Abstract
Key message
Two methods, PCR and amplicon labeling based, were developed and successfully applied to reliably detect CRISPR/Cas9 induced indels in rice.
Abstract
The use of CRISPR/Cas9 has emerged as a powerful nuclease-based genome editing tool in several model organisms including plants for mutagenesis by inducing precise gene editing through efficient double strand DNA breaks (DSBs) at the target site and subsequent error-prone non-homologous end joining (NHEJ) repair, leading to indel mutations. Different molecular methods including enzymatic mismatch cleavage (EMC), high-resolution melting curve analysis (HRMA) and conventional polymerase chain reaction (PCR) combined with ligation detection reaction (LDR) have been developed to quick identify CRISPR/Cas9 induced mutations. However, their intrinsic drawbacks limit their application in the identification of indel mutants in plants. Here we present two methods (one simple PCR based and the other amplicon labeling based) for effective and sensitive detection of CRISPR/Cas9 induced indels in rice. In PCR-based method, targets were amplified using two pairs of primers for each target locus and visualized on gel electrophoresis, while in amplicon labeling-based method, targets were amplified using tri-primers (with one a universal 6-FAM 5′-labelled) and detected by DNA capillary electrophoresis. Both methods can accurately define indel sizes down to ± 1 bp, and are amenable for high throughput analysis, therefore, will significantly facilitate the identification of indel mutants generated by CRISPR/Cas9 for further functional analysis and breeding in rice and other plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
CRISPR/Cas9 system has been established as a powerful nuclease-based genome editing tool in plants to generate new plant genotypes by introducing precisely targeted double strand breaks that are resolved by endogenous repair pathways (Feng et al. 2014). In contrast to traditional Agrobacterium mediated T-DNA transgenic method that depends on random recombination or integration in plants, site specific mutation via CRISPR/Cas9 system is achieved by constructing a single guide RNA (sgRNA) that directs the Cas9 nuclease to genomic targets to create double strand breaks (DSBs), which are repaired predominantly by error-prone non-homologous end joining (NHEJ) machinery, ultimately leads to indel mutations, most frequently indel mutants with only 1 bp variation (Pan et al. 2016; Ren et al. 2016; Zhu et al. 2017). These small variations of nucleotides may cause loss of function through frame-shift mutation, however, such small variations in the genome cannot be detected by agarose gel electrophoresis (Denbow et al. 2017), which impedes the screening for such mutants and subsequent functional analysis. There is an urgent need to develop effective and robust detection methods for plant research.
Several methods have been developed to screen or identify genome editing including CRISPR/Cas9 induced indel mutations at the target locus in other systems rather than plants. The most frequently used one is the enzymatic mismatch cleavage (EMC) technique, which takes advantages of enzymes that are able to cleave hetero-duplex DNA at mismatches formed by single or multiple nucleotides (Vouillot et al. 2015). Because its cleavage efficiency is affected by many factors such as the sequence, the number of mismatched nucleotides, and the flanking sequence between the two DNA strands, this method is more suitable for detection of large indels (Zhu et al. 2014; Vouillot et al. 2015). In addition, although it is easy to operate, its sensitivity, however, is quite low (Huang et al. 2012). Furthermore, it cannot discriminate homozygous mutants from wild-type, neither heterozygous mutants from biallelic mutants (Harayama and Riezman 2017). High-resolution melting curve analysis (HRM) method offers more advantages over EMC. HRM is a fluorescence-based technique that measures the Tm of a particular PCR product, and identifies the mutant by analyzing the melting behavior for hetero-duplex and homo-duplex DNA fragments (Thomas et al. 2014). It is rapid, unrestrictive, and suitable for detecting low level chimeric mutants, but it needs specific instruments, and is not suitable for large indel (> 100 bp) detection (Thomas et al. 2014). Conventional polymerase chain reaction (PCR) combined with ligation detection reaction (LDR) is an alternative easy and quick method for routine genotyping, however, because it detects indel on agarose gel electrophoresis, its sensitivity to detect mutants with only a few base pairs genetic variations is weak (KC et al. 2016). Other reported methods include Indel Detection by Amplicon Analysis (IDAA) (Lonowski et al. 2017), qPCR (Yu et al. 2014), cloning and Sanger sequencing, digital PCR (Findlay et al. 2016; Mock et al. 2016), and restriction fragment length polymorphism (RFLP) (Kim et al. 2014), some of them are expensive (such as digital PCR and qPCR), time consuming (for instance, cloning and Sanger sequencing) and less sensitive (for example, PCR-LDR and RFLP). Besides EMC (Nekrasov et al. 2013; Shan et al. 2014), HRM (Denbow et al. 2017), qPCR (Peng et al. 2018) and Sanger sequencing (Feng et al. 2014), other methods, such as annealing at critical temperature PCR (ACT-PCR) (Hua et al. 2017), mutation sites-based specific primers PCR (MSBSP-PCR) (Guo et al. 2018) and cleaved amplified polymorphic sequence (CAPS) (Kohata et al. 2018), have been recently developed in plants. Although those methods are proved to be applicable, each has several limitations including time- and/or labor-consuming (ACT-PCR and MSBSP-PCR), cost (qPCR), low detection specificity (CAPS) or in the case of MSBSP-PCR, the inability to distinguish homozygous from heterozygous mutations.
