Abstract
Key message
A sodium hydrogen exchanger (NHX) gene from the date palm enhances tolerance to salinity in Arabidopsis plants.
Abstract
Plant sodium hydrogen exchangers/antiporters (NHXs) are pivotal regulators of intracellular Na+/K+ and pH homeostasis, which is essential for salt stress adaptation. In this study, a novel orthologue of Na+/H+ antiporter was isolated from date palm (PdNHX6) and functionally characterized in mutant yeast cells and Arabidopsis plants to assess the behavior of the transgenic organisms in response to salinity. Genetically transformed yeast cells with PdNHX6 were sensitive to salt stress when compared to the empty vector (EV) yeast cells. Besides, the acidity value of the vacuoles of the transformant yeast cells has significantly (p ≤ 0.05) increased, as indicated by the calibrated fluorescence intensity measurements and the fluorescence imagining analyses. This observation supports the notion that PdNHX6 might regulate proton pumping into the vacuole, a crucial salt tolerance mechanism in the plants. Consistently, the transient overexpression and subcellular localization revealed the accumulation of PdNHX6 in the tonoplast surrounding the central vacuole of Nicotiana benthamiana leaf epidermal cells. Stable overexpression of PdNHX6 in Arabidopsis plants enhanced tolerance to salt stress and retained significantly higher chlorophyll, water contents, and increased seed germination under salinity when compared to the wild-type plants. Despite the significant increase of Na+, transgenic Arabidopsis lines maintained a balanced Na+/K+ ratio under salt stress conditions. Together, the results obtained from this study imply that PdNHX6 is involved in the salt tolerance mechanism in plants by controlling K+ and pH homeostasis of the vacuoles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinization is one of the main abiotic stresses that limits the survival and productivity of plants (Munns and Gilliham 2015). Various plant species respond differently to the saline environment, and their stress tolerance mechanisms depend on the overall genetic makeup. Therefore, plants are classified as either halophytes or glycophytes. Halophytes are salt-tolerant plants; whereas, glycophytes are salt-sensitive plants (Himabindu et al. 2016; Mishra and Tanna 2017). In glycophytes, a high cytosolic K+/Na+ ratio is essential for optimal metabolic functions under salt stress conditions (Shabala and Pottosin 2014). Hence, the K+/Na+ balance is achieved by reducing the noxious accumulation of sodium ions (Na+) and by enhancing the cellular uptake of potassium ions (K+) (Munns and Tester 2008). Non-selective cation channels and Na+–K+ transporters are the primary regulators of the intracellular Na+ and K+ homeostasis (Tester and Davenport 2003).
Plant sodium hydrogen exchangers/antiporters (NHXs) are closely related to the NHE family of mammalian Na+/K+ exchangers (Bassil et al. 2012). The NHX/NHE family has been classified into two groups based on their subcellular localization: plasma membrane and intracellular (Brett et al. 2005). Furthermore, all the plant intra-cellular NHX have been subdivided into two subgroups, namely class-I and class-II; class-I proteins localize in the vacuolar membrane, whereas class-II proteins localize in the pre-vacuolar compartments (PVC) and other non-vacuolar endosomal compartments (Jiang et al. 2010). NHX gene family has been extensively studied in different plant species under salinity stress conditions (Ford et al. 2012; Gong et al. 2014; Guan et al. 2011; Li et al. 2011; Liu et al. 2012; Mishra et al. 2014; Xu et al. 2013). The intracellular transmembrane NHXs play a vital role in pH, Na+ and K+ homeostasis (Barragán et al. 2012; Reguera et al. 2015). Based on the subcellular location, they contribute to two main salt stress tolerance strategies; vacuolar sequestration of Na+ (Bonales-Alatorre et al. 2013) and/or extrusion of cytosolic Na+ to the extracellular space (Hamam et al. 2016). Besides, the NHX proteins are involved in stomatal function, growth, and development, as well as in protein and vesicle trafficking (Barragán et al. 2012; Reguera et al. 2015).
NHX6 is a concomitant isoform of NHX5 localized in the endosomal membranes of the Golgi, trans-Golgi network (TGN), and PVC (Jiang et al. 2010; Reguera et al. 2015). These two endosomal NHXs were previously demonstrated to regulate intracellular processes of cell expansion, vacuolar protein biogenesis, trafficking, pH, and osmotic pressure regulation (Li et al. 2011; Reguera et al. 2015). Apart from this, the molecular characterization of AtNHX5 and AtNHX6 genes in nhx5 and nhx6 double mutant Arabidopsis lines enhanced the root growth, facilitated K+ transport and pH homeostasis in transgenic plants (Wang et al. 2015).
The date palm has evolved as a relatively abiotic stress-tolerant tree; hence, this plant has recently attracted considerable attention for being a good source of information regarding the salt adaptation mechanisms from physiological (Al Kharusi et al. 2017, 2019; Jana et al. 2019; Yaish 2015), and molecular aspects (Al-Harrasi et al. 2018; Patankar et al. 2016, 2018, 2019b; Yaish et al. 2015, 2017; Zaid and De Wet 1999).
As a primary investigation step, researchers apply a common strategy in which channel and transporter coding genes are functionally characterized in model plants and are consequently used to improve salinity tolerance in various crops. For example, overexpression of the Arabidopsis NHX5 (AtNHX5) enhanced salt- and drought tolerance in paper mulberry by promoting the accumulation of osmolytes, which counteracts the osmotic stress resulting from abiotic stresses (Li et al. 2011). Similarly, overexpression of the Arabidopsis AtNHX1 gene in tomato plants enhanced their tolerance to salt stress by retaining high intracellular K+ level and increasing proline and sugar accumulation in the cytosol (Leidi et al. 2010). Another report showed that overexpression of a vacuolar NHX1 gene from mung bean (VrNHX1) in Arabidopsis increased salt tolerance by increasing the K+/Na+ ratio, enhancing proline accumulation and by reducing the accumulation of malondialdehyde (MDA) (Mishra et al. 2014).
A previous RNA sequence analysis showed that PdNHX6 gene was induced in the leaf tissues of date palm seedlings when subjected to salinity (Yaish et al. 2017). Therefore, we conducted molecular and functional characterization of this gene in yeast cells and the Arabidopsis model plant to gain a better understanding of the role played by PdNHX6 in salt tolerance mechanisms. Our results showed that overexpression of PdNHX6 in a mutant yeast strain (BYT458) impaired the growth under salinity stress and led to a reduction in the vacuolar pH value. However, transgenic Arabidopsis plants overexpressing PdNHX6 showed enhanced tolerance to salt stress, which was associated with a higher chlorophyll accumulation, relative water content (RWC), and seed germination rates when compared to non-transgenic control plants. Additionally, transgenic Arabidopsis plants were able to neutralize the increased Na+ concentration under salt stress and maintained a balanced intracellular K+ level.
