Abstract
To further understand the regulatory mechanism for anther dehiscence in rice, we carried out transcriptome analysis for the following two tissues: the anther wall and pollen at the anthesis stage. With the anatomical meta-expression data, in addition to these tissues, the differentially expressed genes (DEGs) between the two tissues were further refined to identify 1,717 pollen-preferred genes and 534 anther wall-preferred genes. A GUS transgenic line and RT-qPCR analysis for anther wall-preferred genes supported the fidelity of our gene candidates for further analysis. The refined DEGs were functionally classified through Gene Ontology (GO) enrichment and MapMan analyses. Through the analysis of cis-acting elements and alternative splicing variants, we also suggest the feature of regulatory sequences in promoter regions for anther wall-preferred expression and provide information of the unique splicing variants in anther wall. Subsequently, it was found that hormone signaling and the resulting transcriptional regulation pathways may play an important role in anther dehiscence and anther wall development. Our results could provide useful insights into future research to broaden the molecular mechanism of anther dehiscence or anther wall development in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is an important staple crop consumed by more than half of human population (Muthayya et al. 2014). Because of rapid population growth and climate change, improving crop production is of utmost importance. Owing to its importance for pollen maturation and sensitivity to climate change, the anther, a well-characterized male reproductive organ, is among the foci of studies related to crop yield (De Storme and Geelen 2014). Most of the research on anther development has been conducted to elucidate the mechanism of pollen development, in relation to the sporophyte tissue, tapetum, or intrinsic control mechanism of pollen as it develops from pollen mother cells to mature pollen (Zhang et al. 2008). In addition, many studies on anther development have focused on pollen development related to the anther wall, especially the tapetum, the innermost layer of the anther wall. However, molecular understanding of anther wall development has been very limited.
Anther dehiscence is the final stage of pollen release and is an important stage in plant reproduction and fertilization (Wilson et al. 2011). This process consists of several biological processes, where the signals are regulated by various factors, such as abiotic stress and water status (Xie et al. 2019). The importance of the anther wall was identified through studies in which anther wall dehiscence did not occur among rice mutants exhibiting the male sterility phenotype (Steiner-Lange et al. 2003; Song et al. 2018). So far, it has been found that the plant hormone auxin plays an important role not only in flower development but also in anther dehiscence. During rice anthesis, OsFTIP7 regulates the timing of auxin-mediated anther dehiscence (Song et al. 2018). In addition, AUXIN RESPONSE FACTOR17 (OsARF17) directly regulates the MYB108-encoding transcription factor for anther dehiscence (Xu et al. 2019). miR167 negatively regulates ARF6 and ARF8 (Zheng et al. 2019b). If miR167 regulation of anther growth is hindered, anther dehiscence is delayed by prolonged desiccation. Research on rice dehiscence has been conducted, but the identification of an effective gene candidate or the related biochemical pathways have not yet been reported. To date, the transcriptomes for anthers and pollens, according to the developmental stages, have been produced by various groups, and web-based expression analysis tools using the transcriptome data have been developed, thus establishing the foundation for accelerating related research. Among them, RiceAntherNet (https://www.cpib.ac.uk/anther/riceindex.html) (Lin et al. 2017) and Rice Male Gamete Expression Database (RMEDB, http://ricephylogenomics-khu.org/RMEDB/home.php) (Chandran et al. 2020) are representative examples of these studies. However, there have been no reports on transcriptome analysis of the anther wall during the flowering stage.
In this study, we carried out transcriptome data analysis for the following two tissues: anther walls and pollen during the anthesis process. Through transcriptome comparison between the two tissues, we identified the first 11,441 genes showing more than twofold upregulation in the anther wall compared to pollen. Among them, 534 genes that have low expression in other organs or tissues, but high expression in the anther wall, were selected for further analyses. Through Gene Ontology (GO) enrichment and MapMan analyses, biological processes, important regulatory and metabolic pathways related to the anther wall were identified. In addition, a regulatory network model for anther wall development was suggested to establish a basis for accelerating future research.
Results
Genome-wide identification of anther wall-preferred genes in rice
To investigate global gene candidates associated with the anther wall of rice, pollen and anther walls were collected from spikelets during pollination. In total, 17,525 differently expressed genes (DEGs) between the two tissues were identified through RNA-Seq analysis. With the conditions of p-value < 0.05 and log2FC > 1 or < − 1, 11,441 genes showed higher expression in the anther wall than in pollen, and 6,084 genes were measured to have higher expression in pollen than in the anther wall (Fig. 1a). These expression data were visualized using a volcano plot (Fig. 1b). Tissue-preferred genes were selected from pollen and anther wall among DEG to analyze transcriptional regulation mechanisms in tissues, excluding genes involved in other processes or housekeeping genes. For this purpose, raw data of the callus, shoot, leaf, root and developing seeds were collected from NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) (Table S1). These data, along with the pollen and anther wall data from our study, were normalized as described (Hong et al. 2020). We then carried out K-means clustering (KMC) analysis for 12 groups, which was the optimal number for KMC analysis as determined by the gap statistic method (Tibshirani et al. 2001). Subsequently, we identified 534 anther wall-preferred genes and 1,717 pollen-preferred genes (Fig. 1c). Heatmap data of refined pollen-preferred and anther wall-preferred genes are depicted in Fig. 1d.