CRISPR/Cas9 system has been the preference of recent researches to target towards improving the yield and quality of rice (Shan et al. 2014; Mazumdar et al. 2016), the important staple food crop in the world, over genetically modified (GM) approaches. One reason behind that is the capacity of the CRISPR/Cas9 system to generate transgene free rice mutants, which helps to avoid regulatory issues as in GM rice. Although recently released decision from the Court of Justice of the European Union, that organisms obtained by mutagenesis including genome editing tools are GMOs and are in principle subject to the obligations laid down by the GMO Directive, had non-negligible negative effects on rice society, new plant breeding techniques (NPBT) including genome editing tools, such as CRISPR/Cas9, will continue to contribute to both basic research and applied application to create new improved traits in rice. Because most of the CRISPR/Cas9 generated mutants in plants are small indels that are inheritable (Zhang et al. 2014), development of effective and economic identification methods would enable cost effective screening for desired indels in pool of rice mutants at the early stages, which would, in turn, significantly facilitate further functional analysis or breeding purpose.
In this study, taking the advantages of several known indel information CRISPR/Cas9 generated mutants in rice, we developed two methods, one simple PCR based and the other amplicon labelling based, for fast and sensitive detection of indel mutations in rice. The sensitivity, precision and reliability of the two developed methods for the identification of indels were assayed using CRISPR/Cas9 induced mutants targeting different genes. Our results demonstrated that both methods can detect indels with high sensitivity down to ± 1 bp, and are amenable for high throughput analysis.
Materials and methods
Plant materials
CRISPR/Cas9 induced rice (Oryza sativa) mutants on several genes including SD1 were used in this study (Supplementary Table 1). Wild-type (9522 cultivar rice) was used as a control for PCR based and amplicon labeling-based methods. All mutants and wild-type were grown in the paddy field of Shanghai Jiao Tong University, Shanghai, China.
Genomic DNA extraction
Genomic DNA from leaf tissues were extracted as previously described with minor modifications (Murray and Thomson 1980). 30 mg liquid nitrogen fresh frozen leaf tissue of different rice materials was ground in the presence of liquid nitrogen, then incubated with lysis buffer (1.5 X CTAB) and RNase at 65 °C for 60 min. The liquid phase was collected after centrifugation and extracted again with phenol: chloroform and trichloromethane and mixed with same volume of isopropyl alcohol to precipitate the genomic DNA. The pellet was washed twice with 70% ethanol and then dissolved in 60 µL ddH2O. The quality and quantity of the extracted genomic DNA was evaluated using both the NanoDrop 1000 UV/Vis Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) to obtain values of OD260/OD280 and OD260/OD230, and the electrophoresis on 1% (w/v) agarose gel in 0.5 × TBE with GelRed staining. All extracted genomic DNA was stored at − 20 °C until using in the experiments.
Primer and probe design
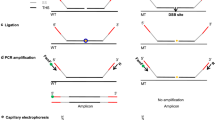
All oligonucleotide primers for both the PCR-based method and the amplicon labeling-based method were designed using Primer Premier 5.0 and synthesized by Invitrogen Co., Ltd. (Shanghai, China). For PCR-based methods, the outer pair of primers were designed to prime outside the MHS (PoF/R), and the inner pair of primers were designed to flank the MHS (PiF/R) at the 3´ most nucleotide of the MHS (Fig. 1a). For amplicon labeling-based method, tri-primers were designed based on target sequence (the FAM labeled forward primer Fam F has the identical sequence to non-labeled forward primer F, Fig. 2a). Sequences of all oligonucleotide primers used in this study are listed in Supplementary Table 1.