Materials and methods
In silico analysis of protein sequence
Deduced amino acid sequences of the NHX family of the date palm and NHX6 of other plant species were retrieved from the National Center of Biotechnology Information (NCBI) database. The retrieved sequences were aligned using Clusta1W (Feng and Doolittle 1987), and the phylogenetic trees were constructed using the Neighbor-Joining (N-J) algorithm, with 1000 bootstrap replicates, implemented within the MEGA 7 program (Kumar et al. 2016). NCBI conserved domain database (CDD) was used to assign the conserved domains to the NHX6 family of different species (Marchler-Bauer et al. 2014). The theoretical isoelectric point (pI) and molecular weight (Mw) of the PdNHX6 were predicted through the ExPASy (ProtScale) tool (Gasteiger et al. 2003). A putative promoter sequence of 2000-bp upstream PdNHX6 was analyzed for identifying the transcription factor binding sites (TFBSs) using the PlantPAN 2.0 database (https://PlantPAN2.itps.ncku.edu.tw/) (Chow et al. 2015). The distribution of TFBSs involved in abiotic stress within this region compared with other general TFBSs is represented in a pie diagram. Protein transmembrane topology and signal peptides were predicted from amino acid sequences using Protter database (https://wlab.ethz.ch/protter/start/) (Omasits et al. 2013).
Plant materials and growth conditions
Date palm (cv. Khalas) seeds were thoroughly washed with tap water, followed by surface sterilization using 70% ethanol. The seeds were further washed several times with sterile distilled water and then soaked in water overnight at 37 °C. The next day, the seeds were mixed with sterilized moist vermiculite and incubated at 37 °C in the dark for one week. The germinated seeds were transferred to 2-L pots containing peat moss and sand mixture (1:1, v/v). The plants were grown under the controlled environmental conditions of 30 °C, 16 h light/8 h dark photoperiod cycle, and watered to field capacity. The salt stress treatment was applied using a 300-mM NaCl solution, as previously described (Al Harrasi et al. 2017).
Quantitative PCR (qPCR) analysis and molecular cloning of PdNHX6 gene
Total RNA was extracted from the roots and leaves of the date palm seedlings, treated with DNAse I, and used to synthesize cDNA. The qPCR gene expression analysis of PdNHX6 was carried out using a primer pair (NHX6FA and NHX6RA) (Supplementary Table S1), as previously described [32]. Briefly, the qPCR reaction mixture contained 5-μL SsoAdvanced Universal SYBR® Green Supermix (Bio-Rad, USA), 0.1 μL of each primer (10 pmol), 2 μL of 20-fold diluted cDNA and 2.8 μL of nuclease-free water in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA) at 95 °C for 20 s followed by 40 cycles of 95 °C for 3 s, and 60 °C for 30 s. The cDNA of PdNHX6 was amplified from salt-treated root samples by PCR and the primer pair: NHX6FB and NHX6RB (Supplementary Table S1). The resultant PCR product was cloned into pGEM-T Easy vector (Promega, Madison, USA) and the DNA construct was confirmed by DNA sequencing. The positive clone was subsequently amplified using NHX6FC and NHX6RC primer pair, including the attB1 and attB2 DNA sites. The PCR product was subsequently cloned into an entry pDONR™/Zeo vector (Invitrogen, Carlsbad, USA), which was then used to introduce PdNHX6 into different expression vectors for functional analysis using gateway cloning approach (Thermo Fisher Scientific, USA).
Transient expression of PdNHX6 in Nicotiana benthamiana leaf epidermal cells
The cDNA of PdNHX6 was cloned in-frame along with the N-terminal yellow fluorescent protein (YFP), into pSITE-nEYFP-C1 expression vector (TAIR stock ID: CD3-1648). The construct was verified for in-frame fusion by DNA sequencing, and the plasmid was further amplified in E. coli cells, followed by plasmid extracted using GeneJET plasmid miniprep kit (Thermo Scientific, Germany), as per the manufacturer’ protocol.
Leaves of Nicotiana benthamiana (3–4 weeks old) were co-infiltrated with Agrobacterium harboring pSITE-nEYFP-C1:: PdNHX6 as well as a tonoplast marker plasmid, VAC-RK (TAIR stock ID: CD3-975). The epidermal cells were observed three days after infiltration under a confocal laser scanning microscope (FV3000 Olympus) with FV3000 software for image processing. The expression of PdNHX6 and the tonoplast marker were detected using YFP filter set (Ex: 514 nm/Em: 520–560 nm) and RFP filter set (Ex: 561 nm/Em: 600–650 nm), respectively.
Heterologous expression of PdNHX6 in yeast
PdNHX6 cDNA was cloned into the yeast plasmid pYES-DEST52 (Thermo Fisher Scientific, USA), under the control of galactose inducible GAL1 promoter. The function of the recombinant vector (PdNHX6) compared to the empty vector (EV) was assayed in Saccharomyces cerevisiae mutant strain (BYT458; genotype: BY4741; ena1-5Δ::loxP nha1Δ::loxP vnx1Δ::loxP) (Petrezselyova et al. 2013), which was kindly provided by Professor Hana Sychrova, Czech Republic. Both PdNHX6 and EV were introduced into the yeast cells by PEG-lithium acetate method and using Yeastmaker™ yeast transformation system 2 (Clonetech Laboratories Inc., Mountain View, California, USA) as per the manufacturer’ instructions. The genetically transformed yeast cells were selected based on the auxotrophic selectable marker URA3 gene, which enhances the growth of transformed colonies on solid synthetic medium (SSM) lacking uracil. The salt stress tolerance ability of PdNHX6 yeast cells compared to EV was tested on medium supplemented with 100 mM NaCl using yeast spot assay (Patankar et al. 2018).
Similarly, the behavior of the PdNHX6 and EV yeast cells was tested in SD liquid culture supplemented with 50-mM NaCl. The intracellular Na+ and K+ levels in yeast cells were measured as previously described (Patankar et al. 2019a). In this experiment, three independent yeast colonies were used to assess the response of PdNHX6 and EV to the salinity stress condition.
Estimation of vacuolar pH of the yeast cells harboring PdNHX6
Vacuolar pH of the transformed yeast cells was measured and compared with EV control cells using 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF-AM) (Molecular Probes, Eugene, Oregon, USA), to understand the role of PdNHX6 in salinity tolerance mechanism. A vacuolar pH calibration curve was prepared as previously described (Ali et al. 2004), and used to calculate the vacuolar pH of EV and PdNHX6 transformed yeast cells. Briefly, yeast cells (six replicates each) were cultured in SD medium (pH 5.0) for two days, then pelleted and washed twice with distilled water. The cells were incubated with 50-mM BCECF-AM at 30 ºC for 40 min and resuspended in the same medium after three washes and immediately read at 490 nm in Epoch Microplate Spectrophotometer (BioTek, Winooski, USA) supported by Gen5 software (version 2.07). Fluorescence intensity (FI) was measured at 490-nm excitation wavelength, and then the background values of BCECF-free culture were subtracted from FI and normalized to cell densities to calculate the vacuolar pH of PdNHX6 and EV. For the vacuolar pH imaging, slides were coated with 0.1-mg/ml concanavalin A (Sigma-Aldrich, St. Louis, MO, USA), incubated overnight at 30 ºC, and then washed twice to remove excess concanavalin A. For the yeast cultures, the EV and PdNHX6 labeled with 50-mM BCECF were spotted on concanavalin A spot, incubated at 30 ºC for 10 min and washed twice with distilled water. A drop of SD medium was added to yeast cells and then visualized under the fluorescence microscope.