Pollen-preferred and anther wall-preferred genes were identified from DEGs. a Number of DEGs between pollen and the anther wall. DEGs were selected under the conditions of log2FC(pollen/anther wall) > 1 or < −1, and p-value < 0.05. b Volcano plot of DEGs between pollen and the anther wall. Upregulated genes in pollen are indicated by red dots, and downregulated genes in pollen are indicated by blue dots (i.e., upregulated genes in the anther wall). c The clusters resulted from K-means clustering are shown. The 12 panels above are clusters of the anther wall preferred gene, and the 12 panels below is clusters of the pollen preferred gene. The number of genes lying in each cluster is mentioned. Multiple black lines represent each gene showing its expression values in six tissues: shoot, root, seed, leaf, pollen, and anther wall. In each cluster, the pink line represents the mean of expression values of all the genes. d Log2 normalized read counts were used for heat map generation. Blue indicates a low level of the log2 normalized read count, and yellow indicates a high level. Bar in flower image = 1 mm; bar in anther image = 500 µm; bar in pollen image = 200 µm
Validation of anther wall-preferred genes via RT-qPCR analysis and histochemical assay
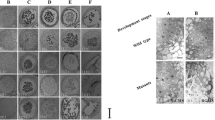
To verify the anther wall-preferred expression identified in our transcriptome analysis, we performed RT-qPCR analysis, and used a promoter trap system. First, we determined the relative expression levels of the six selected anther wall-preferred (AW) genes using RT-qPCR in six tissues: shoot, root, seed, leaf, pollen, and anther wall. The six AW genes (AW12/LOC_Os09g20940, AW14/LOC_Os08g38370, AW28/LOC_Os08g34900, AW29/LOC_Os03g30150, AW53/LOC_Os03g60210, and AW60/LOC_Os01g51550) showed high anther wall expression but were not significantly expressed in other tissues (Fig. 2a). In addition, the AW genes were more precisely verified using the promoter-GUS transgenic line. We have previously described this promoter trap technique using T-DNA that carries the promoter-less GUS reporter gene in japonica rice (Jeon et al. 2000). In this study, we identified a promoter trap line (2A-20288) for the AW19 gene (LOC_Os07g10700) and confirmed that the line contained a T-DNA insertion in the fourth exon of the AW19 gene (Fig. 2b). In addition, all lines with the T-DNA band showed GUS expression in the anther wall through co-segregation analysis of T-DNA insertion and GUS expression (Fig. 2c). RT-qPCR results showed that the AW19 gene was preferentially expressed in the anther wall (Fig. 2d) similar to the GUS expression analyses in seedlings and flowers. GUS signal was displayed in the anther wall and was not detected in other tissues (Fig. 2e). Hence, these results enhance the reliability of our transcriptional data for diverse analyses.
Validation of anther wall-preferred genes with RT-qPCR and histochemical assay. a Expression of anther wall-preferred AW genes in Oryza sativa analyzed via RT-qPCR in six tissues of rice: shoot, Root, Seed, Leaf, Pollen, and Anther wall. Rice ubiquitin 1 (OsUbi1, LOC_Os03g13170) was used as an internal control. The y-axis shows the expression level relative to OsUbi1, while the x-axis shows the samples used for analyses. Error bars represent the standard errors of three biological replicates. Significant differences are indicated by asterisks; **p < 0.01 and ***p < 0.001. Data were analyzed by employing one-way analysis of variance with repeated measures using Tukey’s pairwise comparison test. b Schematic diagram of the T-DNA line of the AW19 gene with GUS. c Polymerase chain reaction (PCR) analysis for genotyping using two primers sets (F/R and R/N). Genotyping results showed co-segregation of T-DNA insertion and GUS expression. d RT-qPCR for the AW19 gene in six tissues: shoot, root, seed, leaf, pollen, and anther wall. Rice ubiquitin 1 (OsUbi1, LOC_Os03g13170) was used as an internal control. The y-axis shows the expression level relative to OsUbi1, and the x-axis shows the samples used for the analyses. Error bars represent the standard errors of three biological replicates. Significant differences are indicated by asterisks; *p < 0.01. e Expression pattern of AW19-GUS transgenic rice plants at seedling and flower stages. Bar in seedling image = 1 cm; bar in anther image = 200 µm; bar in flower image = 1 mm
Evaluation of the functional significance for AW genes using literature searches
To search for effective candidate genes and biochemical pathways for anther dehiscence or anther wall development, genes with a known function that are preferentially expressed on the anther wall were identified through the funRiceGenes website (https://funricegenes.github.io/) (Yao et al. 2018). In Table 1, there are 12 genes that were functionally characterized. Of them, four (OSWRKY42, OSDREB1F, OsERF83, and SERF1) are transcription factors, two are transporters (OsBOR4 (Tanaka et al. 2013) and OsKAT1 (Obata et al. 2007)), one is a kinase (OsHXK1), and all are involved in signaling and transcriptional regulation pathways. Of them, OsHXK1 is the only one involved in the initiation of tapetal PCD, whereas most others are related to ROS regulation, abiotic stress response, and biotic stress response (Zheng et al. 2019a). Of the remaining genes, two (DPW and XYH) are related to anatomical structure morphogenesis, one (HTH1) is related to metabolism, one (ODCb (Quinet et al. 2010)) is related to biosynthesis, and one (OsFBX322 (Hwang et al. 2020)) is related to protein degradation. Of these, DPW (Shi et al. 2011a) and HTH1 (Xu et al. 2017) are involved in regulating male sterility. On the basis of the functional characterization data of AW genes, the functions of three genes (OsHXK1 (Zheng et al. 2019a), DPW, and XYH (Sharma et al. 2012)) are involved in anther wall development and regulation of male sterility. Thus, we expect that AW genes aside from the three known genes mentioned above can be useful candidates for elucidating anther dehiscence and anther wall development.