The development of PCR-based method and its application in the identification of rice CRISPR/Cas9 mutants in rice. a Simplified schematic presentation of the principle and the gel electrophoresis result of the PCR-based method. Multiple PCR was performed using a pair of inner (PiF/R) and outer (PoF/R) primers, and the resulting PCR products were analyzed on 2% agarose gel. PAM, proto-spacer adjacent motif; xxx, any three nucleotides upstream of PAM. b Sensitivity test of developed PCR-based method. A serial dilution of mixed samples containing different ratios of a known CRISPR/Cas9 induced 1 bp deletion SD1 mutant (Mt) genomic DNA to wild-type (Wt) genomic DNA were used for amplicon amplification using SD1 Po and Pi primer pairs, respectively, and resulting amplification products were visualized on 2% agarose gel electrophoresis. Lane M, Trans 2K DNA marker; lane 1, NTC (negative control); lanes 2–10, 60 ng DNA mixtures with Mt to Wt ratios of 0%, 10%, 20%, 40%, 50%, 60%, 80%, 90% and 100%, respectively. c Relative quantification of the band intensities (relatives to wild-type) of Pi PCR products of mixed samples by ImageJ. d Analysis of amplified PCR products of rice SD1 mutants generated by CRISPR/Cas9 using developed PCR-based method on 2% agarose gel electrophoresis. d#, deletion with #bp; i#, insertion with #bp. e Results of Sanger sequencing of corresponding PCR products in d
The development of amplicon labelling-based method and its application in the identification of rice CRISPR/Cas9 mutants. a Simplified schematic of the principle and the result determination of the amplicon labeling-based method. Three primers include a pair of target allele specific forward (F) and reverse (R) primers and an additional universal 6-carboxyfluorescein (6-FAM) 5′-labelled allele specific forward primer (FamF) were used to amplify the PCR products in both wild-type (Wt) and mutants (Mt), and all fluorophore labelled amplicon of the targets containing indels were analyzed by fragment analyzer. The x-axis and y-axis represent amplicon size in base pairs and relative fluorescence units (RFU), respectively. DSB site, double strand break site; gDNA, corresponding DNA of the guide RNA; MHS, mutational hot spot regions. b Sensitivity test of the developed amplicon labelling method. A serial dilution of mixed samples containing 20%:80%, 10%:90%, 1%:99% and 0.1%:99.9% ratios of CRISPR/Cas9 induced SD1 mutant (Mt, 4 bp deletion) genomic DNA to wild-type (Wt) genomic DNA were used. The height of detected fluorophore labelled amplicon peak increased for wild-type, but decreased for mutant alleles as the ratio of Mt:Wt in the mixture decreased. Red and green stars denote Mt allele and Wt allele, respectively. c Fragment analysis of the fluorophore labelled amplicon of targets amplified using SD1 tri-primers in a fragment analyzer. d#, deletion with # bp; i#, insertion with # bp. d Results of Sanger sequencing of corresponding PCR products in c. e Zygosity analysis of developed amplicon labelling method. Targets were amplified using SD1 tri-primers, and resulting fluorophore labelled amplicons were analyzed in a fragment analyzer. The red line indicates the amplicon peak of wild-type. Wt, Wild-type; d4-HM, 4 bp deletion homozygous mutant; i5-HM, 5 bp insertion homozygous mutant; HE, 5 bp insertion heterozygous mutant. f Chromatographic presentation of the Sanger sequencing results in wild-type, HM and HE mutants identified by SD1 tri-primers. (Color figure online)
PCR-based method
For amplification of SD1 target, PCR was carried out in a 20 µL reaction mixture containing 1 × PCR buffer, 1 × Q-solution (Qiagen, Germany), 0.2 µM dNTPs, 5 µM primer, 1 unit of HotStarTaq DNA Polymerase (Qiagen, Germany) and 60 ng genomic DNA. The reaction mixtures were heated to 95 °C for 15 min followed by 35 cycles of amplification at 94 °C for 30 s, 55 °C for 30 s, and a final stage at 72 °C for 45 s. For amplification of other targeted genes, PCR was carried out in a 25 µL reaction mixtures containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 µM primer, 1.25 units of Taq DNA Polymerase (TaKaRa Biotechnology Co., Ltd.) and 60 ng genomic DNA. The reaction mixtures were heated to 94 °C for 5 min, followed by 35 cycles of amplification at 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and a final stage at 72 °C for 7 min. The amplified PCR products were analyzed on 2% agarose gel.
