Abstract
Key message
Using quantitative assays for autophagy, we analyzed 4 classes of atg mutants, discovered new atg2 phenotypes and ATG gene interactions, and proposed a model of autophagosome formation in plants.
Abstract
Plant and other eukaryotic cells use autophagy to target cytoplasmic constituents for degradation in the vacuole. Autophagy is regulated and executed by a conserved set of proteins called autophagy-related (ATG). In Arabidopsis, several groups of ATG proteins have been characterized using genetic approaches. However, the genetic interactions between ATG genes have not been established and the relationship between different ATG groups in plants remains unclear. Here we analyzed atg2, atg7, atg9, and atg11 mutants and their double mutants at the physiological, biochemical, and subcellular levels. Involvement of phosphatidylinositol 3-kinase (PI3K) in autophagy was also tested using wortmannin, a PI3K inhibitor. Our mutant analysis using autophagy markers showed that atg7 and atg2 phenotypes are more severe than those of atg11 and atg9. Unlike other mutants, atg2 cells accumulated several autophagic vesicles that could not be delivered to the vacuole. Analysis of atg double mutants, combined with wortmannin treatment, indicated that ATG11, PI3K, and ATG9 act upstream of ATG2. Our data support a model in which plant ATG1 and PI3K complexes play a role in the initiation of autophagy, whereas ATG2 is involved in a later step during the biogenesis of autophagic vesicles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cells possess enzymes to break down a variety of macromolecules. These enzymes are important for nutrient recycling and the elimination of toxic and damaged molecules. To prevent nonspecific degradation, catabolic activities in eukaryotic cells are tightly regulated and isolated either in specialized protein complexes (such as the proteasome) or in membrane-bound lytic compartments (the vacuole in plants and yeasts or the lysosome in metazoans).
Autophagy is a membrane trafficking route by which cytoplasmic materials are delivered to the vacuole/lysosome for degradation (Carlsson and Simonsen 2015; Michaeli et al. 2016). Autophagy is initiated at the phagophore, a membrane cisterna that subsequently expands to sequester a portion of the cytoplasm. The sequestering activity of the phagophore is completed when the growing border closes via membrane scission to generate a double-membrane structure called the autophagosome. After the autophagosome matures, its outer membrane fuses with the vacuolar membrane to release the autophagic body into the vacuolar lumen. Finally, the autophagic body is rapidly degraded by acid hydrolases in the vacuole. To observe autophagic bodies in plant cells, the low pH of the vacuole needs to be increased by treatment with concanamycin A (ConA), an inhibitor of vacuolar proton pumps.
Autophagy is controlled and executed by autophagy-related (Atg) proteins encoded by a set of core Atg genes which are conserved between yeasts and metazoans. These genes are typically classified into 5 groups (Fig. 1a), based on the phenotypic classes and functions of the proteins they encode (Shibutani and Yoshimori 2014). The first group (Fig. 1a, blue), containing Atg1, Atg13, and Atg17 (or its homolog Atg11), encodes subunits of a protein kinase complex, while proteins encoded by the second group (Vps34, Vps15, Vps30/Atg6, and Atg14) are subunits of the class III phosphatidylinositol 3-kinase (PI3K) complex I (Fig. 1a, green). In yeast and mammals, these two protein complexes are known to recruit downstream Atg proteins to the phagophore. Autophagy initiation also involves Atg9, which encodes the only transmembrane protein among the core Atg proteins (Fig. 1a, orange). The function of Atg9 is unclear but may encompass the transport of vesicles to nucleate the phagophore. The fourth group of Atg genes consists of Atg2 and Atg18 (Fig. 1a, magenta). Atg18 is a phosphatidylinositol 3-phosphate (PI3P) effector that interacts with Atg2. The precise functions of Atg2 and Atg18 are as yet unknown. Finally, the fifth group contains Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16 (Fig. 1a, yellow), all of which are essential for the conjugation of a ubiquitin-fold protein, Atg8, to phosphatidylethanolamine (PE). Atg8–PE, the product of the conjugation reaction, is a useful marker of autophagic membranes, since it is found on phagophores, autophagosomes, and autophagic bodies. Atg8–PE is proposed to be involved in almost all the steps of autophagosome dynamics, particularly later steps such as phagophore expansion and closure and autophagosome fusion. Atg8–PE also plays a role in cargo selection during various types of selective autophagy through direct interactions with cargo or via interaction with autophagy receptors.
