Abstract
Key message
By using the organelle glue technique, we artificially manipulated organelle interactions and controlled the plant metabolome at the pathway level.
Abstract
Plant cell metabolic activity changes with fluctuating environmental conditions, in part via adjustments in the arrangement and interaction of organelles. This hints at the potential for designing plants with desirable metabolic activities for food and pharmaceutical industries by artificially controlling the interaction of organelles through genetic modification. We previously developed a method called the organelle glue technique, in which chloroplast–chloroplast adhesion is induced in plant cells using the multimerization properties of split fluorescent proteins. Here, we generated transgenic Arabidopsis (Arabidopsis thaliana) plants in which chloroplasts adhere to each other and performed metabolome analysis to examine the metabolic changes in these lines. In plant cells expressing a construct encoding the red fluorescent protein mCherry targeted to the chloroplast outer envelope by fusion with a signal sequence (cTP-mCherry), chloroplasts adhered to each other and formed chloroplast aggregations. Mitochondria and peroxisomes were embedded in the aggregates, suggesting that normal interactions between chloroplasts and these organelles were also affected. Metabolome analysis of the cTP-mCherry-expressing Arabidopsis shoots revealed significantly higher levels of glycine, serine, and glycerate compared to control plants. Notably, these are photorespiratory metabolites that are normally transported between chloroplasts, mitochondria, and peroxisomes. Together, our data indicate that chloroplast–chloroplast adhesion alters organellar interactions with mitochondria and peroxisomes and disrupts photorespiratory metabolite transport. These results highlight the possibility of controlling plant metabolism at the pathway level by manipulating organelle interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Living cells modify the arrangement and interaction of organelles to optimize cellular processes, enabling efficient signal transduction and metabolite exchange. Plant cells in particular dynamically change the arrangement and interaction of organelles in response to fluctuating environments. A typical example is the relocation of organelles with chloroplasts in response to light. Chloroplasts move toward weak light and away from strong light to optimize photosynthetic efficiency (Wada 2013). Activation of photosynthesis is accompanied by physical interactions of mitochondria and peroxisomes with chloroplasts (Islam et al. 2009; Oikawa et al. 2015; Midorikawa et al. 2022). As the enzymes responsible for the biosynthesis of photorespiratory metabolites are localized in chloroplasts, mitochondria, and peroxisomes (Eisenhut et al. 2019), the association of these organelles is thought to facilitate photorespiratory metabolite exchange.
Fluorescent proteins are powerful tools that are used in a wide range of applications in life science research. Many wild-type fluorescent proteins naturally occur as multimers, but they are engineered to monomeric forms because multimerization affects the localization and function of fluorescently tagged proteins (Shaner et al. 2005). However, membrane proteins tagged with monomerized fluorescent proteins occasionally alter organelle membranes, probably due to multimerization (Saito et al. 2011; Costantini et al. 2012; Segami et al. 2014).
The bimolecular fluorescence complementation (BiFC) assay is based on the structural complementation of N- and C-terminal fragments of a fluorescent protein. These two fragments are separately fused to other proteins and fluoresce when brought together to visualize and detect protein interactions in living cells (Hu et al. 2002; Kodama and Hu 2012). In our previous study, we fused the N-terminal fragment of monomeric Venus (mVenus) with the N-terminal 50 amino acids of Arabidopsis (Arabidopsis thaliana) OUTER ENVELOPE MEMBRANE PROTEIN 7 (OEP7) and co-expressed this construct with the DNA sequence of the free C-terminal fragment of mVenus in Egeria densa cells (Ichikawa et al. 2022). The co-expression forced a physical interaction between chloroplasts, possibly because the C-terminal fragments form bridges between the N-terminal fragments on the outer envelope via hydrophobic interactions. We named this method the organelle glue technique, a system that artificially controls organellar interactions. The forced physical interaction between chloroplasts likely changes the functional interactions between chloroplasts and other organelles and alters cellular metabolic activity.
Many crops have been genetically modified to alter metabolite production by knocking out or ectopically expressing genes encoding biosynthetic enzymes (Kumar et al. 2020). However, no studies have attempted to alter metabolite production in plants by manipulating organelle arrangement. Developing techniques for metabolic modifications that target organelle interactions rather than biosynthetic genes will provide new possibilities for producing genetically modified crops. In this study, we performed metabolome analysis on transgenic Arabidopsis plants with enhanced chloroplast–chloroplast physical interactions via our organelle glue technique using a chloroplast outer envelope-targeted fluorescent protein. We determined that photorespiration-related metabolites that are transported between chloroplasts, mitochondria, and peroxisomes accumulated in the transgenic plants. This result demonstrates the possibility that metabolite production can be modified by manipulating organelle interactions.