Heterologous expression of PdNHX6 in Arabidopsis
PdNHX6 cDNA was cloned into the binary plant expression vector pEarleyGate 203 (TAIR stock ID: CD3-689), under the control of cauliflower mosaic virus CaMV 35S promoter. A sequence confirmed clone of EarleyGate 203:: PdNHX6 was introduced into Agrobacterium tumefaciens LBA4404 strain (Invitrogen, Carlsbad, USA) using electroporation. The resultant construct was confirmed by PCR using a gene-specific and plasmid-specific primer (Supplementary Table 1). Subsequently, the construct was used to genetically transform Arabidopsis thaliana (ecotype Columbia Col-0) through the floral dip method (Clough and Bent 1998). The floral dip was repeated after two weeks to increase the transformation efficiency. Seeds were collected after maturation (T0) and replanted for selection using 0.01% glufosinate ammonium solution (BASTA®) (Bayer, Bielefeld, Germany). The survived plantlets (T1) were genotyped for the presence of the transgene by PCR using the 35S forward primer, a gene-specific reverse primer (NHX6RA), gene-specific forward (NHX6FA) and a OCS terminator reverse primers (Supplementary Table S1). T2 Seeds were collected and grown on half-strength Murashige and Skoog (MS) medium plates supplemented with 15-mg/ml BASTA® and only the transgenic lines, which showed the 3:1 Mendelian genetic segregation ratio were selected and planted in soil. The seeds of T3 selected lines were further tested for 100% survival on MS medium plates, supplemented with 15-mg/ml BASTA representing the independent homozygous transgenic lines. In this study, three independent homozygous lines were considered for the forthcoming experiments. To confirm the expression of PdNHX6 in Arabidopsis transgenic lines, a semi-quantitative RT-PCR was carried out. Total RNA was extracted from the WT and transgenic Arabidopsis plants (T3 generation) using a RNeasy Plant Mini Kit (Qiagen, Germantown, USA), as per the manufacturer’s instructions. After DNAse I treatment (Qiagen, USA) of the isolated RNA, cDNA was synthesized using a SuperScript IV First-Strand Synthesis System (Invitrogen, Carlsbad, USA), as per the manufacturer’s instructions. Semi-quantitative RT-PCR of PdNHX6 was performed using a primer pair NHX6FA and NHX6RA (Supplementary Table S1). The Arabidopsis actin 2 (accession number AT3G18780) was used as a reference gene, which was amplified by a primer pair ACT2F and ACT2R (Supplementary Table S1).
In vitro assay of root length, fresh and dry weights
Cold-stratified seeds from WT and three transgenic lines were germinated on half-strength MS-agar plates for four days. The seedlings were then transferred to half-strength MS-agar plates (control) or MS-agar plates supplemented with 100-mM NaCl (salt stress) for two weeks. Thereafter, the root length, fresh and dry weights of the WT and transgenic lines were measured. The measurement was carried out for four biological replicates, each of which had four technical replicates.
In vitro seed germination rate test
Surface-sterilized seeds from WT and three transgenic lines were sown on MS-agar plates (control) and MS-agar plates supplemented with 100-mM NaCl (salt stress). The plates were kept under the controlled growth conditions; 22 °C temperature, 60% relative humidity, and 16-h light/8-h dark photoperiod cycle. The germinated seeds were counted each day for seven days. The germination was considered by visible radical emergence. Four replicates and 60 seeds of each line per replicate were used in the experiment.
Growth of PdNHX6 transgenic lines grown under salinity stress
The WT and PdNHX6 transgenic seeds were germinated on half MS plates for four days; thereafter, the plantlets were transferred to the potting mixtures in 0.5-L pots. The plantlets were incubated under the controlled growth conditions, which were mentioned previously. During the first three weeks of the experiment, all plants were watered to field capacity with distilled water. After that, the plants were watered with a 200-mM NaCl solution (salinity stress treatment) every four days for two weeks. However, plants of the control treatment were continually watered with distilled water. Each treatment group included four technical replicates and consisted of three biological replicates. After the elapsed period of the salinity stress treatment, the plants recovered from their respective stress by watering them with distilled water to determine their recovery ability. The soil electrical conductivity (EC) of each treatment group was measured using an Em50 Digital Data Logger (Decagon Devices, WA, USA).
Assaying physiological parameters of PdNHX6 transgenic lines
The response of the PdNHX6 transgenic plants to salinity was compared with the WT plants in terms of phenotypic changes as well as physiological parameters. The physiological parameters, including relative water content (RWC), total chlorophyll, Na+, and K+ concentration were analyzed. RWC of the leaves was measured using the following equation: Leaf RWC (%) = [(fresh weight − dry weight)/(turgid weight − dry weight)] × 100 (Mullan and Pietragalla 2012). Chlorophyll was extracted from fresh leaf tissues using 80% acetone; then, the concentration was calculated as previously described (Arnon 1949).
The concentration of Na+ and K+ ions was measured from 14-day-old Arabidopsis plants grown on half-strength MS medium (control) or in plates supplemented with 100-mM NaCl (salt stress). Furthermore, the plants were dried, and Na+ and K+ ions were extracted using 0.5-M nitric acid followed by two days incubation on a room temperature-shaker at 100 rpm. The samples were filtered, and the concentrations of the soluble Na+ and K+ ions were measured using a Systronics flame photometer 128 (Systronics, India), following a previously published protocol (Munns et al. 2010). Three biological replicates, each with four technical replicates, were used in this analysis.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics 21 package. One-way analysis of variance (ANOVA) test was used to compare the differences between the mean. The significance (p < 0.05) between means of the tested variables was determined by Duncan’s Multiple Range Test (DMRT).
Results
NHX2 and NHX6 are the predominant members of NHX family in date palm genome
In silico analysis, based on the available information in GenBank, had identified 11 members of NHX family in the date palm genome (Fig. 1a). The analysis showed that date palm genome encodes four isoforms each of NHX2 and NHX6 genes, while there was only one isoform each for NHX1, NHX4, and NHX8. The analysis also showed that the NHX1, NHX2, and NHX6 members were similar, as they have clustered together within the same branch on the phylogenetic tree, while the NHX6 isoforms and NHX8 have clustered separately, away from the groups. In this report, we present PdNHX6 (Accession number XM_008812982.2) as the first gene which was successfully cloned among this family. The coding sequence of this gene consists of 1521 bp, which codes for 506-amino acid protein (55.43 kDa) with an average isoelectric point (pI) of 5.65. Phylogenetic analysis of NHX6 protein sequences from different plant species revealed that PdNHX6 was closely related to the African oil palm (Elaeis guineensis) and pineapple (Ananas comosus) (Fig. 1b).