Proteins encoded by core Atg genes and phenotypic analysis of representative atg mutants. a Diagram of core Atg proteins conserved among yeast, animals and plants. Five groups of core Atg proteins are color-coded: Atg1 kinase complex (blue), PI3K complex I (green), Atg9 (orange), Atg2–Atg18 complex (magenta), and Atg8/12 conjugation system (yellow). Protein names are based on yeast nomenclature and any proteins exclusive to yeast are not shown. For example, Atg29 and Atg31 are not indicated here, although they are components of the yeast Atg1 complex containing Atg17. Arrows indicate enzyme reactions involving Atg proteins. Vps34 is a lipid kinase for phosphatidylinositol (PI), producing phosphatidylinositol 3-phosphate (PI3P). Atg7 is an E1-like activase for both Atg8 and Atg12. Atg3 and Atg10 are E2-like conjugases for Atg8 and Atg12, respectively. Atg12 is conjugated to Atg5 and resulting Atg12–Atg5 conjugate interacts with Atg16, thereby enhancing another conjugation reaction between Atg8 and phosphatidylethanolamine (PE). Atg4, a limiting protease, has a dual function: cleaving C-terminal residues to allow Atg8–PE conjugation and cleaving PE from Atg8–PE to release and recycle Atg8 from autophagic membranes. b, c Graphs illustrating differential sensitivity of atg mutants to carbon starvation. Survival rate data plotted in b were used to determine Relative Survival Index (RSI; see Table 1 for the calculation) in c. At least 4 independent seed populations per genotype were tested in 8 trials. d Differential effects of Arabidopsis atg mutations on pexophagy, as assessed by anti-catalase immunoblot. Each lane has protein extracts from 5 mg FW (fresh weight) of 2-week-old seedlings with indicated genotypes. Intensity of catalase protein bands was normalized by that of Rubisco large subunit. Bars in the graph show mean ± SE calculated from 9 biological replicates (**P < 0.01; NS, not significantly different; Student’s t test). (Color figure online)
The main biochemical roles of core Atg genes appear to be conserved between yeast and mammalian cells. Hierarchical analysis of Atg genes in yeast (Suzuki et al. 2007) and of their homologs in mammals (Itakura and Mizushima 2010) also indicated that the basic hierarchy of core Atg genes in mammals is similar to that in yeast, yet there are yeast- and mammalian-specific factors involved in autophagosome dynamics. In addition, the detailed interactions among and regulation of the core Atg proteins may vary in different taxa. For example, yeast Atg9 was recently shown to be a substrate of Atg1 kinase (Papinski et al. 2014), and a mammalian Atg1 homolog, ULK1, phosphorylates not only mAtg9 homolog (Zhou et al. 2017) but also Bec-1/mAtg6 (Russell et al. 2013) and Vps34 homologs (Egan et al. 2015). Yeast Atg9 interacts with Atg2 and Atg18 (Wang et al. 2001), whereas such interactions have not been reported in mammals.
