic line #20. j qRT-PCR analysis of transgenes in Col-0 and transgenic Arabidopsis lines overexpressing PlacTFL1a/b and PlacBFT genes. k Numbers of rosette leaves before bolting and days of bolting time in Col-0 and transgenic lines. Line #10 displayed the strong phenotypes, which we cannot obtain seeds and progenies from it that was so excluded from the statistical analysis. Bars: 10 mm (a–f), 1 mm (g–i)
qRT-PCR analysis of endogenous flowering-related genes in 21-day-old seedlings of Arabidopsis wild-type and 35S:PlacTFL1-like transgenic lines. The numbers after the genes indicate transgenic lines of the three 35S:PlacTFL1-like genes, respectively. Data represent the mean ± SE from three biological replicates, and AtEF1α was used as internal control. WT, wild-type seedlings. The asterisks indicate significant differences compared with the WT plants (P < 0.05)
Overexpression of PlacTFL1a in petunia represses flowering and promotes branching
Due to the similar and strong phenotypes in Arabidopsis overexpressing PlacTFL1a/b and PlacBFT, we further investigated the function of PlacTFL1a by ectopic expression in petunia. After verified by PCR amplification of the transgene, twenty-five independent T0 transgenic plants were obtained, but only three of them (#3, #6, and #23) showed evident expression via RT-PCR detection (Fig. 7a). Phenotypic observation and statistics analysis indicated that #6 and #23 lines showed obviously late flowering with more leaves and branches (Fig. 7b, c). In particular, the transgenic individual #6 maintained vegetative growth for more than one year and did not flower after several generations of cutting propagation, whereas wild-type W115 plants usually blossom within two months after planted (Fig. 7c). Finally, no seeds and sexual progenies can be obtained from #6 that was so excluded from the statistical analysis (Fig. 7b). Transgenic line #23 showed moderate phenotype of delayed flowering. It is worth mentioning that the main shoots of #23 usually return to vegetative growth after forming the first flower (Fig. 7c, d). RT-PCR analysis revealed that plants showing severe phenotype had higher expressed levels of the transgene compared with the plants exhibiting moderate or weak phenotype (Fig. 7), indicating that the transgene is responsible for the phenotypic alterations.
Phenotype analysis of transgenic petunia plants ectopically expressing PlacTFL1a gene. a RT-PCR analysis of transgenes in wild-type petunia (W115) and transgenic lines. b Numbers of branches and leaves on the principal shoot before flowering in W115 and T1 transgenic lines. c W115 (left) and 35S:PlacTFL1a transgenic lines #23 (middle) and #6 (right) after cultivated for two months. d T0 35S:PlacTFL1a transgenic lines #23 (left) and #6 (right) after transplanted for seven months. *Significantly difference compared to Col-0 (P < 0.05). Bars: 10 cm
To study the underlying mechanism of repressed flowering in PlacTFL1a-overexpressing plants, expression of petunia FT-like gene (PhFT) and AP1 homologues (PFG, FBP26, and FBP29 genes) were investigated in apical buds and leaves of the transgenic line #6 and wild-type ‘W115’. The results indicated that PhFT gene was up-regulated in apical buds of 35S:PlacTFL1a transgenic plant, whereas PFG, FBP26 and FBP29 genes were down-regulated, especially FBP26 was significantly repressed in both apical buds and leaves (Fig. 8).
qRT-PCR analysis of endogenous flowering-related genes in 35S:PlacTFL1a transgenic line (#6). a, b The expression levels of PhFT, PFG, FBP26, and FBP29 in apical buds (a) and leaves (b) of W115 (WT) and 35S:PlacTFL1a transgenic line. PhEF1α was used as internal control; the asterisks indicate significant differences compared with the WT plants (P < 0.05)
Interactions between London plane TFL1-like and FD-like proteins
A yeast two-hybrid analysis was performed to evaluate whether the TFL1-like proteins of Platanus can interact like the most situation in other species with its FD-like proteins. The coding sequences of PlacTFL1a/b, PlacBFT, PlacFDL and PhFDL1/2 genes were cloned into the pGADT7 (prey) and pGBKT7 (bait) domains, respectively, and used for interactional analysis. The results indicated that only PlacTFL1a had a weak interaction with PlacFDL in P. acerifolia (Fig. 9). Based on the ability of overexpressing PlacTFL1a to repress flowering in petunia, the interactions between P. acerifolia TFL1-like proteins and the FD-like proteins of petunia were also investigated. As a result, no interaction was detected between PlacTFL1a and petunia FD-like proteins (Fig. 9). However, the interaction of PlacBFT and PhFDL1 was detected in one direction (Fig. 9).