The phylogenetic analysis of the date palm sodium hydrogen exchangers (NHX) protein family (a), and the phylogenetic relationship between PdNHX6 (red font) and its isoforms from different plant species (b). The phylogenetic trees were constructed using the neighbor-joining algorithm. The numbers shown on the nodes represent the percentage of 1000 replicates in the bootstrap analysis (colour figure online)
Prediction of PdNHX6 transmembrane topology displayed 11 transmembrane domains (Fig. 2a), a region where PdNHX6 shares high similarities with other orthologs (Supplementary Fig. S1). This region is commonly conserved among NHX family and known as sodium/hydrogen exchanger 3. Apart from this, the conserved region consists of three highly conserved acidic residues at D132, E156, and D161 (Fig. 2b), which was previously proven to be essential for K+ transport (Wang et al. 2015). In addition to the transmembrane domains, PdNHX6 encodes signal peptides at the N-terminus of the protein sequence, which may mark the protein for secretion. Therefore, this protein might exist as a membranous or/and soluble protein. The protein sequence of PdNHX6 has 78% identity with the Arabidopsis NHX6, AtNHX6 (NP_178079.2), where it showed differences in protein sequence along the conserved region (Fig. 2b, blue arrows). Interestingly, when the topology of the AtNHX6 was investigated, the result showed the presence of 12 transmembrane domains however, unlike PdNHX6, no signal peptide was predicted (Supplementary Fig. S2). This observation may imply a variation in function between PdNHX6 and AtNHX6.
The putative topology of 11 transmembrane domains and signal peptide predicted in PdNHX6 using Protter database (a). Multiple sequence alignment of the deduced amino acid sequence of date palm (PdNHX6) and seven NHX6 isoforms from different plant species (b). Yellow boxes within the Na+/H+ exchanger three conserved domains (red box), are the conserved acidic residues of PdNHX6; D132, E156, and D161. These residues were predicted based on alignment with AtNHX6, which had been demonstrated to be essential for K+ and pH homeostasis. Blue arrows indicate differences in protein sequence among the conserved regions (colour figure online)
The putative promoter of PdNHX6 codes for various TFBSs associated with abiotic stresses
A putative promoter region of 2000 bp located upstream of PdNHX6 coding sequence was computationally searched for the presence of the TFBSs. Among the 1583 putative TFBSs revealed by the analysis, 38% TFBSs were previously known to be abiotic stress-responsive, such as the bHLH, bZIP, AP2/ERF, WRKY, MYB, trihelix, NAC, and ZF-HD (Supplementary Fig. S3). These TFBSs were previously known for their role in the regulation of the expression of drought and salinity responsive genes (Dai et al. 2007; Krishnamurthy et al. 2019; Liu et al. 2014; Xu et al. 2018; Yaish et al. 2010; Zhang et al. 2009).
Expression of PdNHX6 was upregulated in leaf tissue when exposed to salinity
The expression level of PdNHX6 was detected in root and leaf tissues of date palm seedlings under control and salinity stress conditions (Fig. 3). While PdNHX6 was not significantly (p < 0.05) affected in the roots when exposed to salt stress, it was noticed that PdNHX6 was significantly upregulated by almost 17-fold in the leaf tissues under the same conditions.
Quantitative PCR (qPCR) expression analysis of PdNHX6 in the roots and leaves of date palm seedlings under control and salinity stress conditions. Bars represent the mean ± SE of three independent biological replicates. Different letters on top of the bars represent a significant difference at p < 0.05
Expression of PdNHX6 in yeast impaired growth under control and salinity stress conditions
The effect of PdNHX6 was tested in yeast mutant strain cells (BYT458), which contains nonfunctional vacuolar and plasma membrane transporter system (Petrezselyova et al. 2013). Yeast cells overexpressing PdNHX6, as well as empty vector (EV) transformant cells (negative control) were grown on SSM medium (Control), or SSM supplemented with 100-mM NaCl (Fig. 4b). Under both control and salt stress conditions, the PdNHX6 cells showed impaired growth compared to the EV cells (Fig. 4b).
Overexpression of PdNHX6 in a salt-sensitive yeast strain (BYT458). Schematic presentation of the yeast expression vector construct (a). The transgenic PdNHX6 and the empty vector (EV) yeast cells were spotted on solid media SSM (control), and SSM supplemented with 100-mM NaCl (salt stress) to assay the response of the transgenic yeast (b). The EV was used as a negative control in the experiment. The growth response of transgenic PdNHX6 and EV yeast cells in a liquid medium, LSM (control) (c), and LSM supplemented with 50 mM NaCl (d). Each OD value represents the mean ± SE of three independent biological replicates and the statistical significance at p < 0.05 is indicated by an asterisk (*)
Similarly, the growth rates of PdNHX6 transgenic yeast were significantly (p < 0.05) reduced in liquid cultures using LSM or LSM supplemented with 50-mM NaCl, when compared with the EV cells (Fig. 4c, d). The 50-mM NaCl was used as salt stress because a concentration of NaCl had drastically inhibited the growth of these mutant yeast cells.
Determining the intracellular accumulation of Na+ and K+ ions of EV and transgenic yeast cells could partially help predict the modulated response to the salt stress upon the transgene expression. The concentration of ions was measured in yeast cells grown in LSM, supplemented with 0-mM or 25-mM NaCl (Fig. 5). The 25-mM NaCl was selected because it is the highest non-lethal concentration for the mutant yeast cells. Although both EV and transgenic yeast grew at similar rates, the level of Na+ ions was significantly (p < 0.05) higher in the transgenic yeast compared to EV cells under salt stress conditions (25-mM NaCl) (Fig. 5a). The concentration of K+ ions was also measured, and it was found that the accumulation of K+ in transgenic yeast was significantly (p < 0.05) higher than the EV under both control and salinity conditions (Fig. 5b). These observations may support the hypothesis that PdNHX6 regulates Na+ and K+ ions uptake in yeast under salt stress.
Intracellular Na+ (a) and K+ (b) concentration in PdNHX6 and EV yeast cells grown in LSM (control) and LSM supplemented with 25 mM NaCl. The bars represent the mean concentration ± SE of three independent biological replicates. Different alphabets on top of the bars represent a significant difference at p < 0.05
PdNHX6 is involved in homeostatic vacuolar regulation of pH in yeast cells
It was previously shown that NHX6 is involved in vacuolar protein trafficking and pH regulation (Li et al. 2011; Reguera et al. 2015). To test this ability of PdNHX6, the vacuolar pH for both EV and PdNHX6 transformed yeast was measured using a calibration curve (Supplementary Fig. S4). Acidic vacuolar pH was detected for EV and PdNHX6 yeast cells. However, a significantly (p < 0.05) lower pH was detected in the vacuole of PdNHX6 transformed yeast when compared to the EV yeast (Fig. 6a). This observation supports the notion that PdNHX6 activates H+ pumping into the vacuole and hence involved in pH regulation under salinity stress. Likewise, fluorescence images showed lower acidic vacuolar pH values for PdNHX6 yeast compared to the EV, as indicated by pH scale bar (Fig. 6b).