Functional classification of anther wall- and pollen-preferred genes using GO enrichment and MapMan analyses
To explore the biological function of the AW genes in rice, GO enrichment for AW genes was performed with a hypergeometric p-value < 0.05, query number > 2, and fold enrichment value > 2. The top 10 GO terms were identified, and the results were compared with those of pollen-preferred genes (Fig. 3).
Gene Ontology (GO) enrichment showing the top 10 GO terms for the biological process category of two groups of genes: pollen-preferred and anther wall-preferred genes. GO terms were selected under twofold enrichment value and < 0.05 hyper p-value. The color of the dots depicts the fold enrichment value: red indicates more enriched GO terms, and blue indicates less enriched GO terms. The size signifies the statistical significance presented as the −log10 (p-value)
The GO terms enriched in the AW genes are as follows: the GO fold enrichment value of the stimulus response (GO:0050896) was 12.1; auxin-mediated signaling pathway (GO:0009734), 4.9; lipid metabolic process (GO:0006629), 4.6; response to oxidative stress (GO:0006979), 2.8; transcription (GO:0006350), 2.4-fold; regulation of transcription (GO:0045449), 2.3; and regulation of transcription, DNA-dependent (GO:0006355), 2.1. By contrast, the GO terms enriched in the pollen-preferred gene were as follows: the GO fold enrichment value of the biological process (GO:0008150) was 13.1; cell wall modification (GO:0042545), 7.5; actin cytoskeleton organization (GO:0030036) 5.9; one-carbon metabolic process (GO:0006730), 5.2; sexual reproduction (GO:0019953), 5; cation transport (GO:0006812), 4.8; cellulose biosynthetic process (GO:0030244), 4.3; cell wall organization (GO:0007047), 3.7; potassium ion transport (GO:0006813), 2.4; and protein homooligomerization (GO:0051260), 2.4.
In addition, MapMan analysis was performed to functionally classify and visualize the pollen- and AW genes. A total of 1,717 pollen-preferred genes and 534 wall-preferred genes were uploaded to the regulation (Fig. 4a), biotic stress (Fig. 4b), receptor-like kinases (Fig. 4c), and transcription (Fig. 4d) overviews of the MapMan toolkit. MapMan data could support the previous report claiming that receptor-like kinases (RLKs) and calcium signaling play an important role in pollen tube growth and germination (Guo et al. 2009; Steinhorst et al. 2013). Furthermore, ethylene responsive factor (ERF)-related genes, acting as major repressors under various stress conditions, and hormones, including auxin, ethylene, and cytokinin-related genes, were identified in the AW genes. Interestingly, the GO term of the auxin-mediated signaling pathway was identified in the anther wall preferred genes. In the MapMan regulation overview, the identification of the genes involved in IAA metabolism also supports a close relationship between auxin and anther wall development. These results suggest that the anther wall might be associated with the auxin response, ERF-mediated abiotic stress response, and signaling pathways mediated by RLKs and calcium.
Analysis of the cis-acting element of promoter regions and functional analysis of regulatory factors
Cis-acting element analysis can help us understand the transcriptional regulation of DEGs between the anther wall and pollen. In addition, the mechanism of transcriptional regulation on anther dehiscence might be predicted by analyzing transcription factors interacting with these elements.
The Multiple Expectation–maximization for Motif Elicitation (MEME) suite was used to detect conserved motifs that might affect gene regulation in the 2 kb upstream promoter regions of the DEGs. We compared the promoter regions of the two groups (pollen-preferred genes and AW genes) and subsequently identified five statistically significant cis-acting regulatory element sequences (CREs) in each group. Next, the CREs was sent to TomTom to find transcription factors that can bind to known CREs in the Arabidopsis genome. In the case of pollen, transcription factors involved in gibberellin biosynthesis (FUS3, DDF1, and DDF2) and circadian rhythm (CDF5, LHY1, RVE2, RVE5, RVE6, RVE8, AGL25, LCL5, and CBF2) were identified. In the case of the anther wall, transcription factors involved in ethylene signaling, auxin response, and brasinosteroid biosynthesis were identified (Fig. 5a).
Cis-acting element analysis. a Three motifs that conserved in each of the two groups (pollen- and anther wall-preferred genes) using Multiple Expectation maximization for Motif Elicitation (MEME). Position-specific frequency matrices for the motifs identified by MEME were submitted to TomTom (Arabidopsis database) to identify matching transcription factors. The most significant transcription factors for each motif are shown. b Gene Ontology (GO) enrichment showing the top 10 GO terms for the two groups of transcription factors found in a. GO terms were selected under the fold enrichment value using TAIR (https://www.arabidopsis.org/tools/go_term_enrichment.jsp)
To explore the biological function of the transcription factors identified in the above analysis, the TAIR bulk GO annotation retrieval tool was used to analyze the top five GO terms for the transcription factors found in each of the two groups (Fig. 5b). GO analyses of transcription factors for the regulation of the pollen-preferred group revealed strong enrichment for GO terms related to the gibberellin biosynthetic process (100; GO:0010371), regulation of circadian rhythm (22.94; GO:0042752), circadian rhythm (13.65; GO:0007623), and rhythmic process (13.65; GO:0048511). By contrast, in the case of transcription factors for the regulation of the AW group, we identified strong enrichment for GO terms related to the ethylene-activated signaling pathway (145.45; GO:0009873), cellular response to ethylene stimulus (123.07; GO:0071369), response to ethylene (67.21; GO:0009723), phosphorelay signal transduction system (96.25; GO:0000160), and hormone-mediated signaling pathway (22.27; GO:0009755). This result indicated that ethylene response factors, phosphor-relay signaling, and hormone-mediated signaling may play an important role in the transcriptional regulation of anther wall development, and pollen is transcriptionally controlled by GA and circadian rhythm.