Like in yeast and animals, plant Atg genes are important for autophagy (Hanaoka et al. 2002; Michaeli et al. 2016). Mutations in core ATG genes in Arabidopsis cause defective autophagy and hypersensitivity to various types of nutrient, and to abiotic and biotic stress factors. Biochemical properties of many core ATG proteins were similar to their yeast and mammalian homologs. For example, in vitro reconstitution of Arabidopsis ATG8 conjugation demonstrated that ATG7 can act as an E1-like activating enzyme for ATG8 (Fujioka et al. 2008). Another example is that Arabidopsis ATG18a binds to PI3P and PI(3,5)P2 (Oxley et al. 2013). At least one gene from each core ATG group was characterized using viable Arabidopsis mutants, except the PI3K complex I group. Homozygous atg6 mutants cannot be retrieved, because ATG6 is essential for pollen germination (Fujiki et al. 2007). In yeast and mammals, Vps34, Vps15, and Vps30/Atg6 are also components of PI3K complex II, which has a main function in endosomal trafficking. Atg14 is an autophagy-specific component for yeast PI3K complex I (Fig. 1a), but no Atg14 homologs have been identified in plants. The PI3K inhibitor wortmannin (Wm) has been used to test the requirement of PI3K in autophagy (Merkulova et al. 2014; Zhuang et al. 2013). However, autophagy is not clearly inhibited until plants are treated with Wm for 4 h (Shin et al. 2014), and an indirect and off-target effect of Wm cannot be excluded after such a prolonged treatment.
It has been assumed that plant ATG proteins are organized, similar to yeast and mammalian ATG pathways; however, experimental evidence is lacking to support this assumption. In spite of a recent report of interaction between ATG9 and ATG5 in Arabidopsis during a specific type of induced autophagy (Zhuang et al. 2017), there have not been comprehensive studies comparing all groups of core ATG genes and investigating their genetic interactions during autophagy.
Here we tested genetic interactions of core ATG genes in Arabidopsis. We compared phenotypes of atg11, atg9, atg2, and atg7 single mutants and their double mutants. We observed over-accumulation of autophagic vesicles in atg2 mutants, which was suppressed by either Wm, atg11, or atg9. Based on these genetic data, we concluded that PI3K, ATG11 and ATG9 act upstream of ATG2 in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Transgenic Arabidopsis thaliana ProUBQ10::GFP-ATG8a (Kim et al. 2013) and T-DNA insertional mutants atg2-1 (Inoue et al. 2006), atg7-2 (Chung et al. 2010), atg9-3 (Shin et al. 2014), atg11-1 (Li et al. 2014), and atg18a-2 (Lenz et al. 2011), were previously described. Primers used for genotyping are listed in Supplementary Table S1. Arabidopsis seeds were surface-sterilized and stored at 4 °C for 3 days. The seeds were germinated in liquid Murashige and Skoog (MS) medium (1 × MS macronutrient salt with vitamins, 1% [w/v] sucrose, pH 5.7) or in solid MS medium containing 0.25% (w/v) Phytagel. Seedlings were incubated at 20–22 °C under a long-day condition (16 h-light/8 h-dark photoperiod).
For nitrogen starvation experiments, liquid MS medium was replaced by either fresh liquid MS or MS-N medium (MS micronutrient solution plus 3 mM CaCl2, 1.5 mM MgSO4, 1.25 mM KH2PO4, and 5 mM KCl, pH 5.7), and seedlings were further incubated for 48 h (Chung et al. 2010).
For carbon starvation experiments (Chung et al. 2010), seeds were germinated in solid MS–Suc medium (1 × MS macronutrient salt with vitamins, pH 5.7, 0.25% [w/v] Phytagel). After seedlings were grown for 14 days, plates containing seedlings were wrapped with aluminum foil and incubated for 6, 8, 10, or 12 days. Before wrapping, plates were inspected to mark seedlings showing retarded growth, which were excluded from analysis. After 7 days of recovery under the long-day condition, seedlings developing new leaves were considered to have survived starvation.
Confocal microscopy, image processing and quantification
Hypocotyl and root tissue were observed using a Zeiss 510 laser confocal microscope (Carl Zeiss). A 488-nm excitation line and a BP500-530IR emission filter were used to detect GFP-ATG8a signal. Confocal microscope images were processed with ImageJ (National Institutes of Health, NIH). Measure Tool was used to quantify fluorescence intensity. To analyze GFP-ATG8a puncta, the images were converted 8-bit and Analyze Particles Tool was used to identify a punctate signal.