Amplicon labeling-based method
To generate amplicon labeling yields, PCR was carried out in a 25 µL reaction mixture containing optimized 0.5 µM universal 6-FAM 5′-labelled forward primer, 0.05 µM forward primer, 0.5 µM reverse primer, 2 × Gflex PCR buffer, and Tks Gflex DNA Polymerase (TaKaRa Biotechnology Co., Ltd.). For amplification of SD1 target, the touchdown thermocycling program started at an initial heating at 94 °C for 2 min, followed by initial annealing at 65 °C ramping down by 1 °C per cycle to 55 °C for ten cycles, and an additional 25 cycles of amplification at 98 °C for 10 s, 55 °C for 15 s, and 68 °C for 40 s, and a final stage at 68 °C for 40 s. For amplification of LOC_Os06g04420 target, the touchdown thermocycling program started at 94 °C for 2 min, followed by initial annealing at 69 °C ramping down by 1 °C per cycle to 59 °C for 10 cycles, and an additional 25 cycle of 98 °C for 10 s, 59 °C for 15 s, and 68 °C for 40 s, and a final stage at 68 °C for 40 s. The amplification of PCR products were examined on 2% agarose gel. For fragment analysis, 1 µL amplified PCR products were mixed with 0.5 µL LIZ500 size standard (ABI/Life Technologies, USA) and analyzed on ABI3010 sequenator (ABI/Life Technologies, USA).
Results
Identification of CRISPR/Cas9 generated rice mutants by PCR-based method
CRISPR/Cas9 induced mutation is predictable, because most of indels occur at the double strand breaks (DSBs) sites, namely mutational hot spot regions (MHS), generally within 4 base pairs upstream of the proto-spacer adjacent motif (PAM) (5′–3′). In Zebrafish, 42.67% of CRISPR/Cas9 induced mutations affect all the four base pairs, 17.48% affect 3 of them, 14.91% affect 2 of them and 20.31% affected 1 of them (Yu et al. 2014). Based on this observation, if the 3′ end of primers are designed to exactly cover these MHSs, theoretically, the CRISPR/Cas9 induced indels could be detected, because mismatches in the 3′ end of the primer will greatly reduce or completely loss the PCR amplification efficiency. This principle has been widely employed to detect point mutation in various systems (Rigat et al. 1992; Orum et al. 1993). Considering all the above-mentioned phenomenon, in this study, we designed two pairs of primers aiming to screen for indels induced by CRISPR/Cas9 in rice. One pair of primers referred as outer primer (PoF/R) were designed to primer outside of the MHS region, and the other pair of primers so called inner primer (PiF/R) were designed to flank at the 3′ most nucleotide of the MHS, specifically the PiF primer was designed till the 4th nucleotide upstream of the PAM. Therefore, both wild-type and mutant alleles will be amplified by outer primer pairs, while inner primer pairs can amplify only wild-type allele (Fig. 1a). To simplify the PCR, the PiR and PoR sequence are identical (Fig. 1a). To develop and optimize this system in plant, genomic DNA from wild-type and a CRISPR/Cas9 induced mutant targeting SD1 with known indel information in rice were used. The results showed that by such a PCR amplification, CRISPR/Cas9 induced indels in rice can be easily detected (Supplementary Fig. 1a, b) as does in Zebrafish (Yu et al. 2014).
To validate the sensitivity or efficiency of developed PCR-based method for the indel mutant identification, a serial of mixed DNA samples containing different ratios of wild-type genomic DNA to mutant genomic DNA from a known 1 bp deletion homozygous rice SD1 mutant were used. The results showed that as the ratios of mutant DNA in the mixture increased, the amplification efficiency of inner primer decreased accordingly (Fig. 1b). Notably, when the ratio of wild-type DNA in the mixture reduced to zero (in the case of lane 10, a homozygous mutant), Pi/R primer pair produced no band (Fig. 1b). Using ImageJ software, we managed to quantify the relative band density of amplicons amplified using Pi/R primer pair in tested samples to that of wild-type. The results indicated that as compared with that of wild-type, all mixed samples (heterozygous mutants) showed significantly reduced Pi band intensity while homozygous line sample had zero Pi band intensity (Fig. 1c). These results suggest that, on one hand, the developed PCR-based method can qualitatively and quantitatively detect CRISPR/Cas9 induced mutants, and on the other hand, it can distinguish homozygous from heterozygous lines.
To further evaluate its efficiency in the qualitative detection of CRISPR/Cas9 induced deletion mutants, the developed PCR method was then applied to detect additional 4 available CRISPR/Cas9 induced SD1 deletion mutants. Clearly, this PCR-based method can effectively detect the deletion mutants, because even if the mutant contained only one nucleotide deletion, no PCR amplification was observed when Pi primer pairs were used (Fig. 1d). To confirm the PCR results, all four amplified PCR products using the Po/R primer pair were Sanger sequenced and proved to be homozygous indel mutations (Fig. 1e). The developed PCR-based method was additionally applied to identify CRISPR/Cas9 induced deletion mutants targeting several other genes in rice, all were approved to be successful and confirmed by Sanger sequencing (Supplementary Fig. 1c–e). Altogether, the abovementioned results indicated that the developed PCR-based method is efficient, accurate and highly sensitive method for the identification of CRISPR/Cas9 induced deletion mutants down to 1 bp in rice.