Difference in shape of alternative splicing forms of pollen and the anther wall
To better understand the transcript dynamics in the anther wall dehiscence process, we analyzed the difference in the alternative splicing forms of pollen and the anther wall using rMATS (Shen et al. 2012). Differential splicing analysis between pollen and anther wall RNA detected 355 significant alternative splice events in 283 genes during anther wall development when compared to pollen development. To explore the biological function of these genes with alternative splicing events in rice, GO enrichment was performed under hyper p-value < 0.05, FDR < 0.05, and IncLevelDifference > 0.1 or < − 0.1, and the top 10 GO terms were identified (Fig. 6a). The highest enriched GO term in genes with alternative splicing events is the phospholipid biosynthetic process (30.48; GO:0008654). To test the fidelity of the alternative splicing events detected, we randomly selected two known genes to confirm their differential expression between pollen and the anther wall by semi RT-qPCR. Sashimi plot analysis revealed that OsGI (LOC_Os01g08700) and OsRMT1 (LOC_Os04g51400) have different splicing forms by intron intention usage between pollen and anther walls (Fig. 6b). We designed primers for semi RT-qPCR for the regions marked with arrows in Fig. 6c. As a result, as in gDNA, long bands with intron intention were identified in the anther wall, and in the case of pollen, only short bands by spliced intron were identified (Fig. 6d). These results indicate that there might be different posttranscriptional regulation patterns between the anther wall and pollen tissue, and this difference in alternative splicing forms might provide new clues to understand the anther dehiscence process or anther wall development in rice.
Differential splicing analysis in pollen and the anther wall of rice. a Gene Ontology (GO) enrichment showing the top 10 GO terms for alternatively spliced genes. b Representative images of Sashimi plots depicting alternative splicing patterns in the anther wall (red) and pollen (yellow). Read counts for each sample are indicated on the y-axis. c Gene structure of the region where splicing occurs. The arrow indicates the primer position. d Gel images of semi RT-qPCR products for the OsGI gene and OsRMT1 gene. Actin was used as the cDNA and gDNA marker
Discussion
In this study, we explored the regulatory mechanisms in anther development and its dehiscence through the comparative analysis of the transcriptome between two tissues, namely, anther wall, and pollen. We elucidated 1,717 pollen- and 534 AW DEGs and performed validation and functional enrichment of these genes. Finally, we revealed a complex regulatory mechanism in anther development and its dehiscence in the context of hormone signaling, transcriptional regulations related to abiotic stress, and lipid metabolism.
Previous studies revealed that these hormones are involved in anther dehiscence. For example, auxin controls the timing of anther dehiscence. In the auxin-perception mutants afb1-3 and triple mutant tir1afb2afb3, anther dehiscence occurs earlier than that in wild-type Arabidopsis (Cecchetti et al. 2013). OsFTIP7 regulates the timing of anther dehiscence by mediating auxin (Song et al. 2018). Ethylene is also important in anther wall dehiscence as ethylene insensitivity caused the delay in anther dehiscence in tobacco (Rieu et al. 2003). The importance of ethylene perception in anther dehiscence has also been demonstrated in a loss-of-function mutant study for the ethylene receptor gene. The antisense suppression of PhETR2, encoding an ethylene receptor in petunia, led to anther dehiscence before anthesis, indicating that PhETR2 regulates the timing of anther dehiscence (Wang and Kumar 2007). Jasmonic acid (JA) is also reported to be a hormone that affects anther dehiscence. The Defective in Anther Dehiscence (DAD1) gene encodes the chloroplastic phospholipase A1 that catalyzes the initial step of JA biosynthesis. Mutation of DAD1 resulted in anther dehiscence defects (Ishiguro et al. 2001). Cytokinin (CK) regulation plays an important role in the anther dehiscence of rice. Arabidopsis histidine-containing phosphotransferase proteins, which are mediators in CK signaling, negatively modulated the thickening of the secondary cell wall of the anther endothecium (Jung et al. 2008). MapMan analysis revealed that 23 AW genes were identified in the category of hormone signaling and metabolism: eight for auxin, eight for ethylene, two for JA, and two for CK (Table S5). Furthermore, the results from the KEGG enrichment analysis of the AW genes supported the relationship between hormones and anther dehiscence (Fig. S1). As anther dehiscence relies on the normal development of the anther and complex mechanisms, such as anther wall decay and various hormones, further studies are needed to unveil how the AW genes are involved in hormone signaling and metabolism during anther development and its dehiscence.