Measurement of vacuolar pH of PdNHX6 and EV yeast cells using fluorescent BCECF-AM dye based on the calibration curve. The fluorescence intensity (FI) was measured at 490-nm excitation wavelengths, and the background values of BCECF-free culture was subtracted from FI and normalized to cell densities to calculate the vacuolar pH of PdNHX6 and EV (a), Calculated pH represents the mean ± SE of six independent biological replicates. The p value on top of the bars represents a significant difference between the pH of PdNHX6 and EV. Accumulation of BCECF-AM dye in the vacuoles of PdNHX6 and EV yeast cultured (b). Bright-field (top) and fluorescent (bottom) images at an excitation wavelength of 440 nm were captured at 40X magnification. Scale bar = 100 μm, and pH scale bar on the right side of images shows color-based pH range
PdNHX6 was transiently expressed in Nicotiana benthamiana leaves. Subcellular localization results showed that PdNHX6 was accumulated in tonoplast (Fig. 7bI). This was confirmed by the co-localization with tonoplast-mCherry marker (Fig. 7bII). The PdNHX6-YFP fluorescence showed excellent overlap with mCherry fluorescence signal in the merged image (Fig. 7bIII). This serves as additional support for the involvement of PdNHX6 in vacuolar pH regulation in the plant cells.
Subcellular localization of PdNHX6 in Nicotiana benthamiana leaf epidermal cells. Schematic representation of the plasmid construct (pSITE-nEYFP-C1), consisting of the nEYFP fused PdNHX6, expressed under the control of a constitutive CaMV 35S promoter (a). The accumulation of nEYFP-PdNHX6, as well as mCherry [tonoplast marker (vac-rk CD3‐975)], in the leaf epidermal cells, were observed under a confocal laser scanning microscope (b). PdNHX6 expression was detected in the tonoplast membrane indicated in white arrows. Images were captured under YFP channel (Ex: 514 nm/Em: 520–560 nm) (I), RFP channel (Ex: 561 nm/Em: 600–650 nm) (II) and the merged channel of both (III). Scale bar = 10 µm
Expression of PdNHX6 in Arabidopsis plants confers tolerance to salinity stress
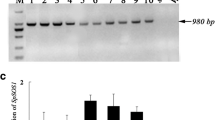
Transgenic Arabidopsis plants were genotyped using PCR to confirm the genetic transformation and stability of PdNHX6 (Supplementary Fig. S5). Transgenic Arabidopsis lines were selected for further studies only if their T2 progeny segregated on a 3:1 ratio, to ensure the presence of only a single transgene within the genome. Subsequently, three independent homozygous lines were obtained (T3), and the expression of PdNHX6 was verified by semi-quantitative RT-PCR (Fig. 8b, Supplementary Fig. S6).
Generation and molecular characterization of PdNHX6 transgenic Arabidopsis lines. Schematic representation of the plasmid construct overexpressing PdNHX6 fused with the Myc epitope (a). Expression analysis of three independent homozygous PdNHX6 transgenic Arabidopsis lines (TL1, TL2, and TL3) and the WT using semi-quantitative RT-PCR and shown on an ethidium bromide-stained 1% agarose gel. Amplification products of Arabidopsis actin gene (AtACT2) (AT3G18780) was used as the internal control (b) The effect of PdNHX6 on intracellular Na+ (c) and K + (d) homeostasis in Arabidopsis plantlets grown on MS-agar plates (shaded bars) and MS-agar plates supplemented with 100-mM NaCl (dotted bars). The bars represent the mean concentration ± SE of three independent biological replicates. The bars with different letters are significantly different at p < 0.05
The growth of the transgenic lines was assayed on MS medium, along with WT plants, to evaluate the effect of PdNHX6 expression on root length, fresh and dry weights under control (Supplementary Fig. S7a) and salinity treatment (Supplementary Fig. S7b) conditions. There were insignificant differences between the root lengths of the transgenic and WT plants when grown under control and NaCl treatment, however, the dry weight of the transgenic seedlings was slightly higher than those of WT under both treatments, and the fresh weight of the transgenic lines TL1 and TL2 was also marginally higher but only under the salt stress condition (Supplementary Fig. S5b).
K+ and Na+ concentrations were measured in the WT and transgenic plants subjected to the control and salt treatments (Fig. 8c, d), to study the effect of PdNHX6 on ion homeostasis under salt stress. The Na+ concentration was significantly (p < 0.05) increased in the WT and transgenic plants in response to salinity (Fig. 8c), however, a significant (p < 0.05) reduction in K+ concentration under salt stress was detected only in the WT plants (Fig. 8d). Nevertheless, the decrease in K+ concentration of transgenic plants was insignificant compared to the control condition. This observation may reflect the ability of the transgenic plants to maintain the ionic balance under salinity stress, even with the increased level of Na+ (Fig. 8c).
For further evaluation of the role of PdNHX6, Arabidopsis plants of three independent homozygous transgenic lines, together with WT, were exposed to salinity on soil (Fig. 11a, b). At the end of the treatment, the EC of the control and salt-treated soils were 0.90 ± 0.45 (mean ± SD) and 44.9 ± 1.2 dS/m, respectively. Transgenic plants treated with salinity showed better viability in comparison to the WT (Fig. 9a), and the transgenic lines were able to withstand the salt stress, especially TL2 plants, which remained vigorous and intact until the end of the treatment (after two weeks) (Fig. 9b). The total chlorophyll content was measured and compared with the WT plants to evaluate the photosynthetic efficiency of the transgenic plants. The transgenic lines showed a higher chlorophyll level than the WT under control and salinity treatment conditions (Fig. 9c); while, the increase in the chlorophyll content was significant (p < 0.05) in two transgenic lines (TL1 and TL2) when exposed to the salt stress (Fig. 9c). Similarly, TL1 and TL2 showed a significantly (p < 0.05) higher RWC, compared to the WT plants, under the salinity condition (Fig. 9d).
Assaying the salinity tolerance ability of PdNHX6 transgenic Arabidopsis lines in vivo. The soil-grown transgenic PdNHX6 lines exposed to salinity (200-mM NaCl) after 7 (a) and 14 (b) days of treatment. The chlorophyll content (c) and relative water content (d) of WT and three homozygous PdNHX6 transgenic Arabidopsis lines, exposed to salinity stress
The effect of salinity treatment was also monitored for individual plants until the maturation stage (Fig. 10). The PdNHX6 transgenic lines showed enhanced tolerance to salinity stress compared to the WT (Fig. 10a, b). Although these transgenic plants showed a higher salinity tolerance in comparison with WT, these plants could not recover after two weeks of salinity exposure (Fig. 10c, d).
Assaying the salinity tolerance ability of PdNHX6 transgenic Arabidopsis lines in vivo. The soil-grown transgenic PdNHX6 lines and the WT exposed to salinity (200-mM NaCl) after 7 (a) and 14 (b) days of treatment. The transgenic lines showed tolerance to salt stress and remained green even after 2 weeks of salt stress treatment. To compare the recovery ability of the transgenic and the WT plants after salt treatment, the plants were irrigated with distilled water for an additional 2 weeks (c, d). After the first week of recovery period (c), TL1 and TL2 plants showed better survival than the WT
The germination rate of the transgenic line seeds changed under salinity
To evaluate the effect of salinity treatment on seed quality, the germination rate of WT and transgenic lines seeds was tested on plain MS plates (control) as well as on MS plate supplemented with 100-mM NaCl (Fig. 11a, b). The seeds were incubated on the medium for seven days, and the germination rate was calculated each day.