Discussion
Phylogenetic evolution of the FT/TFL1-like genes in basal eudicots
Previous phylogenetic analyses indicated that the PEBP gene family undergone two ancient duplications that generated three clades: MFT-like, FT-like and TFL1-like (Karlgren et al. 2011; Liu et al. 2016b). The first duplication giving rise to the MFT-like and FT/TFL1-like clades was suggested to take place in the common ancestor of seed plants or even earlier; the second duplication happened before the divergence of seed plants, resulting in the FT-like and TFL1-like clades (Liu et al. 2016b). After the separation of gymnosperms and angiosperms, further duplications occurred in each group. In angiosperms, an early whole-genome duplication (WGD) event has produced duplicate genes in each clade, and so most angiosperms contain approximately a half-dozen PEBP genes (Table S3). According to the evolutionary history of FT/TFL1 gene family, basal eudicots like P. acerifolia should possess MFT-like, FT-like and TFL1-like homologues. Previously, our research group have identified two Platanus FT-like genes PaFT and PaFTL that exhibited the function of promoting flowering in transgenic Arabidopsis or tobacco plants (Zhang et al. 2011; Cai et al. 2019). And we isolated here another three genes of the family, which belong to TFL1-like clade (Figs. 1, 2).
In accordance with previous studies, our phylogenetic tree divided the PEBP proteins from 13 species—into MFT-like, FT-like, and TFL1-like groups. MFT-like clade was further separated into two subgroups, and two or more genes are present in most species except Aquilegia coerulea, Arabidopsis, Fragaria vesca and Populus nigra that may have lost one copy (Fig. 2). In Platanus, two MFT-like genes were speculated, which was supported by the transcriptome data wherein two distinct MFT-like transcripts were found (data not shown). TFL1-like clade was also grouped into two classes, BFT-like and TFL1-like, both of which consist of genes from all included basal angiosperm, basal eudicots, and core eudicots (Fig. 2), indicating that the duplication generating these two lineages should occur in the common ancestor of angiosperms. The members from rice are present only in TFL1-like other than BFT-like lineage, suggesting that monocots may have lost the BFT-like genes. Furthermore, the phylogenetic tree shows that the genes from core eudicots in the TFL1-like lineage form two subgroups, ATC-like and TFL1-like (Fig. 2), indicating that TFL1-like clade experienced another duplication during the evolution of core eudicots. Whereas, some species like Populus and Jatropha have lost the TFL1-like genes.
In basal eudicots, Nelumbo nucifera and Platanus belong to the same order, Proteales (Byng et al. 2016). In contrast to 7 PEBP members in Platanus (two MFT-like, three TFL1-like, and two FT-like), N. nucifera contains more FT/TFL1 genes, including four MFT-like, four FT-like and four TFL1-like genes (Fig. 2), which should be resulted from a recent whole-genome duplication in N. nucifera (Ming et al. 2013; Wang et al. 2013). Another species of the basal eudicot, A. coerulea, contains one MFT-like, three TFL1-like, and three FT-like genes, indicating it has lost one MFT-like member and experienced further duplication of the TFL1-like and FT-like genes.