Materials and methods
Plant materials and growth conditions
Sterilized Arabidopsis (Arabidopsis thaliana) seeds were plated on one-half-strength Murashige and Skoog medium with 0.5% (w/v) gellan gum and 1% (w/v) sucrose and grown under continuous white light (50 μmol photons m−2 s−1) at 22 °C. The ecotype Columbia-0 (Col-0; CS60000) was used as the wild type. Sterilized Nicotiana benthamiana seeds were plated on the same medium as described above and grown at 25 °C under a 16-h-light/8-h-dark cycle. The Arabidopsis and N. benthamiana seedlings were transferred to soil (potting mix:vermiculite = 1:3) at 13 days after sowing. Subsequent growth conditions are the same as above (22 °C under continuous light for A. thaliana, 25 °C under a 16-h-light/8-h-dark cycle for N. benthamiana).
Plasmid construction and transformation
The DNA sequences for cTP-mCherry, cTP-mVenus, and the peroxisome marker po-mCerulean were cloned into the pENTR1a vector or the pDONR207 vector as described in previous studies (Osaki and Kodama 2017; Ichikawa et al. 2022). For the cTP-sfGFP construct, the DNA fragment encoding the N-terminal 50 amino acids of Arabidopsis OEP7 was PCR amplified using pENTR1a-cTP-mVenus as a template with the primer set 5′-AAC CAA TTC AGT CGA CAT GGG AAA AAC TTC GGG A-3′ and 5′-ACC GCC GCT ACC GCC GTC ATC GGG GTC TTT GGT-3′. The DNA fragment encoding sfGFP was PCR amplified using a synthetic double-stranded DNA fragment encoding sfGFP (Eurofins Genomics) as a template with the primer set 5′-GGC GGT AGC GGC GGT ATG GTG AGC AAG GGC GAG-3′ and 5′-AAG CTG GGT CTA GAT ATC TTA CTT GTA CAG CTC GTC-3′. The two fragments were joined by recombinant PCR with the primer set 5′-AAC CAA TTC AGT CGA CAT GGG AAA AAC TTC GGG A-3′ and 5′-AAG CTG GGT CTA GAT ATC TTA CTT GTA CAG CTC GTC-3′. The DNA fragment encoding cTP-sfGFP was cloned into the pENTR1a vector at the SalI and EcoRV sites using the In-Fusion HD Cloning Kit (Clontech) according to the manufacturer’s instructions. For the mCherry-sfGFP construct, a linear pENTR1a-sfGFP fragment was amplified by inverse PCR using pENTR1a-cTP-sfGFP as a template with the primer set 5′-GTC GAC TGA ATT GGT TCC-3′ and 5′-GGA GGC TCT GGA GGC ATG GTG AGC AAG GGC GAG-3′. The DNA fragment encoding mCherry was PCR amplified using pENTR1a-cTP-mCherry as a template with the primer set 5′-AAC CAA TTC AGT CGA CAT GGT GAG CAA GGG CGA G-3′ and 5′-GCC TCC AGA GCC TCC CTT GTA CAG CTC GTC CAT-3′. The two DNA fragments were circularized using the In-Fusion HD Cloning Kit. To express the cloned genes under the control of the Cauliflower mosaic virus 35S (CaMV35S) promoter, the cTP-mVenus, mCherry-sfGFP, and po-mCerulean fragments were cloned into the pGWB602 vector and the cTP-sfGFP and cTP-mCherry fragments were cloned into the pGWB508 vector (Nakagawa et al. 2007) by LR reaction (Invitrogen). The vector encoding the mitochondria marker mt-GFP (Nelson et al. 2007) was obtained from the Arabidopsis Biological Resource Center (stock number CD3-987).
Transient expression and transformation
Constructs encoding the fluorescently tagged proteins were transiently expressed in N. benthamiana leaves using Agrobacterium tumefaciens—mediated infiltration as described previously (Ishikawa et al. 2022). Transgenic Arabidopsis plants expressing cTP-sfGFP or cTP-mCherry driven by the CaMV35S promoter were generated using the floral dip method (Clough and Bent 1998).