Seed germination assay of PdNHX6 transgenic Arabidopsis lines and WT. Seed germination on control MS plates and MS plates with 100 mM NaCl (salt stress) on day 3 (a) and day 6 (b). The same number of seeds were used in each treatment and the germination rate of seeds grown under normal growth condition (c) and salinity stress condition (d) over 7 days was calculated. The results shown are the mean ± SE of four independent biological replicates and the statistical significance at p < 0.05 is indicated by asterisks (*)
Most of the WT and the transgenic seeds were able to germinate on control plates but showed different germination rates based on the genotype. For example, after 24 h from stratification, the germination rate of TL1 seeds was significantly (p < 0.05) higher than the wild type. Nevertheless, a significant (p < 0.05) increase in the germination rate of TL1 and TL2 seeds, in comparison to the WT, was detected on the second and third days (Fig. 11c). However, most of the seeds used in this experiment have germinated by the sixth day of the experiment (Fig. 11b, c). Seeds grown under salt treatment conditions showed different germination patterns (Fig. 11a, b). While the WT seeds started germination only after 24 h, the transgenic seeds germinated earlier (Fig. 11d). For example, 12% of TL2 seeds were germinated within 24 h compared with 0% of the WT seeds. Besides, the germination rate of TL1 seeds was significantly (p < 0.05) higher than WT seeds on day five, and both TL1 and TL2 seeds showed the highest germination rate until the seventh day.
Discussion
Plant tolerance to salt stress can be achieved by three major strategies; Na+ or Cl− exclusion and secretion, Na+ or Cl− compartmentalization in the tissues, and osmotic tolerance (Munns and Tester 2008). Transport of water and ions (primarily Na+, K+, and Cl−) into and throughout the plants is the fundamental action to these mechanisms (Craig Plett and Møller 2010), where the antiport activity of Na+/H+ and K+/H+ exchangers in the tonoplast vesicles is considered as the first model of secondary active transport procedure in plants (Rodríguez-Rosales et al. 2009). In this study, we functionally characterized PdNHX6 as the first gene from the date palm NHX family.
In silico analysis revealed that PdNHX6 shared the Na+/H+ exchanger 3 domain, which was characterized as monovalent cation: proton antiporter-1 (CPA1), with other members of NHX6 plant orthologs. This domain can catalyze the electroneutral exchange of monovalent Na+ for H+ (Ma et al. 2015). Furthermore, computational analysis of the PdNHX6 gene sequence revealed the existence of various putative TFBSs recognized as abiotic stress-responsive elements within the promoter region. Among these TFBSs, bHLH is abundantly distributed within the putative PdNHX6 promoter region. A recent study indicated that under salinity, the expression of the Arabidopsis AtNHX1 and AtNHX6 genes is regulated by two bHLH TFs (AtMYC2 and AtbHLH122) (Krishnamurthy et al. 2019). Additionally, binding of AST1 transcription factor to a trihelix site in the promoter regions of AtNHX2, AtNHX3, and AtNHX6 enhanced salinity tolerance in Arabidopsis (Xu et al. 2018). The trihelix site was also identified in this study within the putative PdNHX6 promoter. This finding may suggest a similar mode of interaction between PdNHX6 and the functional NHX previously characterized in other plant species, upon exposure to the saline environment. The expression level of this gene, as determined by qPCR, significantly increased in the leaf tissues under salt stress; however, this expression was insignificantly altered in the root tissues when grown under the same environmental conditions. This finding may suggest that PdNHX6 is extruding Na+ from the leaf tissues via the phloem. However, this proposed mechanism requires further experimental justifications.
Overexpression of PdNHX6 in Arabidopsis plants enhanced tolerance to salt stress. Transgenic lines exposed to salinity showed higher photosynthetic efficiency in terms of retaining more chlorophyll, as well as higher RWC than the control WT plants. Similar results were obtained when other date palm genes of salinity tolerance enhancement capabilities such as the aquaporin gene PdPIP1;2 (Patankar et al. 2019a) and the metallothionein 2A (PdMT2A) (Patankar et al. 2019b) were introduced into Arabidopsis.
The subcellular localization results showed that PdNHX6 was accumulated in the tonoplast membrane. While previous studies in different plant species showed that the NHX6 is accumulated in the endosomal membranes of the Golgi, TGN, and PVC (Bassil et al. 2012; Jiang et al. 2010; Reguera et al. 2015; Wang et al. 2015), this is the first study to show that NHX6 can be a tonoplast-located antiporter. In fact, Na+ influx has increased under salt stress conditions in both PdNHX6 transgenic and WT plants.
Nevertheless, the transgenic plants could maintain a balanced K+ level compared to the WT, which showed a reduced intracellular K+ level under salt stress conditions. A previous study showed that there are three conserved acidic residues (D165, E189, and D194) in AtNHX6, which are responsible for K+ transport and plant growth (Wang et al. 2015). Computational amino acid sequence analysis showed that these residues are also present within the PdNHX6 sequence, which may perform a similar role in date palm salt tolerance mechanism.
Overexpression of PdNHX6 in yeast mutant strain did not improve yeast cell growth under both control and salt stress. Conversely, a previous study showed that overexpression of the Arabidopsis AtNHX6 in AXT3 mutant yeast strain had enhanced the growth under salinity stress conditions (Wang et al. 2015). Nevertheless, our data revealed the involvement of PdNHX6 in Na+/K+ and pH regulation in the yeast cells. Similar to the results obtained from the analysis of the transgenic Arabidopsis, the transgenic yeast cells have accumulated higher Na+ levels than the EV strain under salt stress conditions. However, the K+ accumulation was significantly higher than the EV under both control and salinity stress conditions. This notion is consistent with the commonly known function of NHXs by which this protein usually regulates the K+ transport under normal growth conditions (Jiang et al. 2010); while, it is involved in Na+ and K+ transport under salt stress (Barragán et al. 2012; Reguera et al. 2015). Given the notion that PdNHX6 regulates the vacuolar pH in yeast cells, suggests that PdNHX6 might facilitate the exchange of Na+/H+ across the vacuolar membranes. While these NHX6 proteins are known for endosomal pH regulation function by preventing acidification through the H+ leakage out of these compartments (Qiu 2016), the increase in acidity of the vacuole of the transgenic yeast indicated by our data could be more related to the vacuolar K+ transportation, which was confirmed by the increase in the K+ level in the transgenic yeast. The increased level of H+ in the vacuole generates a proton gradient that might be used for K+ exchange through other tonoplast channels, such as two-pore K+ channels (TPK1) (Latz et al. 2013). This retains a higher cytosolic K+ level, a mechanism which is essential for the salinity tolerance in plants. In addition, the pH homeostasis is required for proper protein trafficking to the vacuole as well as proteolytic processing of storage proteins (Reguera et al. 2015). Therefore, this might be another mechanism by which PdNHX6 may enhance salinity tolerance in date palm. A previous study revealed that NHX5 and NHX6 play a crucial role in the processing of seed storage proteins (Qiu 2016), a mechanism that leads to a reduction in the nitrogen and amino acid requirements during the germination process and early growth of the seedlings (Dragwidge et al. 2018). Consistently, the results obtained from this study showed that the overexpression of the PdNHX6 resulted in earlier germination of the transgenic seeds. In addition, NHX5 and NHX6 can regulate plant development through an auxin-dependent mechanism (Dragwidge et al. 2018; Fan et al. 2018), a mechanism, which may affect seed development and germination under stress.