Divergent expression and function of the TFL1-like genes in P. acerifolia
In general, gene expression pattern has significant relationship with their functions. The expression patterns of TFL1-like genes show obviously changes among different members and/or plant species, and their functions also exhibit significant divergence and diversity (Wickland and Hanzawa 2015). For instance, Arabidopsis TFL1 transcripts are present in vegetative and inflorescence meristems to repress flowering and maintain inflorescence indeterminacy (Bradley et al. 1997; Serrano-Mislata et al. 2016), its paralog BFT is expressed in the shoot apical meristem, young leaf, and axillary inflorescence meristem (Yoo et al. 2010), whereas another paralog ATC was only detected in the hypocotyl of young plants, and not in the inflorescence meristem (Mimida et al. 2001). Apple TFL1-like genes MdTFL1 and MdTFL1a are expressed in the vegetative tissues in both the adult and juvenile phases; MdCENa (ATC ortholog) is mainly expressed in fruit receptacles, cultured tissues, and roots, while MdCENb is silenced in most organs (Mimida et al. 2009). The three TFL1-like genes in Jatropha curcas also show distinct expression patterns: JcTFL1a and JcTFL1c are mainly expressed in the roots of juvenile plants, whereas JcTFL1b transcripts are abundantly accumulated in the fruits and stems (Li et al. 2015, 2017).
Like most TFL1-like genes in other species, the three TFL1-like genes of London plane are preferentially expressed in vegetative tissues, but they have distinct spatiotemporal expression patterns. PlacTFL1a was widely expressed in vegetative organs of both juvenile and adult plants, including stems, leaves, apical buds, VBs, and the vegetative tissues of MBs (Figs. 3, 4). The expression of PlacTFL1a in SBs increased gradually prior to the inflorescence initiation (from April to May), but dramatically decreased during the inflorescence differentiation period (June), suggesting that PlacTFL1a play a crucial role in maintaining the vegetative growth and repressing the reproductive development of London plane, which is further supported by the highest expression level of PlacTFL1a in the VBs at July when the inflorescences are developing (Fig. 4). Based on this hypothesis, we speculate that higher expression level of PlacTFL1a should be present in the VBs at June, but at that moment the subpetiolar buds maintaining vegetative status could not be distinguished visibly from those undergoing flower bud differentiation, and so not detected. It is interesting that the expression levels of PlacTFL1b is significantly lower than PlacTFL1a and only weak expression is detected in a few tissues (Figs. 3, 4), although they are very closely related in terms of the coding sequences, with the identity of 91.7% at nucleotide level. In general, functional evolution of genes depends on two aspects: the change of gene coding sequences and alteration of gene expression patterns. Overexpression of PlacTFL1a and PlacTFL1b in Arabidopsis resulted in comparable phenotypic changes (Fig. 5), indicating that PlacTFL1b has retained its function in point of protein sequence but may has lost most functions in London plane due to expression degeneration after duplication, similar to above-mentioned apple MdCENb (Mimida et al. 2009). Expression pattern of PlacBFT is also significantly different from that of PlacTFL1a, with predominant expression in stems and roots, and weak in growing SBs (Fig. 3), suggesting PlacBFT may have undergone subfunctionalization during evolution.
Unlike some TFL1-like genes that are strongly expressed in developing inflorescences, such as Arabidopsis TFL1 (Bradley et al. 1997), Antirrhinum CEN (Bradley et al. 1996), and HvTFL1s in rubber tree (Bi and Tahir 2019), no TFL1-like genes of London plane were expressed evidently in inflorescences with various developmental stages (Fig. 3; Fig. S2). Given that the expression of TFL1-like genes in the inflorescences is related to their functions in the control of inflorescence architecture (Bradley et al. 1996, 1997; Nakagawa et al. 2002; Fernandez et al. 2010; Perilleux et al. 2019), we speculate that the TFL1-like genes of London plane may not involve in inflorescence development. To verify their functions, the three TFL1-like genes of Platanus were further investigated by transgenic studies in Arabidopsis and petunia. Overexpression of each gene delayed or repressed flowering, increased the number of leaves and nodes in transgenic plants compared to their wild-type counterparts, as reported in other species that constitutively express TFL1-like genes, confirming their highly functional conversation in flowering regulation among different plant species.