Through the GO and MapMan analyses, transcriptional regulation related to stress response was shown to play an important role in anther wall development. Among the AW genes in our study, four genes were identified as transcription factors involved in the abiotic stress response (Table 1). OsWRKY42 and SERF1 were shown to be involved in the salt stress response (Schmidt et al. 2014; Han et al. 2014). The AP2/ERF family of transcription factors, of which SERF1 is a member, is involved in gene regulation in response to abiotic stresses in plants, especially the DREB subfamily (Lata and Prasad 2011). DREB1F, a member of the DREB subfamily, is also reported to increase tolerance to salt, drought, and low temperatures (Wang et al. 2008). Finally, OsERF83 is also involved in the response to drought stress. As there are various studies showing the importance of abiotic stresses and its transcriptional regulation in the anther dehiscence process (Sakata and Higashitani 2008; Jagadish et al. 2010; Zinn et al. 2010; Jung et al. 2021), the two members of the AP2/ERF family, namely, DREB1F and SERF1, could be interesting targets for interpreting a crosslink between anther dehiscence and abiotic stress during the anther maturation process.
Through cis-acting element enrichment analysis of AW genes, it is proposed that the regulation of hormone and phosphorelay signaling have a major role in anther development and dehiscence (Fig. 5). This result is consistent with previous reports that signal integration in abiotic stress occurs by phosphorelay signaling and several hormones function as key regulators of stress tolerance (Zwack and Rashotte 2015; Skalak et al. 2021). In particular, the expression of AW genes might be controlled by many ERF transcription factors. Motifs preserved in the promoter region of the AW gene matched the binding motif of AP2/ERF transcription factors, such as DEAR2, PUCHI, and ERFs, and 56 of the 67 transcription factors identified belong to the AP2/ERF Family (Fig. 5a, Table S6). The AP2/ERF family of transcription factors have been reported as key regulators in various stress responses, in which they also respond to hormones to improve plant survival during stress conditions (Xie et al. 2019). Therefore, it can be speculated that the AP2/ERF transcription factors and related pathways have important roles in anther dehiscence or anther wall development. In addition, the AP2/ERF transcription factors and various hormone signaling factors identified in this study can be major targets for studying mechanisms for anther wall dehiscence and development during abiotic stress.
In the anther wall, lipid metabolism is the most common functional group determined by both MapMan and GO enrichment analyses. Lipid metabolism, mainly lipid biosynthesis and lipid transport, are important for reproductive development, such as anther development and dehiscence in plants (Xiao et al. 2014). In particular, lipids are primarily synthesized in a tapetum cell, which constitutes the innermost layer of the anther wall and transfers to pollen or other anther wall layers to promote anther development (Wan et al. 2020). The DPW gene selected as an anther wall preferential gene with a known function (Table 1) was reported to participate in the primary fatty alcohol synthesis step during anther development and mutations in DPW display defective anther development and male sterility (Shi et al. 2011b). Furthermore, through alternative splicing analysis, we suggest that many genes involved in the phospholipid biosynthetic process have different alternative splicing forms between the anther wall and pollen tissue (Fig. 6a). In previous reports, alternative splicing events are involved in the secondary metabolism of tea plants (Qiao et al. 2019). In addition, the Lipin1 gene, which regulates phospholipid synthesis, plays a role in maintaining lipid metabolism homeostasis by regulating phospholipid levels through alternative splicing in liver cells (Wang et al. 2021). However, the mechanism of lipid metabolism or biosynthesis in rice anther development is largely unknown, and lipid level control by alternative splicing has not yet been reported in plants. Although we still need to further confirm the activity and functionality of alternative splicing isoforms between the anther wall and pollen, we assume that the alternative splicing of genes involved in lipid biosynthesis may play an important role in maintaining lipid metabolism during anther development.
In this study, we prepared a working model in anther development and dehiscence (Fig. 7). This model emphasizes two important transcriptional regulatory mechanisms in the rice anther wall. We believe that the suggested model could prove useful for the future examination of the anther wall development and dehiscence mechanisms.

modified from KEGG (Fig. S1). Lipid metabolism control by alternative splicing might also be associated with this process
A summary model of the anther dehiscence process. Hormone signaling related to the abiotic stress response occurs in anther development and dehiscence. Blue boxes show the mapped anther wall-preferred DEGs in plant hormone signaling pathways, as
Methods
Plant materials and growth conditions
Japonica rice (Oryza sativa spp. japonica) variety, Dongjin, was cultivated as the wild-type plant under a day/night cycle of 16 h/8 h at 28 °C/22 °C with approximately 80% humidity for two weeks in a growth chamber and was transplanted to a living modified organism-controlled paddy field (Kyung Hee University, Yongin, South Korea). Anther wall and pollen samples were collected from the paddy field using the RNAlater stabilizing solution (Invitrogen) at the anthesis stage.
Transcriptome analysis and identification of DEGs
Anthers were collected during anthesis and vortexed vigorously to separate pollen from the anther wall. The RNA was extracted using RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocols and passed a quality control process. Then, sequencing libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Part #15031047 Rev. E) and sequenced by Macrogen, Inc., with the Illumina NovaSeq 6000 platform.
FASTQ files were cleaned using Cutadapt software (v2.3 (Martin 2011)) and mapped to a reference rice genome, MSU7, using HISAT2 aligner (v2.1.0 (Kim et al. 2019)). The read counts were extracted with FeatureCount (v1.6.3 (Liao et al. 2014)), and DEGs were statistically tested using DESeq2 R package (v1.30.1 (Love et al. 2014)). The DEGs between pollen and the anther wall were selected with > 1 or < − 1 log2 fold-change values and p-value < 0.05. For the selected DEGs, the log2 fold-change data were visualized using a volcano plot. The raw data were deposited to the ArrayExpress repository (accession number: E-MTAB-7974 for pollen and E-MTAB-11005 for anther wall).