Immunoblot analysis
Total proteins were extracted from N. benthamiana leaves transiently expressing an mCherry-sfGFP fusion protein 1.5 days post-infiltration or transgenic Arabidopsis shoots 11 days after sowing. The 15 mg of N. benthamiana leaves were ground with 250 µL of 2 × Laemmli sample buffer with 350 mM dithiothreitol. Arabidopsis shoots were ground with 10 µL of 2 × Laemmli sample buffer (with or without 350 mM dithiothreitol) per 1 mg. After boiling at 95 °C for 5 min and centrifugation at 20,000 × g for 10 min, 5 µL of supernatant and its serial fivefold dilutions were analyzed by immunoblot analysis according to a previous study (Ishikawa et al. 2015). sfGFP was detected using anti-GFP (Roche Diagnostics) at a dilution of 1:3000 and anti-mouse secondary antibody (Thermo Fisher Scientific) at a dilution of 1:3000. mCherry was detected using anti-RFP mAb HRP-DirecT (Medical & Biological Laboratories) at a dilution of 1:2000.
Confocal microscopy
Live-cell imaging was performed with an SP8X confocal microscope system (Leica Microsystems). Arabidopsis and N. benthamiana leaves were cut into 3-mm squares and mounted between two coverslips with water. sfGFP, mVenus, and mCherry were excited with 484-, 498-, and 554-nm lasers from the white light laser source and detected at 494–545, 510–545, and 565–636 nm by hybrid detectors, respectively. Chlorophyll autofluorescence was excited with 484-nm laser and detected at 655–743 nm by a photomultiplier tube detector. All images were taken with HC PL APO CS 63 × water-immersion lenses in photon counting mode with the time-gating system (Kodama 2016) at 100 Hz/1024 × 1024 pixels. Chloroplasts were visualized by chlorophyll autofluorescence.
Metabolite profiling
Metabolite profiling was performed as described previously (Kusano et al. 2007) with minor modifications. Frozen aerial parts of Arabidopsis were crushed and extracted in 25 mg fresh weight of tissues per milliliter of extraction medium (CHCl3:MeOH:H2O 1:3:1, v/v/v) including 10 stable isotope reference compounds (final concentration, 15 ng/µL for injection). After centrifugation for 10 min at 15,100 × g, 200 μL of the supernatant was evaporated to dryness in a Savant SPD2010 SpeedVac Concentrator (Thermo Electron Corporation, Waltham, MA, USA). Extracts were derivatized by 30 μL of methoxyamine hydrochloride (20 mg/mL in pyridine) for methoximation. After 21.5 h of derivatization at room temperature, the sample was trimethylsilylated for 1 h using 30 μL of N-Methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) at 37 °C with shaking. Then, 30 μL of n-heptane was added following silylation. All derivatization steps were performed in a vacuum glove box (128 VSC 1000; Sanplatec, Osaka, Japan) filled with 99.9995% (G3 grade) dry nitrogen. Gas chromatography time-of-flight mass spectrometry (GC–TOF–MS) analysis was performed as described previously (Kusano et al. 2007). For metabolite annotation, we used the national institute of standards and technology (NIST) mass search software, Golm Metabolome Database (Kopka et al. 2005), and our custom library based on standard compound analysis. Seven individual wild-type plants or transgenic lines were processed by the above method, and all annotated metabolites were analyzed using MetaboAnalyst v. 5.0 (https://new.metaboanalyst.ca/MetaboAnalyst/).
Results
Ectopic accumulation of mCherry targeted to the chloroplast outer envelope induces chloroplast–chloroplast adhesion
We fused superfolder green fluorescent protein (sfGFP), mVenus, or mCherry with the N-terminal 50 amino acids of chloroplast OEP7 via a linker sequence (cTP-sfGFP, cTP-mVenus, and cTP-mCherry). These three fluorescent proteins are GFP (for sfGFP and mVenus) or DsRed (for mCherry) derivatives that are reported to have a short maturation time and reduced dimer formation (Shaner et al. 2004; Pédelacq et al. 2006; Kremers et al. 2006). Then, we transiently expressed these constructs in epidermal cells of Nicotiana benthamiana true leaves. When we expressed cTP-sfGFP or cTP-mVenus, chloroplasts were normally dispersed throughout the cells (Fig. 1a). On the other hand, when we expressed cTP-mCherry, chloroplasts adhered to each other and aggregated (Fig. 1a). This suggests that full-length mCherry localized at the chloroplast outer envelope membrane can be used for the organelle glue technique. The chloroplast aggregations appeared to have a fused outer envelope and protrude a longer stromule. At the center of the chloroplast aggregations, a chloroplast thickly covered with outer membranes and stromules emits strong fluorescence. Co-expressing cTP-mCherry with genes encoding a mitochondria marker protein (mt-GFP) and a peroxisome marker protein (po-mCerulean) revealed that mitochondria and peroxisomes were embedded in the chloroplast aggregations (Fig. 1b).