Potential mechanism of PlacTFL1a function
In model plants, TFL1 functions via directly repressing flowering-related genes, such as AP1, FUL, and LFY (Bradley et al. 1997; Ratcliffe et al. 1999; Hanano and Goto 2011). In contrast to PlacTFL1a, the expression of London plane AP1 homologs (FUL-like genes, PlacFLs) increased in SBs at the stage of inflorescence initiation (June) and maintained their expression level during the inflorescence developing process (Zhang et al. 2019), suggesting that PlacTFL1a inhibits reproductive development and flowering probably through repressing the expression of FUL-like genes in Platanus, which is consistent with the results reported in pear in which the expression of TFL1-like genes (PpTFL1-1a and PpTFL1-2a) rapidly decrease in reproductive meristems followed by upregulation of PpAP1 and PpFUL genes (Bai et al. 2017). Furthermore, significant downregulation of PFG, FBP26, and FBP29 genes were detected in 35:PlacTFL1a transgenic petunia plants that displayed the phenotype of severely repressed flowering (Fig. 8). It has been reported previously that knockdown of PFG and FBP26 genes (two FUL orthologs) represses the transition from vegetative to reproductive development in petunia, resulting in a phenotype exactly similar to the 35:PlacTFL1a transgenic plant #23 in our study (Immink et al. 1999), which indicates that 35:PlacTFL1a represses flowering at least partially through regulating the expression of AP1/FUL-like genes in petunia. All these results support a probably conserved regulatory mechanism between TFL1 and AP1/FUL-like genes in flowering regulation. However, our transgenic individual #6 showed drastically non-flowering phenotype even after several generations of propagation by cutting lasting for approximate two years, which is much more late flowering than the PFG and FBP26 down-regulated plants, even than a quadruple mutant of all the petunia AP1/FUL-like genes, pfg fbp26 fbp29 euap1 (Morel et al. 2019), suggesting that 35:PlacTFL1a must have regulated other flowering-related genes besides the AP1/FUL-like genes. Indeed, we found a FT-like gene (PhFT) was significantly up-regulated in the leaves of 35S:PlacTFL1a transgenic line #6 (Fig. 8). However, FT and its orthologs in most plant species were proved to function as florigens that promote flowering (Wickland and Hanzawa 2015). Interestingly, our recent study indicated that PhFT (corresponding to PhFT1 therein) might function as a repressor of flowering in petunia, because its overexpression in Arabidopsis resulted in significantly late flowering (Wu et al. 2019). The functions of repressing flowering have also been reported for several FT-like genes in other species, such as BvFT1 in sugar beet (Pin et al. 2010), HaFT1 in sunflower (Blackman et al. 2010), NtFT1/2/3 in tobacco (Harig et al. 2012), and SlSP5G(2/3) in tomato (Cao et al. 2015). In summary, PlacTFL1a represses flowering in petunia might through activating the flowering repressor PhFT1 and inhibiting the flowering promotors AP1/FUL-like genes, and some other unknown regulators if any.
It is well known that TFL1 repress flowering probably via interacting with FD to compete with FT (Hanano and Goto 2011; Ho and Weigel 2014; Zhu et al. 2020). Besides Arabidopsis, TFL1-like proteins interacting with FD homologues have been identified in Rosa chinensis, kiwifruit and so on (Varkonyi-Gasic et al. 2013; Randoux et al. 2014; Kaneko-Suzuki et al. 2018). The results of yeast two-hybrid analysis demonstrated that only PlacTFL1a has weak interaction with PlacFDL, while both PlacTFL1b and PlacBFT have no interaction with PlacFDL (Fig. 9). Even so, ectopic expression of both PlacTFL1b and PlacBFT in Arabidopsis still can delay flowering. Two hypotheses could be used to explain this result: one possibility is that PlacTFL1b and PlacBFT are able to interact with other FD-like members in London plane, as well as Arabidopsis FD protein; alternatively, interaction between Platanus TFL1-like protein and FD-like protein is not necessary for its function in repressing flowering. The latter assumption is supported by the fact that overexpression of PlacTFL1a in petunia results in repressed flowering, but no interaction between PlacTFL1a as well as PlacTFL1b and petunia FD-like proteins (PhFDL1 and PhFDL2) was detected (Fig. 9), while PlacBFT does interact with one of the petunia FD-like protein PhFDL1. A recent genome-wide ChIP-seq analysis demonstrated that TFL1 may interact with other DNA-binding proteins, besides FD, to regulate the expression of downstream genes (Goretti and Silvestre 2020). In summary, our results confirmed the function of Platanus TFL1-like genes in repressing flowering, but probably via a distinct regulatory mechanism.