Heatmap analysis
The Multiple Experiment Viewer (MeV) program was used for the statistical analysis and visualization of the primary selected DEGs of pollen and the anther wall, which resulted in the identification of 6,084 pollen-preferred genes and 11,441 wall-preferred genes. K-means clustering (KMC) analysis was performed for genes using the Euclidean distance algorithm. Subsequently, 1,717 genes preferred by pollen and 534 genes preferred by the anther wall were selected among 12 clusters, which was set based on the gap statistic method (Tibshirani et al. 2001). Heatmap images for pollen-preferred and AW genes were generated using the MeV program. Detail information about public transcriptome data is in Supplementary Table 1.
Functional classification using GO and MapMan analyses
GO terms were downloaded from ROAD (http://ricephylogenomics.khu.org/ROAD_old/analysis/go_enrichment.shtml) to characterize the functions of transcripts belonging to the selected gene list (Moon et al. 2020). In total, 1,717 pollen-preferred genes and 534 AW genes were uploaded, and GO terms with hyper p-value < 0.05, fold enrichment > 2, and a query number > 2 were selected. The top 10 GO terms based on fold enrichment were used for further analysis. A plot was generated to visualize the data using the ggplot2 R package (v3.3.5).
MapMan analysis was performed to visualize the biological processes and pathways for the 1,717 pollen-preferred genes and 534 AW genes as previously described (Zhang et al. 2020). We uploaded the locus ID of these genes to the MapMan toolkit and analyzed their regulation, metabolism, cell function, and biotic stress overviews.
Alternative splicing analysis
The RNA-Seq data of pollen and the anther wall were analyzed for splicing alterations. To identify such events, replicate Multivariate Analysis of Transcript Splicing (rMATS v4.1.0) was used to determine junctional reads (Shen et al. 2012). Differential alternative spliced genes with hyper p-value < 0.05, FDR < 0.05, and |IncLevelDifference|> 0.1 were selected and visualized with rmats2sashimiplot (v 2.0.2). To validate the splicing alterations, semi RT-qPCR analysis was performed by designing primers of two loci in the neighboring exons where splicing was predicted (Table S4).
Histochemical assay for AW genes
We generated more than 100,000 potential promoter trap lines that retain the promoter-less GUS gene near the right border of T-DNA (Jeon et al. 2000). We screened the 60 AW genes in Table S2. A promoter trap line (2A-20288) for the AW19 gene (LOC_Os07g10700) was identified through co-segregation analysis of T-DNA insertion and GUS expression. The primers used for T-DNA genotyping were 5′-TGCTACCTCCCTTGACAGTG-3′ (206 bp downstream from the ATG start codon of the AW19 gene), 5′-AGTTTCTGACATCTAAGCGTTG-3′ (1536 bp downstream from the ATG start codon of AW19 gene), and 5′-AACGCCTGATCAATTCCACAG-3′ (80 bp downstream from the ATG start codon of the GUS gene). Histochemical GUS-staining was performed as described previously (Moon et al. 2019). The stained flowers and pollen grains were photographed with a BX61 microscope (Olympus).
RT-qPCR analysis
Tissue samples, including pollen and anther wall (Oryza sativa cv. Dongjin), grown in paddy fields were frozen in liquid nitrogen and homogenized with TissueLyser II (Qiagen). Total RNA was extracted using RNAiso Plus according to the manufacturer’s protocol (Takara). cDNA was synthesized using a SuPrimeScript RT premix from GeNet Bio (Vo et al. 2015). RT-qPCR was performed with a Rotor-Gene Q RT-qPCR cycler (Qiagen) using the following thermal cycling procedure: 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 1 min. To evaluate tissue-preferred expression patterns by RT-qPCR, we used rice ubiquitin 1 (OsUbi1, LOC_Os03g13170) as an endogenous control to normalize the variance in the amount of sample cDNA (Morgante et al. 2011). Gene-specific RT-qPCR primers were designed for a specific region of each gene (Table S3), and the accuracy and efficiency of each primer set were verified through PCR amplification of the gDNA to optimize the PCR conditions and melting curve. The fold-change was calculated using the comparative 2−∆∆Ct method (Pfaffl 2001). Three biological replicates were analyzed, and each reaction was performed at least in triplicate.
Cis-acting element analysis
To search the candidate regions for cis-acting regulatory elements (CREs), we used the motif finding program MEME (v5.3.3) (Bailey et al. 2009). Cis-acting element analysis was performed by comparing the 2 kb upstream region of 1,717 pollen-preferred genes and 534 AW genes. We searched for three sequences for each group with a minimal width of 4 bp and maximal width of 10 bp using the other as control groups. These sequences were further analyzed using the TomTom tool, which enables comparison with databases of known CREs (Arabidopsis transcription factor families) (Gupta et al. 2007).
Literature search
Functionally characterized genes were retrieved from the funRiceGenes database to identify functions of anther wall preferred genes (Yao et al. 2018). The result is shown in Table S5.