Enhanced chloroplast–chloroplast physical interactions in cTP-mCherry-expressing cells. a Confocal microscope images of N. benthamiana leaves transiently expressing constructs encoding cTP (the N-terminal 50 aa of OEP7) fused with sfGFP, mVenus, or mCherry (cTP-sfGFP, cTP-mVenus, or cTP-mCherry, respectively). CHL, chlorophyll autofluorescence. Epidermal cells were observed 1.5 days after infiltration. Bars, 10 µm. b Mitochondria and peroxisome localization in the presence or absence of cTP-mCherry. Constructs encoding the mitochondria marker (mt-GFP) or the peroxisome marker (po-mCerulean) were transiently expressed with or without cTP-mCherry. Fluorescent signals are also seen in the cytosol due to mistargeting of mt-GFP and po-mCerulean. Epidermal cells of N. benthamiana leaves were observed 1.5 days after infiltration. Bars, 10 µm. c Immunoblot analysis of fluorescent proteins fused with cTP. Total proteins were extracted from N. benthamiana leaves transiently expressing cTP-sfGFP, cTP-mVenus, or cTP-mCherry 1.5 days after infiltration with Laemmli sample buffer with or without the reducing agent dithiothreitol (DTT). Samples extracted with the sample buffer without DTT were analyzed without boiling. Coomassie brilliant blue (CBB) staining shows blots of Rubisco large subunits (RbcL) as a loading control. Red dots, non-denatured form; red asterisks, denatured form; red line, multimers
We next investigated whether the chloroplast–chloroplast adhesion induced by cTP-mCherry is caused by multimerization of mCherry moieties. We performed immunoblot analysis using denatured and non-denatured total protein extracted from cTP-sfGFP-, cTP-mVenus-, or cTP-mCherry-expressing leaves. We detected cTP-sfGFP and cTP-mVenus only as monomers in denatured and non-denatured total protein, while we detected multimerized cTP-mCherry molecules from non-denatured total protein (Fig. 1c). These data suggest that the multimerization of cTP-mCherry molecules on the outer envelope induced chloroplast–chloroplast adhesion.
cTP-mCherry-expressing Arabidopsis plants display chloroplast–chloroplast adhesion
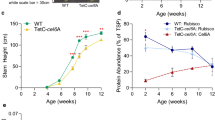
We generated three lines each of transgenic Arabidopsis plants expressing cTP-sfGFP or cTP-mCherry under the control of the CaMV35S promoter and investigated chloroplast–chloroplast adhesion. We observed no chloroplast abnormalities in epidermal cells or mesophyll cells in cTP-sfGFP-expressing leaves (Fig. 2a). However, in cTP-mCherry-expressing leaves, chloroplasts aggregated in epidermal cells (Fig. 2a), as observed in N. benthamiana (Fig. 1a). In mesophyll cells, chloroplasts varied in size and had abnormal morphology (Fig. 2a). To test whether these chloroplast abnormalities were due to cTP-mCherry multimerization, we performed immunoblot analysis using non-denatured total protein extracted from shoots of cTP-mCherry-expressing Arabidopsis seedlings. As was the case with transient expression in N. benthamiana (Fig. 1c), we mostly detected cTP-sfGFP as monomers, whereas a portion of the cTP-mCherry proteins were multimers (Fig. 2b). These results suggested that the interaction of mCherry moieties causes the chloroplast outer envelopes to fuse to each other. Chloroplasts are typically densely packed in mesophyll cells, which may explain why we did not observe increased chloroplast aggregation in cTP-mCherry-expressing cells (Fig. 2a). We suspected that these chloroplast abnormalities were caused by higher accumulation of cTP-mCherry than cTP-sfGFP; therefore, we compared the amounts of cTP-sfGFP and cTP-mCherry in the transgenic plants by immunoblot analysis. We used total protein extracted from N. benthamiana leaves expressing an mCherry-sfGFP fusion protein to quantitatively compare signal intensities detected with different antibodies. The total protein was diluted to ensure whether the antibodies detect antigens in linear proportion to their amount. Quantification using three individual plants for each transgenic line revealed that the amount of cTP-mCherry was about half that of cTP-sfGFP (Fig. 2c, d). This confirmed that the chloroplast–chloroplast adhesion in cTP-mCherry-expressing Arabidopsis plants is caused by the properties of mCherry, not by higher protein accumulation. Next, we examined the effect of the chloroplast abnormalities caused by cTP-mCherry expression on plant growth. cTP-mCherry- and cTP-sfGFP-expressing Arabidopsis plants exhibited similar growth, suggesting that chloroplast–chloroplast adhesion does not affect plant growth in the absence of a stress treatment (Fig. 2e, f). Therefore, in our subsequent experiments, we used the cTP-sfGFP-expressing plants as controls and refer to the cTP-mCherry-expressing plants as chloroplast-glued plants.