Availability of data and materials
All data used in this research are included in this published article and its supplementary information files.
Code availability
Not applicable.
References
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25:605–614
Bai S, Tuan PA, Saito T, Ito A, Ubi BE, Ban Y, Moriguchi T, Wilson Z (2017) Repression of TERMINAL FLOWER1 primarily mediates floral induction in pear (Pyrus pyrifolia Nakai) concomitant with change in gene expression of plant hormone-related genes and transcription factors. J Exp Bot 68:4899–4914
Bai M, Liu J, Fan C, Chen Y, Chen H, Lu J, Sun J, Ning G, Wang C (2021) KSN heterozygosity is associated with continuous flowering of Rosa rugosa Purple branch. Hortic Res 8:26
Bi Z, Tahir AT (2019) Cloning and functional analysis of five TERMINAL FLOWER 1/CENTRORADIALIS-like genes from Hevea brasiliensis. Physiol Plantarum 166:612–627
Bi X, van Esse W, Mulki MA (2019) CENTRORADIALIS interacts with FLOWERING LOCUS T-like genes to control floret development and grain number. Plant Physiol 180:1013–1030
Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20:629–635
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379:791–797
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275:80–83
Byng JW, Chase MW, Christenhusz MJM, Fay MF, Judd WS, Mabberley DJ, Sennikov AN, Soltis DE, Soltis PS, Stevens PF, Briggs B, Brockington S, Chautems A, Clark JC, Conran J, Haston E, Moller M, Moore M, Olmstead R, Perret M, Skog L, Smith J, Tank D, Vorontsova M, Weber A, Grp AP (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181:1–20
Cai F, Shao C, Zhang Y, Bao Z, Li Z, Shi G, Bao M, Zhang J (2019) Identification and characterisation of a novel FT orthologous gene in London plane with a distinct expression response to environmental stimuli compared to PaFT. Plant Biol 21:1039–1051
Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S (2015) Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front Plant Sci 6:1213
Cheng X, Li G, Tang Y, Wen J (2018) Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 145.3:dev158766-dev158766.
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM (2010) Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J 61:545–557
Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15:2742–2754
Goretti D, Silvestre M (2020) TERMINAL FLOWER1 Functions as a mobile transcriptional cofactor in the shoot apical meristem. Plant Physiol 182:2081–2095
Goslin K, Zheng B, Serrano-Mislata A (2017) Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol 174:1097–1109
Guo JL, Yu CL, Fan CY, Lu QN, Yin JM, Zhang YF, Yang Q (2010) Cloning and characterization of a potato TFL1 gene involved in tuberization regulation. Plant Cell Tiss Org 103:103–109
Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23:3172–3184
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102:7748–7753
Harig L, Beinecke FA, Oltmanns J, Muth J, Müller O, Rüping B, Twyman RM, Fischer R, Prüfer D, Noll GA (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J 72:908–921
Hedman H, Källman T, Lagercrantz U (2009) Early evolution of the MFT-like gene family in plants. Plant Mol Biol 70:359–369
Ho WW, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26:552–564
Huang NC, Jane WN, Chen J, Yu TS (2012) Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J 72:175–184
Immink RG, Hannapel DJ, Ferrario S, Busscher M, Franken J, Lookeren Campagne MM, Angenent GC (1999) A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126:5117–5126
Jin S, Nasim Z, Susila H, Ahn JH (2021) Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin Cell Dev Biol 109:20–30
Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka KI (2018) TFL1-like proteins in rice antagonize rice ft-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol 59:458–468
Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U (2011) Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol 156:1967–1977