References
Bailey TL, Boden M, Buske FA et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208. https://doi.org/10.1093/nar/gkp335
Cecchetti V, Altamura MM, Brunetti P et al (2013) Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J 74:411–422. https://doi.org/10.1111/tpj.12130
Chandran AKN, Hong W-J, Abhijith B et al (2020) Rice male gamete expression database (RMEDB): a web resource for functional genomic studies of rice male organ development. J Plant Biol 63:421–430. https://doi.org/10.1007/s12374-020-09267-1
De Storme N, Geelen D (2014) The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant Cell Environ 37:1–18. https://doi.org/10.1111/pce.12142
Guo H, Li L, Ye H et al (2009) Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. PNAS 106:7648–7653. https://doi.org/10.1073/pnas.0812346106
Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8:R24. https://doi.org/10.1186/gb-2007-8-2-r24
Han M, Kim C-Y, Lee J et al (2014) OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol Cells 37:532–539. https://doi.org/10.14348/molcells.2014.0128
Hong W-J, Kim Y-J, Kim E-J et al (2020) CAFRI-rice: CRISPR applicable functional redundancy inspector to accelerate functional genomics in rice. Plant J 104:532–545. https://doi.org/10.1111/tpj.14926
Hwang JE, Hwang S-G, Jung IJ et al (2020) Overexpression of rice F-box protein OsFBX322 confers increased sensitivity to gamma irradiation in Arabidopsis. Genet Mol Biol 43:e20180273. https://doi.org/10.1590/1678-4685-GMB-2018-0273
Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13:2191–2209. https://doi.org/10.1105/tpc.010192
Jagadish SVK, Muthurajan R, Oane R et al (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61:143–156. https://doi.org/10.1093/jxb/erp289
Jeon JS, Lee S, Jung KH et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570. https://doi.org/10.1046/j.1365-313x.2000.00767.x
Jung K, Oh S-I, Kim Y et al (2008) Arabidopsis histidine-containing phosphotransfer factor 4 (AHP4) negatively regulates secondary wall thickening of theanther endothecium during flowering. Mol Cells 25:294–300
Jung SE, Bang SW, Kim SH et al (2021) Overexpression of OsERF83, a vascular tissue-specific transcription factor gene, confers drought tolerance in rice. Int J Mol Sci 22:7656. https://doi.org/10.3390/ijms22147656
Kim D, Paggi JM, Park C et al (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. https://doi.org/10.1038/s41587-019-0201-4
Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62:4731–4748. https://doi.org/10.1093/jxb/err210
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Lin H, Yu J, Pearce SP et al (2017) RiceAntherNet: a gene co-expression network for identifying anther and pollen development genes. Plant J 92:1076–1091. https://doi.org/10.1111/tpj.13744
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Moon S, Chandran A, Kim Y-J et al (2019) Rice RHC encoding a putative cellulase is essential for normal root hair elongation. J Plant Biol 62:82–91. https://doi.org/10.1007/s12374-018-0393-z
Moon S, Hong W-J, Kim Y-J et al (2020) Comparative transcriptome analysis reveals gene regulatory mechanism of UDT1 on anther development. J Plant Biol 63:289–296. https://doi.org/10.1007/s12374-020-09250-w
Morgante CV, Guimarães PM, Martins AC et al (2011) Reference genes for quantitative reverse transcription-polymerase chain reaction expression studies in wild and cultivated peanut. BMC Res Notes 4:339. https://doi.org/10.1186/1756-0500-4-339
Muthayya S, Sugimoto JD, Montgomery S, Maberly GF (2014) An overview of global rice production, supply, trade, and consumption: Global rice production, consumption, and trade. Ann NY Acad Sci 1324:7–14. https://doi.org/10.1111/nyas.12540
Obata T, Kitamoto HK, Nakamura A et al (2007) Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol 144:1978–1985. https://doi.org/10.1104/pp.107.101154
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Qiao D, Yang C, Chen J et al (2019) Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant (Camellia sinensis). Sci Rep 9:2709. https://doi.org/10.1038/s41598-019-39286-z
Quinet M, Ndayiragije A, Lefèvre I et al (2010) Putrescine differently influences the effect of salt stress on polyamine metabolism and ethylene synthesis in rice cultivars differing in salt resistance. J Exp Bot 61:2719–2733. https://doi.org/10.1093/jxb/erq118
Rieu I, Wolters-Arts M, Derksen J et al (2003) Ethylene regulates the timing of anther dehiscence in tobacco. Planta 217:131–137. https://doi.org/10.1007/s00425-003-0976-9
Sakata T, Higashitani A (2008) Male sterility accompanied with abnormal anther development in plants—genes and environmental stresses with special reference to high temperature injury. Int J Plant Dev Biol 2:2
Schmidt R, Schippers JHM, Mieulet D et al (2014) SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice. Mol Plant 7:404–421. https://doi.org/10.1093/mp/sst131
Sharma R, Agarwal P, Ray S et al (2012) Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct Integr Genomics 12:229–248. https://doi.org/10.1007/s10142-012-0274-3
Shen S, Park JW, Huang J et al (2012) MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40:e61–e61. https://doi.org/10.1093/nar/gkr1291
Shi J, Tan H, Yu X-H et al (2011a) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23:2225–2246. https://doi.org/10.1105/tpc.111.087528