Generation of chloroplast-glued Arabidopsis plants. a Confocal microscope images of Arabidopsis leaves expressing cTP-sfGFP or cTP-mCherry under the control of the CaMV35S promoter. True leaves of epidermal cells and mesophyll cells were observed 11 days after sowing. Bars, 10 µm. b Multimerization of cTP-mCherry in the transgenic plants. Immunoblot analysis of total proteins extracted from three lines each of cTP-sfGFP- or cTP-mCherry-expressing Arabidopsis leaves using Laemmli sample buffer with or without the reducing agent dithiothreitol (DTT). Samples extracted with the sample buffer without DTT were analyzed without boiling. Coomassie brilliant blue (CBB) staining shows blots of Rubisco large subunits (RbcL) as a loading control. Red dots, non-denatured form; red asterisks, denatured form; red line, multimers. c, d Comparison of cTP-fluorescent protein accumulation between cTP-sfGFP- and cTP-mCherry-expressing Arabidopsis plants. c Immunoblot analysis of total protein extracted from shoots of three lines each of Arabidopsis expressing cTP-sfGFP or cTP-mCherry. Total protein extracted from N. benthamiana leaves transiently expressing mCherry-sfGFP and its serial fivefold dilutions were used to compare antibody titers. CBB staining shows blots of RbcL as a loading control. d Quantification of cTP-fluorescent protein accumulation based on the immunoblot analysis in c. Different titers of anti-GFP and anti-RFP were adjusted by reactivity to mCherry-sfGFP. Accumulation levels in cTP-sfGFP #1 were set to 1.0. n = 3; data are shown as means ± SD; different letters indicate significant differences, P < 0.05, using Tukey’s multiple comparisons test. e, f Growth of Arabidopsis plants expressing cTP-sfGFP or cTP-mCherry. e Thirteen-day-old cTP-sfGFP- or cTP-mCherry-expressing Arabidopsis plants. Bars, 10 mm. f Quantification of plant length including roots and shoots. No significant difference was found in Tukey’s multiple comparison test. n = 23–31; data are shown as box and whisker plot; P < 0.05, using Tukey’s multiple comparisons test
Chloroplast–chloroplast adhesion leads to accumulation of metabolites involved in the photorespiratory pathway
We presumed that metabolic pathways involving multiple organelles are disrupted in the chloroplast-glued plants. We therefore analyzed the metabolomes of three lines each of the transgenic control plants (cTP-sfGFP#1–3) and the chloroplast-glued plants (cTP-mCherry#1–3) by using GC–TOF–MS. Raw metabolome data, including unannotated metabolites, are available in Table S1. The amounts of 15 metabolites were significantly changed between control plants and the chloroplast-glued plants (Fig. 3a). Among them, 13 metabolites were significantly increased in the chloroplast-glued plants, and four of these metabolites, glycine, homoserine, serine, and glycerate, are related to the photorespiration pathway. Pathway enrichment analysis revealed that the photorespiration-related pathways ‘glycine, serine, and threonine metabolism’ and ‘glyoxylate and dicarboxylate metabolism’ were significantly affected in the chloroplast-glued plants (Fig. 3b, c). The physical distances between chloroplasts, peroxisomes, and mitochondria are thought to be important for activating the photorespiratory pathway (Islam et al. 2009; Oikawa et al. 2015; Midorikawa et al. 2022). In the chloroplast-glued plants, the loss of proper physical and functional interactions between chloroplasts and other organelles may disrupt the photorespiratory pathway and production of its metabolites.