To examine its efficiency in the detection of CRISPR/Cas9 induced insertion and replacement mutants, additional four mutants with 1 and 5 bp insertion and five mutants with 1 bp replacement were used, respectively. While all mutants showed both Po and Pi amplicon bands as wild-type (Supplementary Figs. 2a, 3a), the intensities of the Pi band in all mutants were remarkably weaker than that of wild-type (Supplementary Figs. 2b, 3b), which were consistent with the result of Sanger sequencing (Supplementary Figs. 2c, 3c). Thus, this PCR-based method can identify insertion and replacement mutants down to 1 bp as well in rice.
Identification of CRISPR/Cas9 generated indels in rice by amplicon labelling-based method
The amplicon labelling method was first reported by Oetting et al. (1995) to reveal large indels in PCR products with polyacrylamide gel electrophoresis. Combined with capillary electrophoresis, this method can also resolve small indels down to ± 2 bp and ± 1 bp (Schuelke 2000; Andersen 2003; Zhang et al. 2015). Its application for indel detection in genome edited animal cells has also been reported (Lonowski et al. 2017). Here, the application of this amplicon labelling-based method to detect CRISPR/Cas9 induced mutants in rice was presented. For this purpose, three primers were used to amplify by PCR the fluorophore-labeled amplicons covering the CRISPR/Cas9 edited genomic site. The tri-primers included a pair of target allele specific forward (F) and reverse (R) primers and an additional universal 6-carboxyfluorescein (6-FAM) 5′-labelled primer containing the same sequence as an extension of the target allele specific forward primer (FamF). Fluorescent labelled amplicons obtained from mutant allele with base deletion or insertion could be shorter or longer, respectively, than amplicons obtained from wild-type allele, which can be size discriminated by a DNA sequenator (Gene Mapper) (Fig. 2a). To develop and optimize this tri-primer PCR system, genomic DNAs from wild-type and CRISPR/Cas9 induced SD1 mutants (− 4 bp and + 5 bp) were used for touch down PCR to amplify wild-type amplicons, shorter amplicons containing deletions or longer amplicons harboring insertions that labeled with 6-FAM. The results clearly discriminated insertion and deletion mutants from wild-type in rice (Supplementary Fig. 4a).
To investigate the sensitivity of developed amplicon labelling-based method, a serial of mixed samples containing different ratios of wild-type genomic DNA to mutant genomic DNA from a known rice homozygous mutant with 4 bp deletion were created and PCR amplification was performed. As shown in Fig. 2b, as the ratio of mutant DNA to wild-type DNA in the mixture reduced, the amplicon peak height for the mutant was significantly reduced. The sensitivity was also evaluated using an additional mutant with 5 bp insertion, which showed a similar result (Supplementary Fig. 4b). These results indicated that the developed amplicon labelling-based method is highly effective to detect CRISPR/Cas9 induced indel mutants in rice.
To test whether the developed amplicon labelling-based method is sensitive enough to simultaneously detect multiple mutations at a CRISPR/Cas targeted site, SD1 tri-primers were designed to amplify the mutated amplicons using a pooled DNA samples from five different mutations within the same target loci. The resulting fluorophore labelled amplicon peaks were analyzed, which identified various indels efficiently and accurately, defining indels down to ± 1 bp (Fig. 3c). The Sanger sequencing result of the individually amplified PCR products from these five tested mutants confirmed the identified results revealed by the developed amplicon labelling-based method (Fig. 3d). The developed amplicon labeling-based method was further applied to identify CRISPR/Cas9 induced mutations targeting LOC_Os06g04420 allele, in which all results of 3 identified mutations were also consistent with results from Sanger sequencing (Supplementary Fig. 4c, d).
Notably, this method was effective as well to distinguish homozygous from heterozygous mutants, in which heterozygous mutants (HE) exhibited two peaks while homozygous (HM) mutants exhibited only one peak prior or after the wild-type peak (Fig. 3e). The detected zygosity was consistent with results of Sanger sequencing (Fig. 3f). Abovementioned results indicated that the developed amplicon labelling-based method is sensitive, precise, and effective for CRISPR/Cas9 induced mutant detection. It is amenable for high throughput analysis and also useful for zygosity discrimination.