Skalak J, Nicolas KL, Vankova R, Hejatko J (2021) Signal integration in plant abiotic stress responses via multistep phosphorelay signaling. Front Plant Sci 12:196. https://doi.org/10.3389/fpls.2021.644823
Song S, Chen Y, Liu L et al (2018) OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nature Plants 4:495–504. https://doi.org/10.1038/s41477-018-0175-0
Steiner-Lange S, Unte US, Eckstein L et al (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34:519–528. https://doi.org/10.1046/j.1365-313X.2003.01745.x
Steinhorst L, Kudla J (2013) Calcium-a central regulator of pollen germination and tube growth. Biochim Biophys Acta Mol Cell Res 1833:1573–1581. https://doi.org/10.1016/j.bbamcr.2012.10.009
Tanaka N, Uraguchi S, Saito A et al (2013) Roles of pollen-specific boron efflux transporter, OsBOR4, in the rice fertilization process. Plant Cell Physiol 54:2011–2019. https://doi.org/10.1093/pcp/pct136
Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Ser B (stat Methodol) 63:411–423. https://doi.org/10.1111/1467-9868.00293
Vo KTX, Kim C-Y, Chandran AKN et al (2015) Molecular insights into the function of ankyrin proteins in plants. J Plant Biol 58:271–284. https://doi.org/10.1007/s12374-015-0228-0
Wan X, Wu S, Li Z et al (2020) Lipid metabolism: critical roles in male fertility and other aspects of reproductive development in plants. Mol Plant 13:955–983. https://doi.org/10.1016/j.molp.2020.05.009
Wang Y, Kumar PP (2007) Characterization of two ethylene receptors PhERS1 and PhETR2 from petunia: PhETR2 regulates timing of anther dehiscence. J Exp Bot 58:533–544
Wang Q, Guan Y, Wu Y et al (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67:589–602. https://doi.org/10.1007/s11103-008-9340-6
Wang H, Chan TW, Vashisht AA, Drew BG, Calkin AC, Harris TE, Wohlschlegel JA, Xiao X, Reue K (2021) Lipin 1 modulates mRNA splicing during fasting adaptation in liver. JCI Insight. https://doi.org/10.1172/jci.insight.150114
Wilson ZA, Song J, Taylor B, Yang C (2011) The final split: the regulation of anther dehiscence. J Exp Bot 62:1633–1649. https://doi.org/10.1093/jxb/err014
Xiao Y, Chen Y, Charnikhova T, Mulder PP, Heijmans J, Hoogenboom A, Agalou A, Michel C, Morel J, Dreni L (2014) OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol Biol 86:19–33. https://doi.org/10.1007/s11103-014-0212-y
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in arabidopsis. Front Plant Sci 10:228. https://doi.org/10.3389/fpls.2019.00228
Xu Y, Liu S, Liu Y et al (2017) HOTHEAD-like HTH1 is involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol 58:1238–1248. https://doi.org/10.1093/pcp/pcx063
Xu X-F, Wang B, Feng Y-F et al (2019) AUXIN RESPONSE FACTOR17 directly regulates MYB108 for anther dehiscence. Plant Physiol 181:645–655. https://doi.org/10.1104/pp.19.00576
Yao W, Li G, Yu Y, Ouyang Y (2018) funRiceGenes dataset for comprehensive understanding and application of rice functional genes. GigaScience. https://doi.org/10.1093/gigascience/gix119
Zhang D-S, Liang W-Q, Yuan Z et al (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1:599–610. https://doi.org/10.1093/mp/ssn028
Zhang S, Shi Y, Shan C et al (2020) Analysis of the transcriptome of Polygonatum odoratum (Mill.) druce uncovers putative genes involved in isoflavonoid biosynthesis. J Plant Biol 63:217–228. https://doi.org/10.1007/s12374-020-09246-6
Zheng S, Li J, Ma L, Wang H, Zhou H, Ni E, Jiang D, Liu Z, Zhuang C (2019a) OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc Natl Acad Sci U S A 116:7549–7558. https://doi.org/10.1073/pnas.1817675116
Zheng L, Nagpal P, Villarino G et al (2019b) miR167 limits anther growth to potentiate anther dehiscence. Development 146:174375. https://doi.org/10.1242/dev.174375
Zinn KE, Tunc-Ozdemir M, Harper JF (2010) Temperature stress and plant sexual reproduction: uncovering the weakest links. J Exp Bot 61:1959–1968. https://doi.org/10.1093/jxb/erq053
Zwack PJ, Rashotte AM (2015) Interactions between cytokinin signalling and abiotic stress responses. J Exp Bot 66:4863–4871. https://doi.org/10.1093/jxb/erv172
Funding
This work was supported by grants from the New Breeding Technologies Development Program (PJ01492703 to K.-H.J.), the Rural Development Administration, Republic of Korea, and the National Research Foundation (NRF), Ministry of Education, Science and Technology (NRF-2021M3E5E6025387 to K.-H.J.; NRF-2021R1C1C2091377 to W.-J.H.).
Author information
Authors and Affiliations
Contributions
SM, YJK, and KHJ designed this work; WJH, SKL, SHK, EJK, and JS performed the experiments; WJH, SKL, and SHK analyzed the data; SKL, SHK, and YJK generated the figures and tables; WJH, SKL, SHK, and KHJ wrote the manuscript. All authors have read and agreed to publish the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests for this research.
Additional information
Communicated by Da-Bing Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hong, WJ., Lee, S.K., Kim, SH. et al. Comparative transcriptome analysis of pollen and anther wall reveals novel insights into the regulatory mechanisms underlying anther wall development and its dehiscence in rice. Plant Cell Rep 41, 1229–1242 (2022). https://doi.org/10.1007/s00299-022-02852-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02852-3