Chloroplast–chloroplast adhesion impacts photorespiration-related pathways. a Heat map of metabolite accumulation in cTP-mCherry lines #1–3 (chloroplast-glued plants) and cTP-sfGFP lines #1–3 (transgenic control plants). Metabolites are listed in ascending order of P value for the difference between cTP-mCherry and cTP-sfGFP lines. The yellow square indicates metabolites with P < 0.05. Metabolites involved in the photorespiratory pathway are marked by asterisks. b, c Pathway enrichment analysis of metabolites whose accumulation significantly increased in chloroplast-glued plants. b Bubble chart. Higher-impacted metabolic pathways are indicated by larger red circles. c List of higher-impacted metabolic pathways. p, P values were calculated by Fisher’s exact test. FDR, the adjusted P value using the false discovery rate. Impact, the pathway impact value calculated from pathway topology analysis. Metabolic pathways with P < 0.05 and impact > 0 are listed in the table
Photorespiratory metabolites transported between organelles accumulate in chloroplast-glued plants
The photorespiratory metabolites glycine, serine, and glycerate, which increased in the chloroplast-glued plants, are transported between chloroplasts, peroxisomes, and mitochondria (Fig. 4a). Therefore, we compared the amounts of these metabolites between the wild-type, transgenic control, and chloroplast-glued plants. We observed no significant difference in the amount of glycine, serine, or glycerate between the wild-type and transgenic control plants, but we observed significant increases between the wild-type and chloroplast-glued plants (Fig. 4b). This suggests that in the chloroplast-glued plants, the physical and functional interactions between chloroplasts, peroxisomes, and mitochondria are hindered, which inhibits the transport of metabolites between organelles, resulting in their accumulation.
Chloroplast–chloroplast adhesion causes accumulation of photorespiratory metabolites transported between organelles. a Simplified illustration of the transport of metabolites in the photorespiration pathway between organelles. b Quantification of the amount of glycine, glycerate, and serine in the wild-type (WT, non-transgenic plants), cTP-sfGFP-expressing lines #1–3 (control plants), or cTP-mCherry-expressing lines #1–3 (chloroplast-glued plants). Accumulation levels in the WT were set to 1.0. n = 7; data are shown as means ± SD; asterisks indicate significant differences between the WT, cTP-sfGFP-expressing lines, and cTP-mCherry-expressing lines, P < 0.05, using Dunnett’s multiple comparison test
Discussion
The arrangement of and interaction between organelles are closely related to cellular metabolism. The industrial use of plants for metabolite production is receiving increased attention. Therefore, developing new technologies to manipulate plant metabolism is of great interest. In the present study, we generated chloroplast-glued plants using our previously developed organelle glue technique (Ichikawa et al. 2022), which promotes physical interactions between organelles using the multimerization properties of fluorescent proteins. Peroxisomes and mitochondria were embedded in the aggregated chloroplasts, suggesting changes in the physical and functional interactions between organelles (Fig. 1b). Metabolome analysis revealed that the amounts of the photorespiratory metabolites glycine, serine, and glycerate, which are transported between organelles, significantly increased in the chloroplast-glued plants (Figs. 3, 4). These data highlight the possibility of engineering plant cell metabolomes by manipulating organelle interactions.
Photorespiration consumes O2 and releases CO2 under light conditions and is necessary to replenish the metabolites required for photosynthesis (Hagemann and Bauwe 2016; Timm and Hagemann 2020). On the other hand, it also consumes NADPH and ATP, which are required for photosynthetic CO2 assimilation, thus controlling the rate of photosynthesis. Furthermore, photorespiration is related to resistance to abiotic stresses such as heat stress and drought stress (Wingler et al. 1999; Cui et al. 2016; Cavanagh et al. 2022), which greatly reduce crop yields. Therefore, photorespiration is a major target for crop improvement. However, there are difficulties in the optimization of photorespiration because multiple genes involved in each step are required to be coordinately controlled (Timm and Hagemann 2020). Our results suggested that the introduction of a single transgene can modulate the entire photorespiratory pathway. Adjusting the degree of chloroplast adhesion may be a promising approach to enhance CO2 assimilation rates and tolerance to abiotic stresses by optimizing photorespiration.
Chloroplast–chloroplast adhesion mediated by fluorescent protein multimerization occurred in cTP-mCherry-expressing cells but not in cTP-sfGFP- or cTP-mVenus-expressing cells (Fig. 1a). mCherry originated from the red fluorescent protein (RFP) of the coral Discosoma sp., while sfGFP and mVenus originated from the GFP of the jellyfish Aequorea victoria (Shaner et al. 2004). Although fluorescent protein derivatives were engineered to be monomeric, tagRFP, another RFP derived from Discosoma sp., can multimerize and alter organelle membrane structures when accumulated in animal (Costantini et al. 2012) and plant cells (Saito et al. 2011). RFPs may be more likely than other fluorescent proteins to multimerize when aligned on organelle membranes.