Discussion
As more and more CRISPR/Cas9 induced mutants were generated, effective, accurate and economic screening methods for homozygous and precisely knockout indel mutations became increasingly important not only in basic research, but also in application for breeding. Sequencing was usually the only choice for such a screening in plants during past years (Ma et al. 2015a, b), however, due to the limitation of mutagenesis efficiency and complex characteristics of the generated mutants, the cost of generating a desired indel mutant by CRISPR/Cas9 was far beyond of the characterization of a T-DNA mediated mutant (Ma et al. 2015a, b). Although it is easier to detect the rice mutants simply by PCR and sequencing, and then to decode the zygosity of the individual plant through DSDecode software automatically or manually (Liu et al. 2015; Ma et al. 2015a, b), it is time consuming, and costly, involving PCR amplification, Sanger sequencing, and subsequent decoding of the superimposed sequencing chromatograms of PCR products. Therefore, there is an urgent need to develop quick, effective, and economic methods for this purpose. It is reported that CASP method can be used for analysis of contaminated grains in rice, however, its application in screening for desired indels is not conclusive (Kohata et al. 2018). In this work, taking advantages of traditional PCR, we have developed two methods to effectively detect indel mutations generated by CRISPR/Cas9 in rice. Compared with reported qPCR (Peng et al. 2018) and HRM (Denbow et al. 2017) methods, both methods are economic and simple. Compared with other methods, such as ACT-PCR (Hua et al. 2017), CAPS (Nekrasov et al. 2013; Kohata et al. 2018), EMC (Nekrasov et al. 2013; Shan et al. 2014) and MSBSP-PCR (Guo et al. 2018), both methods are sensitive (down to 1 bp), accurate (consistent with Sanger sequencing result) and consistent (applicable for various genes). Notably, combining with Image J software, the PCR-based method can be used for identification of replacement mutants as well, showing a potential application in the detection of single nucleotide polymorphism (SNP) (Supplementary Fig. 3), and for zygosity analysis (Fig. 1b, c). It’s also worth noting that the amplicon labeling-based method is particularly useful for identification of concomitant mutations (Fig. 2c; Supplementary Fig. 4c) and homozygous mutants (Fig. 2e, f). Thus, methods developed in this study certainly benefit the rice genome editing research community to identify the targeted mutagenesis, especially in a pool of mutations including a high proportion of wild-type, for example, in chimeric lines.
On the other hand, the sensitivity and efficiency of PCR-based method is inevitably affected by the target sequence. Therefore, the developed PCR-based method is less effective or even unsuccessful if the target region contains mono-nucleotide repeats, or the MHS is mutated with identical nucleotide(s) insertion, or mutations occur outside of the predicted MHS. In addition, the developed PCR method is only applicable to identify mutations that are close to and upstream of the PAM. Nevertheless, these limitations of developed PCR-based method can be readily overcome by the amplicon labeling-based method, which can be easily accessible via commercial service.
Furthermore, the exact nucleotide change occurred in a mutant cannot be revealed by either of the two newly developed methods per se, it, however, can be solved by Sanger sequencing when a homozygous mutant is obtained. In this way, not only the time is saved, but the cost for sequencing is also saved. Because all reactions can be done in 96 wells, they both can be used for high throughput analysis. We believe that these two developed indels detection methods would facilitate rice functional studies and breeding using genome editing as a tool. Further studies will exam their applicability in the detection of indels induced by other genome editing systems in rice and other plants.
Author contribution statement
BS carried out most of the experiments and data analysis, and wrote the first draft of the manuscript. LR and YZ assisted in performing experiments, ZD helped in the data analysis, ZX and SJ designed the experiment and revised the manuscript.