In conclusion, the molecular and functional characterization of the PdNHX6 conducted in this study highlighted the importance of this gene in salinity tolerance response of the date palm. Heterologous expression of PdNHX6 enhanced salinity tolerance through the K+ and vacuolar pH homeostasis. The positive effect of these cellular adjustments was apparent on the transgenic plants as there was a significant increase in the chlorophyll, water contents, and seed germination rates. Collectively, our data suggest that this gene is a good candidate to explore the development of salt-tolerant date palm tree…
Author contribution statement
IA-H conceived, designed, performed the experiments, analyze data, and wrote the manuscript; GAJ performed the experiments, HVP revised the manuscript, RA-Y revised the manuscript, SR and PPK performed the subcellular localization of PdNHX6 and revised the manuscript, and MWY designed the experiment, supervised the work, wrote the manuscript, and contributed reagents/materials/analysis tools.
References
Al-Harrasi I, Al-Yahyai R, Yaish MW (2018) Differential DNA methylation and transcription profiles in date palm roots exposed to salinity. PLoS ONE 13:e0191492. https://doi.org/10.1371/journal.pone.0191492
Al Harrasi I, Al-Yahyai R, Yaish MW (2017) Detection of differential DNA methylation under stress conditions using bisulfite sequence analysis. In: Plant stress tolerance. Springer, New York, pp 121–137
Al Kharusi L, Al Yahyai R, Yaish MW (2019) Antioxidant response to salinity in salt-tolerant and salt-susceptible cultivars of date palm. Agriculture 9:8. https://doi.org/10.3390/agriculture9010008
Al Kharusi L, Assaha DV, Al-Yahyai R, Yaish MW (2017) Screening of date palm (Phoenix dactylifera L.) cultivars for salinity tolerance. Forests 8:136
Ali R, Brett CL, Mukherjee S, Rao R (2004) Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J Biol Chem 279:4498–4506
Arnon DI (1949) Copper enzymes in isolated chloroplasts polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15. https://doi.org/10.1104/pp.24.1.1
Barragán V et al (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24:1127–1142
Bassil E, Coku A, Blumwald E (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot 63:5727–5740
Bonales-Alatorre E, Pottosin I, Shabala L, Chen Z-H, Zeng F, Jacobsen S-E, Shabala S (2013) Differential activity of plasma and vacuolar membrane transporters contributes to genotypic differences in salinity tolerance in a halophyte species, Chenopodium quinoa. Int J Mol Sci 14:9267–9285. https://doi.org/10.3390/ijms14059267
Brett CL, Donowitz M, Rao R (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol-Cell Physiol 288:C223–C239
Chow C-N et al (2015) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res 44:D1154–D1160. https://doi.org/10.1093/nar/gkv1035
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Craig Plett D, Møller IS (2010) Na+ transport in glycophytic plants: what we know and would like to know. Plant Cell Environ 33:612–626
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751. https://doi.org/10.1104/pp.106.094532
Dragwidge JM, Ford BA, Ashsnest JR, Das P, Gendall AR (2018) Two endosomal NHX-type Na+/H+ antiporters are involved in auxin-mediated development in Arabidopsis thaliana. Plant Cell Physiol 59:1660–1669. https://doi.org/10.1093/pcp/pcy090
Fan L et al (2018) Na+, K+/H+ antiporters regulate the pH of endoplasmic reticulum and auxin-mediated development. Plant Cell Environ 41:850–864
Feng D-F, Doolittle RF (1987) Progressive sequence alignment as a prerequisitetto correct phylogenetic trees. J Mol Evol 25:351–360. https://doi.org/10.1007/bf02603120
Ford BA, Ernest JR, Gendall AR (2012) Identification and characterization of orthologs of AtNHX5 and AtNHX6 in Brassica napus. Front Plant Sci 3:208
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Gong X, Zhang J, Liu J-H (2014) A stress responsive gene of Fortunella crassifolia FcSISP functions in salt stress resistance. Plant Physiol Biochem 83:10–19
Guan B, Hu Y, Zeng Y, Wang Y, Zhang F (2011) Molecular characterization and functional analysis of a vacuolar Na+/H+ antiporter gene (HcNHX1) from Halostachys caspica. Mol Biol Rep 38:1889–1899
Hamam AM, Britto DT, Flam-Shepherd R, Kronzucker HJ (2016) Measurement of differential Na+ efflux from apical and bulk root zones of intact barley and Arabidopsis plants. Front Plant Sci 7:272
Himabindu Y, Chakradhar T, Reddy MC, Kanygin A, Redding KE, Chandrasekhar T (2016) Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ Exp Bot 124:39–63
Jana GA, Al Kharusi L, Sunkar R, Al-Yahyai R, Yaish MW (2019) Metabolomic analysis of date palm seedlings exposed to salinity and silicon treatments. Plant Signal Behav 14:1663112
Jiang X, Leidi EO, Pardo JM (2010) How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav 5:792–795
Krishnamurthy P, Vishal B, Khoo K, Rajappa S, Loh C-S, Kumar PP (2019) Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis. Plant Cell Rep 38:1–17
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Latz A et al (2013) Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant 6:1274–1289
Leidi EO et al (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61:495–506
Li M, Li Y, Li H, Wu G (2011) Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiol 31:349–357
Liu C et al (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36
Liu L, Zeng Y, Pan X, Zhang F (2012) Isolation, molecular characterization, and functional analysis of the vacuolar Na+/H+ antiporter genes from the halophyte Karelinia caspica. Mol Biol Rep 39:7193–7202. https://doi.org/10.1007/s11033-012-1551-x
Ma Y, Wang J, Zhong Y, Geng F, Cramer GR, Cheng Z-MM (2015) Subfunctionalization of cation/proton antiporter 1 genes in grapevine in response to salt stress in different organs. Hortic Res 2:15031
Marchler-Bauer A et al (2014) CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226
Mishra A, Tanna B (2017) Halophytes: potential resources for salt stress tolerance genes and promoters. Front Plant Sci 8:829
Mishra S, Alavilli H, Lee B-H, Panda SK, Sahoo L (2014) Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance in Arabidopsis thaliana. PLoS ONE 9:e106678. https://doi.org/10.1371/journal.pone.0106678
Mullan D, Pietragalla J (2012) Leaf relative water content physiological breeding II: a field guide to wheat phenotyping. CIMMYT, Mexico, pp 25–27
Munns R, Gilliham M (2015) Salinity tolerance of crops—what is the cost? New Phytol 208:668–673
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, Wallace PA, Teakle NL, Colmer TD (2010) Measuring soluble ion concentrations (Na+, K+, Cl−) in salt-treated plants. In: Plant stress tolerance. Springer, New York, pp 371–382