Chloroplast aggregation in cTP-mCherry-expressing cells is likely induced by multimerization of mCherry. We propose two possibilities for the mechanism by which mCherry multimerization induces chloroplast aggregation. The simplest model is that mCherry anchored on the outer envelope membrane of different chloroplasts might glue the membranes together via mCherry multimerization. However, there is the finding that is contrary to this hypothesis. The ability of fluorescent proteins to alter organelle membranes by multimerization is conventionally assessed by the organized smooth endoplasmic reticulum (OSER) assay, in which fluorescent proteins are fused to ER membrane-anchoring signals and expressed in animal cells (Costantini et al. 2012). A lower OSER score indicates a stronger ability to alter the ER membrane. The OSER score of mCherry was 69.0–95.0%, comparable to those of sfGFP (62.4%) and mVenus (83.9%) (Cranfill et al. 2016; Manna et al. 2018), inconsistent with the ability to connect chloroplasts to each other. The ability of fluorescent proteins to multimerize and alter the membranes may vary depending on the organelle. In cTP-mCherry-expressing cells (Fig. 1a), stromules in chloroplasts appeared to elongate abnormally and connect chloroplasts to each other. Therefore, as a second model, we propose that mCherry multimerization at the outer envelope membrane may induce stromule elongation, resulting in chloroplast–chloroplast adhesion.
Although multimerization of fluorescent proteins is undesirable when analyzing native protein localization and function, this phenomenon has advantages for the organelle glue technique. First, physiological changes can be analyzed while visualizing organelle adhesion. Second, as the multimerization properties of various fluorescent proteins have been investigated in vitro and in vivo, the degree of adhesion between organelles can be adjusted by changing the fluorescent protein. Finally, many studies have experimentally demonstrated that the expression of fluorescent proteins has little to no negative effects on living organisms. Further studies are needed to develop a system in which physical interactions between organelles other than chloroplasts are quantitatively controlled. We plan to further improve the organelle glue technique so that we can fine-tune the arrangement and interaction of organelles in the future.
Data availability
All data in this study are available from the corresponding author upon reasonable request.
References
Cavanagh AP, South PF, Bernacchi CJ, Ort DR (2022) Alternative pathway to photorespiration protects growth and productivity at elevated temperatures in a model crop. Plant Biotechnol J 20:711–721. https://doi.org/10.1111/pbi.13750
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Costantini LM, Fossati M, Francolini M, Snapp EL (2012) Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic 13:643–649. https://doi.org/10.1111/j.1600-0854.2012.01336.x
Cranfill PJ, Sell BR, Baird MA et al (2016) Quantitative assessment of fluorescent proteins. Nat Methods 13:557–562. https://doi.org/10.1038/nmeth.3891
Cui L-L, Lu Y-S, Li Y et al (2016) Overexpression of glycolate oxidase confers improved photosynthesis under high light and high temperature in rice. Front Plant Sci 7:1165. https://doi.org/10.3389/fpls.2016.01165
Eisenhut M, Roell MS, Weber APM (2019) Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol 223:1762–1769. https://doi.org/10.1111/nph.15872
Hagemann M, Bauwe H (2016) Photorespiration and the potential to improve photosynthesis. Curr Opin Chem Biol 35:109–116. https://doi.org/10.1016/j.cbpa.2016.09.014
Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798. https://doi.org/10.1016/S1097-2765(02)00496-3
Ichikawa S, Kato S, Fujii Y et al (2022) Organellar glue: a molecular tool to artificially control chloroplast–chloroplast interactions. ACS Synth Biol. https://doi.org/10.1021/acssynbio.2c00367
Ishikawa K, Miura C, Maejima K et al (2015) Nucleocapsid protein from fig mosaic virus forms cytoplasmic agglomerates that are hauled by endoplasmic reticulum streaming. J Virol 89:480–491. https://doi.org/10.1128/JVI.02527-14
Ishikawa K, Konno R, Hirano S et al (2022) The endoplasmic reticulum membrane-bending protein RETICULON facilitates chloroplast relocation movement in Marchantia polymorpha. Plant J 111:205–216. https://doi.org/10.1111/tpj.15787