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lee C, Kim SJ, Jin S, Susila H, Youn G, Nasim Z, Alavilli H, Chung KS, Yoo SJ, Ahn JH (2019) Genetic interactions reveal the antagonistic roles of FT/TSF and TFL1 in the determination of inflorescence meristem identity in Arabidopsis. Plant J 99:452–464
Li Y, Zhang B (2021) DOTFL1 affects the floral transition in orchid Dendrobium Chao Praya Smile. Plant Physiol 186:2021–2036
Li Z, Liu G, Zhang J, Zhang J, Bao M (2008) Extraction of high-quality tissue-specific RNA from London plane trees (Platanus acerifolia), permitting the construction of a female inflorescence cDNA library. Funct Plant Biol 35:159–165
Li C, Luo L, Fu Q, Niu L, Xu ZF (2015) Identification and characterization of the FT/TFL1 gene family in the biofuel plant Jatropha curcas. Springer, New York, p 2
Li C, Fu Q, Niu L, Luo L, Chen J, Xu ZF (2017) Three TFL1 homologues regulate floral initiation in the biofuel plant Jatropha curcas. Sci Rep 7:43090
Liu G, Li Z, Bao M (2007) Colchicine-induced chromosome doubling in Platanus acerifolia and its effect on plant morphology. Euphytica 157:145–154
Liu Y, Zhang D, Ping J, Li S, Chen Z, Ma J (2016a) Innovation of a regulatory mechanism modulating semi-determinate stem growth through artificial selection in soybean. PLoS Genet 12:e1005818
Liu YY, Yang KZ, Wei XX, Wang XQ (2016b) Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol 212:730–744
Lu S, Li Z, Zhang J, Yi S, Liu L, Bao M, Liu G (2012) Isolation and expression analysis of a LEAFY/FLORICAULA homolog and its promoter from London plane (Platanus acerifolia Willd.). Plant Cell Rep 31:1851–1865
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
McGarry RC, Prewitt SF, Culpepper S, Eshed Y, Lifschitz E, Ayre BG (2016) Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol 212:244–258
Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6:327–336
Mimida N, Kotoda N, Ueda T, Igarashi M, Hatsuyama Y, Iwanami H, Moriya S, Abe K (2009) Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus x domestica Borkh.). Plant Cell Physiol 50:394–412
Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, Li J, Bowers JE, Tang H, Lyons E, Ferguson AA, Narzisi G, Nelson DR, Blaby-Haas CE, Gschwend AR, Jiao Y, Der JP, Zeng F, Han J, Min XJ, Hudson KA, Singh R, Grennan AK, Karpowicz SJ, Watling JR, Ito K, Robinson SA, Hudson ME, Yu Q, Mockler TC, Carroll A, Zheng Y, Sunkar R, Jia R, Chen N, Arro J, Wai CM, Wafula E, Spence A, Han Y, Xu L, Zhang J, Peery R, Haus MJ, Xiong W, Walsh JA, Wu J, Wang ML, Zhu YJ, Paull RE, Britt AB, Du C, Downie SR, Schuler MA, Michael TP, Long SP, Ort DR, Schopf JW, Gang DR, Jiang N, Yandell M, dePamphilis CW, Merchant SS, Paterson AH, Buchanan BB, Li S, Shen-Miller J (2013) Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol 14:R41
Morel P, Chambrier P (2019) Divergent functional diversification patterns in the SEP/AGL6/AP1 MADS-box transcription factor superclade. Plant Cell 31:3033–3056
Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29:743–750
Parcy F, Bomblies K, Weigel D (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129:2519–2527
Périlleux C, Bouché F, Randoux M, Orman-Ligeza B (2019) Turning meristems into fortresses. Trends Plant Sci 24:431–442
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400
Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125:1979–1989
Randoux M, Davière JM, Jeauffre J, Thouroude T, Pierre S, Toualbia Y, Perrotte J, Reynoird JP, Jammes MJ, Hibrand-Saint Oyant L, Foucher F (2014) RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytol 202:161–173
Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ (1998) A common mechanism controls the life cycle and architecture of plants. Development 125:1609–1615
Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126:1109–1120
Repinski SL, Kwak M, Gepts P (2012) The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor Appl Genet 124:1539–1547
Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F (2016) Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development 143:3315–3327
Serrano-Mislata A, Goslin K, Zheng B, Rae L, Wellmer F, Graciet E, Madueño F (2017) Regulatory interplay between LEAFY, APETALA1/CAULIFLOWER and TERMINAL FLOWER1: New insights into an old relationship. Plant Signal Behav 12:e1370164
Si Z, Liu H, Zhu J, Chen J, Wang Q, Fang L, Gao F, Tian Y, Chen Y, Chang L, Liu B, Han Z, Zhou B, Hu Y, Huang X, Zhang T (2018) Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture. J Exp Bot 69:2543–2553
Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci USA 107:8563–8568
Varkonyi-Gasic E, Moss SMA, Voogd C, Wang T, Putterill J, Hellens RP (2013) Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytol 198:732–746
Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G (2011a) Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23:1307–1321
Wang Z, Ye S, Li J, Zheng B, Bao M, Ning G (2011b) Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol 11:109
Wang Y, Fan G, Liu Y, Sun F, Shi C, Liu X, Peng J, Chen W, Huang X, Cheng S, Liu Y, Liang X, Zhu H, Bian C, Zhong L, Lv T, Dong H, Liu W, Zhong X, Chen J, Quan Z, Wang Z, Tan B, Lin C, Mu F, Xu X, Ding Y, Guo AY, Wang J, Ke W (2013) The sacred lotus genome provides insights into the evolution of flowering plants. Plant J 76:557–567
Wang Z, Zhou Z, Liu Y, Liu T, Li Q, Ji Y, Li C, Fang C, Wang M, Wu M, Shen Y, Tang T, Ma J, Tian Z (2015) Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 27:323–336
Wickland DP, Hanzawa Y (2015) The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol Plant 8:983–997
Wu L, Li F, Deng Q, Zhang S, Zhou Q, Chen F, Liu B, Bao M, Liu G (2019) Identification and characterization of the FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family in Petunia. DNA Cell Biol 38:982–995
Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22:1733–1748
Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46:1175–1189
Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH (2004) Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol Cells 17:95–101
Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH (2010) BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J63:241–253
Zhang J, Liu G, Guo C, He Y, Li Z, Ning G, Shi X, Bao M (2011) The FLOWERING LOCUS T orthologous gene of Platanus acerifolia is expressed as alternatively spliced forms with distinct spatial and temporal patterns. Plant Biol 13:809–820
Zhang S, Lu S, Yi S, Han H, Zhou Q, Cai F, Bao M, Liu G (2019) Identification and characterization of FRUITFULL-like genes from Platanus acerifolia, a basal eudicot tree. Plant Sci 280:206–218
Zhang F, Wang Y, Irish VF (2021) CENTRORADIALIS maintains shoot meristem indeterminacy by antagonizing THORN IDENTITY1 in Citrus. Curr Biol 31:2237-2242.e2234
Zhu Y, Klasfeld S, Jeong CW, Jin R, Goto K, Yamaguchi N (2020) TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat Commun 11:5118
Funding
This work was financially supported by the National Natural Science Foundation of China (31272206, 32102425, 31772345) and the Bureau of Forestry and Landscaping of Wuhan Municipality (Grant No. WHGF2022A09, WHGF2019A05). We thank all colleagues in our laboratory for discussions and technical assistance.
Author information
Authors and Affiliations
Contributions
LGF and BMZ designed the experiments. ZSS, ZQ, YXY, LYJ and SMM performed the experiments. ZSS, WJQ, JJ and NCR analyzed the data. ZSS and LGF wrote and revised the manuscript. All authors participated in the research and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Peishan Yi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Zhou, Q., Yang, X. et al. Functional characterization of three TERMINAL FLOWER 1-like genes from Platanus acerifolia. Plant Cell Rep 42, 1071–1088 (2023). https://doi.org/10.1007/s00299-023-03014-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-023-03014-9