References
Andersen PS, Jespersgaard C, Vuust J, Christiansen M, Larsen LA (2003) Capillary electrophoresis-based single strand DNA conformation analysis in high-throughput mutation screening. Hum Mutat 21:455–465
Denbow CJ, Lapins S, Dietz N, Scherer R, Nimchuk ZL, Okumoto S (2017) Gateway-compatible CRISPR-Cas9 vectors and a rapid detection by high-resolution melting curve analysis. Front Plant Sci 8:1171
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang L, Yang DA, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas9-induced gene modifications in Arabidopsis. PNAS 111(12):4632–4637
Findlay SD, Vincent KM, Berman JR, Lynne-Marie P (2016) A digital PCR-based method for efficient and highly specific screening of genome edited cells. PLoS One 11(4):e0153901
Guo J, Li K, Jin L, Xu R, Miao K, Yang F, Qi C, Zhang L, Botella JR, Wang R, Miao Y (2018) A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Methods 14:40
Harayama T, Riezman H (2017) Detection of genome-edited mutant clones by a simple competition-based PCR method. PloS One 12(6):e0179165
Hua Y, Wang C, Huang J, Wang K (2017) A simple and efficient method for CRISPR/Cas9-induced mutant screening. J Genet Genom 44:207–213
Huang MC, Cheong WC, Lim LS, Li MH (2012) A simple, high sensitivity mutation screening using Ampligase mediated T7 endonuclease I and Surveyor nuclease with microfluidic capillary electrophoresis. Electrophoresis 33:788–796
KC R, Srivastava A, Wilkowski JM, Richter CE, Shavit JA, Burke DT, Bielas LS (2016) Detection of nucleotide-specific CRISPR/Cas9 modified alleles using multiplex ligation detection. Sci Rep 6:32048
Kim JM, Kim D, Kim S, Kim JS (2014) Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat Commun 5:3157
Kohata R, Koitabashi K, Kitashiba H, Nishio T (2018) Sensitive mutant detection by concentrating mutant DNA with allelespecific capture and its application to analysis of contaminated grains in rice. Plant Cell Rep 37:865–872
Liu W, Xie X, Ma X, Li J, Chen J, Liu YG (2015) Dsdecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8(9):1431–1433
Lonowski LA, Narimatsu Y, Riaz A, Delay CE, Yang Z, Niola F, Duda K, Ober EA, Clausen H, Wandall HH, Hansen SH, Bennett EP, Frödin M (2017) Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat Protoc 12(3):581–603
Ma X, Chen L, Zhu Q, Liu Y (2015a) Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant 8(8):1285–1287
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015b) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Mazumdar S, Quick WP, Bandyopadhyay A (2016) CRISPR-Cas9 mediated genome editing in rice, advancements and future possibilities. Indian J Plant Physiol 21(4):437–445
Mock U, Hauber I, Fehse B (2016) Digital PCR to assess gene-editing frequencies (GEF-dPCR) mediated by designer nucleases. Nat Protoc 11(3):598–615
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plants Nicotiana benthamiana using Cas9 RNA-guided edonuclease. Nat Biotechnol 31:691–693
Oetting WS, Lee HK, Flanders DJ, Wiesner GL, Sellers TA, King RA (1995) Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30:450–458
Orum H, Nielsen PE, Egholm M, Berg RH, Buchardt O, Stanley C (1993) Single base pair mutation analysis by PNA directed PCR clamping. Nucl Acids Res 21(23):5332–5336
Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765
Peng C, Wang H, Xu X, Wang X, Chen X, Wei W, Lai Y, Liu G, Godwin ID, Li J, Zhang L, Xu J (2018) High-throughput detection and screening of plants modified by gene editing using quantitative real-time polymerase chain reaction. Plant J 95:557–567
Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z (2016) CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci Rep 6:32289
Rigat B, Hubert C, Corvol P, Soubrier F (1992) PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1). Nucl Acids Res 20(6):1433
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments a poor man’s approach to genotyping for research and high-throughput diagnostics. Nat Biotechnol 18:1–2
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9(10):2395–2410
Thomas HR, Percival SM, Yoder BK, Parant JM (2014) High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS One 9(12):e114632
Vouillot L, Thelie A, Pollet N (2015) Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5:407–415
Yu C, Zhang Y, Yao S, Wei Y (2014) A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One 9(6):e98282
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang Y, Catharina S, Camilla H, Lars H, Allan LT, Francesco N, Malene B, Vester C, Morten F, Henrik C, Hans HW, Eric PB (2015) Fast and sensitive detection of indels induced by precise gene targeting. Nucl Acid Res 43:e59
Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D, Meng S, Zhang X, Hu S, Tian Y (2014) An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep 4:6420
Zhu C, Bortesi L, Baysal C, Twyman RM, Fischer R, Capell T, Schillberg S, Christou P (2017) Characteristics of genome editing mutations in cereal crops. Trends Plant Sci 22(1):38–52
Acknowledgements