Islam MS, Niwa Y, Takagi S (2009) Light-dependent intracellular positioning of mitochondria in Arabidopsis thaliana mesophyll cells. Plant Cell Physiol 50:1032–1040. https://doi.org/10.1093/pcp/pcp054
Kodama Y, Hu C-D (2012) Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques 53:285–298. https://doi.org/10.2144/000113943
Kodama Y (2016) Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS ONE 11:e0152484. https://doi.org/10.1371/journal.pone.0152484
Kopka J, Schauer N, Krueger S et al (2005) GMD@CSB.DB: the golm metabolome database. Bioinformatics 21:1635–1638. https://doi.org/10.1093/bioinformatics/bti236
Kremers G-J, Goedhart J, van Munster EB, Gadella TWJ (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45:6570–6580. https://doi.org/10.1021/bi0516273
Kumar K, Gambhir G, Dass A et al (2020) Genetically modified crops: current status and future prospects. Planta 251:1–27. https://doi.org/10.1007/s00425-020-03372-8
Kusano M, Fukushima A, Kobayashi M et al (2007) Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J Chromatogr B Analyt Technol Biomed Life Sci 855:71–79. https://doi.org/10.1016/j.jchromb.2007.05.002
Manna P, Hung S-T, Mukherjee S et al (2018) Directed evolution of excited state lifetime and brightness in FusionRed using a microfluidic sorter. Integr Biol (camb) 10:516–526. https://doi.org/10.1039/c8ib00103k
Midorikawa K, Tateishi A, Toyooka K et al (2022) Three-dimensional nanoscale analysis of light-dependent organelle changes in Arabidopsis mesophyll cells. PNAS Nexus 2:1–25. https://doi.org/10.1093/pnasnexus/pgac225
Nakagawa T, Kurose T, Hino T et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. https://doi.org/10.1263/jbb.104.34
Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51:1126–1136. https://doi.org/10.1111/j.1365-313X.2007.03212.x
Oikawa K, Matsunaga S, Mano S et al (2015) Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat Plants 1:15035. https://doi.org/10.1038/nplants.2015.35
Osaki Y, Kodama Y (2017) Particle bombardment and subcellular protein localization analysis in the aquatic plant Egeria densa. PeerJ 5:e3779. https://doi.org/10.7717/peerj.3779
Pédelacq J-D, Cabantous S, Tran T et al (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. https://doi.org/10.1038/nbt1172
Saito C, Uemura T, Awai C et al (2011) Qualitative difference between “bulb” membranes and other vacuolar membranes. Plant Signal Behav 6:1914–1917. https://doi.org/10.4161/psb.6.12.18061
Segami S, Makino S, Miyake A et al (2014) Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26:3416–3434. https://doi.org/10.1105/tpc.114.127571
Shaner NC, Campbell RE, Steinbach PA et al (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. https://doi.org/10.1038/nbt1037
Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2:905–909. https://doi.org/10.1038/nmeth819
Timm S, Hagemann M (2020) Photorespiration-how is it regulated and how does it regulate overall plant metabolism? J Exp Bot 71:3955–3965. https://doi.org/10.1093/jxb/eraa183
Wada M (2013) Chloroplast movement. Plant Sci 210:177–182. https://doi.org/10.1016/j.plantsci.2013.05.016
Wingler A, Quick WP, Bungard RA et al (1999) The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant, Cell Environ 22:361–373. https://doi.org/10.1046/j.1365-3040.1999.00410.x
Acknowledgements
We thank Ms. Hitomi Takahashi (Utsunomiya University) and Ms. Miho Kitamura (Utsunomiya University) for technical assistance; Dr. Tsuyoshi Nakagawa (Shimane University) for providing vectors (pGWB508 and pGWB602).
Funding
This work was supported by the Japan Science and Technology Agency Exploratory Research for Advanced Technology program (JST-ERATO; Grant No. JPMJER1602) and MEXT KAKENHI (Grant No. 20H05905).
Author information
Authors and Affiliations
Contributions
KI, KN, and YK conceived the research plans and performed most of the experiments including data analysis; MK, MK, and KI performed metabolome analysis; KN and YK supervised the experiments; KI and YK interpreted the results and wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Li Tian.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ishikawa, K., Kobayashi, M., Kusano, M. et al. Using the organelle glue technique to engineer the plant cell metabolome. Plant Cell Rep 42, 599–607 (2023). https://doi.org/10.1007/s00299-023-02982-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-023-02982-2