Omasits U, Ahrens CH, Müller S, Wollscheid B (2013) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886
Patankar HV, Al-Harrasi I, Al-Yahyai R, Yaish MW (2018) Identification of candidate genes involved in the salt tolerance of date palm (Phoenix dactylifera L.) based on a yeast functional bioassay DNA and cell biology
Patankar HV, Al-Harrasi I, Al-Yahyai R, Yaish MW (2019a) Functional characterization of date palm aquaporin gene PdPIP1;2 confers drought and salinity tolerance to yeast and Arabidopsis. Genes 10:390. https://doi.org/10.3390/genes10050390
Patankar HV, Al-Harrasi I, Al Kharusi L, Jana GA, Al-Yahyai R, Sunkar R, Yaish MW (2019b) Overexpression of a metallothionein 2A gene from date palm confers abiotic stress tolerance to yeast and Arabidopsis thaliana. Int J Mol Sci 20:2871
Patankar HV, Assaha DV, Al-Yahyai R, Sunkar R, Yaish MW (2016) Identification of reference genes for quantitative real-time PCR in date palm (Phoenix dactylifera L.) subjected to drought and salinity. PLoS ONE 11:e0166216
Petrezselyova S, Kinclova-Zimmermannova O, Sychrova H (2013) Vhc1, a novel transporter belonging to the family of electroneutral cation–Cl−cotransporters, participates in the regulation of cation content and morphology of Saccharomyces cerevisiae vacuoles. Biochim Biophys Acta (BBA) Biomembr 1828:623–631
Qiu Q-S (2016) AtNHX5 and AtNHX6: Roles in protein transport. Plant Signal Behav 11:e1184810. https://doi.org/10.1080/15592324.2016.1184810
Reguera M et al (2015) pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27:1200–1217
Rodríguez-Rosales MP, Gálvez FJ, Huertas R, Aranda MN, Baghour M, Cagnac O, Venema K (2009) Plant NHX cation/proton antiporters. Plant signal Behav 4:265–276
Shabala S, Pottosin I (2014) Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol Plant 151:257–279. https://doi.org/10.1111/ppl.12165
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Wang L, Wu X, Liu Y, Qiu Q-S (2015) AtNHX5 and AtNHX6 control cellular K+ and pH homeostasis in Arabidopsis: three conserved acidic residues are essential for K+ transport. PLoS ONE 10:e0144716. https://doi.org/10.1371/journal.pone.0144716
Xu H et al (2018) Arabidopsis thaliana trihelix transcription factor AST1 mediates salt and osmotic stress tolerance by binding to a novel AGAG-box and some GT motifs. Plant Cell Physiol 59:946–965. https://doi.org/10.1093/pcp/pcy032
Xu Y et al (2013) Functional characterization of a wheat NHX antiporter gene TaNHX2 that encodes a K+/H+ exchanger. PLoS ONE 8:e78098. https://doi.org/10.1371/journal.pone.0078098
Yaish M (2015) Proline accumulation is a general response to abiotic stress in the date palm tree (Phoenix dactylifera L.). Genet Mol Res 14:9943–9950
Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, Bi Y-M, Rothstein SJ (2010) The APETALA-2-like transcription factor OsAP2–39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet 6:e1001098
Yaish MW, Patankar HV, Assaha DVM, Zheng Y, Al-Yahyai R, Sunkar R (2017) Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genomics 18:1. https://doi.org/10.1186/s12864-017-3633-6
Yaish MW, Sunkar R, Zheng Y, Ji B, Al-Yahyai R, Farooq SA (2015) A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front Plant Sci 6:946
Zaid A, De Wet P (1999) Climatic requirements of date palm. Date palm cultivation. FAO, Roma
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60:3781–3796
Acknowledgements
The authors would like to thank Professor Hana Sychrova, Institute of Physiology Academy of Sciences of the Czech Republic, Prague, Czech Republic, for donating the salt-sensitive mutant S. cerevisiae BYT458 strain, which was used in this study.
Funding
This study is supported by the generous grant number RC/RG-SCI/BIOL/18/01 from the research council (TRC), Oman to MWY.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. All authors revised and approved the final manuscript.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2020_2549_MOESM2_ESM.tif
Supplementary file2 Multiple sequence alignment of the deduced amino acid sequence of date palm (PdNHX6) and seven NHX6 isoforms from different plant species. PdNHX6 protein conserved regions (dark grey-colored highlights) that are shared with other NHX6 members. All the shaded areas expand the common conserved domain of NHX family known as sodium/hydrogen exchanger 3 (TIF 5439 kb)
299_2020_2549_MOESM3_ESM.tif
Supplementary file3 The putative topology of date palm (PdNHX6) and Arabidopsis (AtNHX6), using Protter database. The toplogy of PdNHX6 shows 11 transmembrane domains and signal peptide (a). However, AtNHX6 putative toplogy shows 12 transmembrane domains and no signal peptide was predicted (b) (TIF 2393 kb)
299_2020_2549_MOESM4_ESM.tif
Supplementary file4 The abundance of transcription factor binding sites (TFBSs) within 2000 bp promoter region of PdNHX6 computationally analyzed using PlantPAN 2.0 database (TIF 711 kb)
299_2020_2549_MOESM5_ESM.tif
Supplementary file5 The calibration curve of normalized fluorescent intensity (NI490) plotted against pH ranging from 4.0–8.0 (TIF 582 kb)
299_2020_2549_MOESM6_ESM.tif
Supplementary file6 PCR-based genotyping of PdNHX6 transgenic Arabidopsis plants using different primer pairs; 35S promoter and NHX6RA (a), NHX6FA, and OCS terminator (b) NHX6FA and NHX6RA (c) and 35S promoter and OCS terminator (d) (TIF 996 kb)
299_2020_2549_MOESM7_ESM.tif
Supplementary file7 Expression analysis of three independent homozygous PdNHX6 transgenic Arabidopsis lines (TL1, TL2, and TL3) and the WT using semi-quantitative RT-PCR and shown on an ethidium bromide-stained 1% agarose gel. The Arabidopsis actin 2 (AtACT2) (AT3G18780) was used as a loading control (TIF 3180 kb)
299_2020_2549_MOESM8_ESM.tif
Supplementary file8 Assay of the salinity tolerance ability of PdNHX6 Arabidopsis lines in vitro. The effect of PdNHX6 on root length, fresh weight, and dry weight of Arabidopsis plantlets grown on MS-agar plates (a) and MS-agar plates supplemented with 100 mM NaCl (b). Each genotype is denoted by color code, as shown on top of the scanning images. The bars represent the mean root length ± SE of three independent replicates. The bars with different letters are significantly different at p < 0.05 (TIF 1588 kb)
Rights and permissions
About this article
Cite this article
Al-Harrasi, I., Jana, G.A., Patankar, H.V. et al. A novel tonoplast Na+/H+ antiporter gene from date palm (PdNHX6) confers enhanced salt tolerance response in Arabidopsis. Plant Cell Rep 39, 1079–1093 (2020). https://doi.org/10.1007/s00299-020-02549-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02549-5