This work was supported by grants from the China National Transgenic Plant Special Fund (2016ZX08012-002) and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Laurence Tomlinson.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2019_2392_MOESM1_ESM.pdf
Optimization of PCR-based method and its application in the detection of CRISPR/Cas9 induced mutations. Amplicons of SD1 and other targeted genes were analyzed on 2% agarose gel electrophoresis analysis. a Optimization of PCR-based method using CRISPR/Cas9 induced SD1 mutations. Lane M, Trans 2K DNA marker; Lane 1, NTC (negative control); Lanes 2-5, wild-type (Wt), SD1 mutants with 1 bp, 2 bp, and 4 bp deletion, respectively. b Optimization of PCR-based method using CRISPR/Cas9 induced SD1 mutations. Lane M, Trans 2K DNA marker; Lane 1, NTC; Lanes 2-3, wild-type (Wt), SD1 mutants with 4 bp deletion and 3 bp insertion, respectively. c The application of the developed PCR based method in the detection of CRISPR/Cas9 induced mutations in LOC_Os03g12030. Lane M, Trans 2K DNA marker; Lane 1, NTC; Lane 2, wild-type (Wt); Lane 3, LOC_Os03g12030 mutant with 1 bp deletion. Left panels represent amplicons analyzed on 2% agarose gel electrophoresis, and right panels show Sanger sequencing results of corresponding mutants identified. d The application of the developed PCR based method in the detection of CRISPR/Cas9 induced mutations in LOC_Os09g08130. Lane M, Trans 2K DNA marker; Lane 1, NTC; Lane 2, wild-type (Wt); Lane 3, LOC_Os09g08130 mutant with 2 bp deletion. Left panels represent amplicons analyzed on 2% agarose gel electrophoresis, and right panels show Sanger sequencing results of corresponding mutants identified. e The application of the developed PCR based method in the detection of CRISPR/Cas9 induced mutations in LOC_Os07g26460. Lane M, Trans 2K DNA marker; Lane 1, NTC; Lane 2, wild-type (Wt); Lane 3-4, LOC_Os09g08130 mutants with 2 bp deletion and 2 bp deletion plus 1 bp insertion, respectively. Left panels represent amplicons analyzed on 2% agarose gel electrophoresis, and right panels show Sanger sequencing results of corresponding mutants identified. (PDF 346 KB)
299_2019_2392_MOESM2_ESM.pdf
Detection of insertion mutations in a single SD1 allele induced by CRISPR/Cas9 using developed PCR based method. a Analysis of PCR products amplified using SD1 Po and Pi primers on 2% agarose gel electrophoresis. Lane M, Trans 2K DNA marker; lane 2, wild-type (Wt); lane 3-6, SD1 mutants with insertions of 1A, 1T, 1G and 5A, respectively. b Quantification of the relative band intensities of corresponding Po and Pi PCR products in panel a using ImageJ. c Sanger sequencing results of corresponding PCR products in panel a. (PDF 200 KB)
299_2019_2392_MOESM3_ESM.pdf
Detection of replacement mutations in a single SD1 allele induced by CRISPR/Cas9 using developed PCR based method. a Analysis of PCR products amplified using SD1 Po and Pi primers on 2% agarose gel electrophoresis. Lane M, Trans 2K DNA marker; lane 2, wild-type (Wt); lane 3-7, SD1 mutants with base replacement of T to A and G to A, respectively. b Quantification of the relative band intensities of corresponding Po and Pi PCR products in panel a using ImageJ. c Sanger sequencing results of corresponding PCR products in panel a. (PDF 214 KB)
299_2019_2392_MOESM4_ESM.pdf
Optimization of amplicon labelling based method with SD1 mutants and its application in detection of a single LOC_Os06g04420 mutant induced by CRISPR/Cas9. a The optimization of the amplicon labelling based method. Genomic DNA from wild-type, CRISPR/Cas9 induced SD1 mutants with 4 bp deletion, and 5 bp insertion, respectively, was used for the PCR amplification using tri-primers, respectively, and resulting fluorophore labelled amplicons were analyzed in a fragment analyzer. X-axis and Y-axis represent amplicon size in base pairs and relative fluorescence units (RFU), respectively. Wt, wild-type; d4-HM, 4 bp deletion homozygous mutant; i5-HM, 5 bp insertion homozygous mutant. b Sensitivity test of the developed amplicon labelling method. A serial dilution of mixed samples containing 20%:80%; 10%:90%; 1%:99% and 0.1%:99.9% ratios of CRISPR/Cas9 induced SD1 mutant (Mt) (5 bp insertion) genomic DNA to wild- type (Wt) genomic DNA were used. The height of detected fluorophore labelled amplicon peak increased for wild-type but decreased for mutant alleles as the ratio of Mt:Wt in the mixture decreased. Red and green stars denote Mt allele and Wt allele, respectively. c Detection of mutations in a single LOC_Os06g04420 allele induced by CRISPR/Cas9. Fragment analysis of the fluorophore labelled amplicon of targets amplified using LOC_Os06g04420 tri-primers in a fragment analyzer. d Sanger sequencing results of corresponding mutations identified in c. (PDF 101 KB)
Rights and permissions
About this article
Cite this article
Biswas, S., Li, R., Yuan, Z. et al. Development of methods for effective identification of CRISPR/Cas9-induced indels in rice. Plant Cell Rep 38, 503–510 (2019). https://doi.org/10.1007/s00299-019-02392-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02392-3