Protein analysis
Whole seedlings were homogenized in Laemmli buffer and clarified by centrifugation at 16,000×g for 10 min. Protein extracts were separated by SDS–PAGE and transferred onto Immobilon-P polyvinylidene fluoride membranes (Millipore). Anti-GFP (Roche), anti-histone H3 (Abcam), anti-catalase (Agrisera), anti-VDAC1 (Agrisera), and anti-UGPase (Agrisera) antibodies were diluted at 1:1000. For ATG8 immunoblot analysis, 12% SDS–PAGE containing 6 M urea was used (Chung et al. 2010), and anti-ATG8 antiserum (Thompson et al. 2005) was diluted at 1:500. Protein band intensity was quantified using ImageJ (NIH).
For membrane preparation, whole seedlings were homogenized in TNPI buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM phenylmethyl sulfonyl fluoride, 10 mM iodoacetamide, pH 8.0) at 4 °C. Seedling extract was centrifuged at 2,000×g for 5 min, and supernatant was recovered to obtain total extract (T). After subsequent centrifugation at 100,000 × g for 1 h, pellet containing membrane preparation (M) and supernatant containing soluble proteins (S) were analyzed using SDS–PAGE.
Results
Phenotypic severity varies in different groups of core atg mutants
To test genetic interactions among core ATG genes in Arabidopsis, we chose representatives from four groups of ATG genes: ATG11 from those encoding ATG1 complex; ATG9; ATG2 from ATG2–ATG18 complex and ATG7 from ATG8/12 conjugation system. Using transcript-null alleles of these single-copy genes (Chung et al. 2010; Inoue et al. 2006; Li et al. 2014; Shin et al. 2014), we determined phenotypic classes for each mutant. For the PI3K complex, we took a pharmacological approach, where wild type and mutant seedlings were treated with wortmannin (Wm), a PI3K inhibitor (Merkulova et al. 2014; Zhuang et al. 2013).
Representative mutants of atg7, atg2, atg9, and atg11 were subjected to carbon starvation, a phenotypic assay for autophagy mutants (Fig. 1b). For each mutant, we calculated Relative Survival Index (RSI), which quantifies its tolerance to carbon starvation, relative to wild type (Table 1). RSI of atg7 and atg2 were 13 and 19% of wild type, respectively, whereas those of atg9 and atg11 were more than 60% (Table 1; Fig. 1c), confirming phenotypic data from previous studies (Li et al. 2014; Shin et al. 2014).
To test the effect of core atg mutations on selective autophagy, we performed anti-catalase immunoblot analysis. Two independent, forward genetics studies (Farmer et al. 2013; Shibata et al. 2013) identified new alleles of atg7, atg3, atg2, and atg18a mutants and showed that these mutants accumulated excess peroxisomes and peroxisomal proteins like catalases, indicating that selective autophagy of peroxisomes, or pexophagy, depends on the ATG8 conjugation system and ATG2–ATG18 complex. We found that protein extract from 2-week-old wild-type seedlings contained a much lower catalase content than atg7 and atg2, but only slightly lower than atg9 and atg11 (Fig. 1d).
These differential RSIs and catalase levels among core atg mutants indicate that the loss of either ATG8/12 conjugation or ATG2 function leads to a higher sensitivity to carbon limitation and a more severe defect in pexophagy, compared to the loss of either ATG1/13 or ATG9 function. Consistent with this proposal, phenotypes of atg18a were stronger than those of atg9 and atg11 were (Fig. 1c, d). Because ATG18A is one of eight members in ATG18 multigene family and thus, potentially redundant (Xiong et al. 2005), it was not surprising to find that phenotypes of atg18a were weaker than those of atg2, which is a mutation in a single-copy gene.
Differential autophagic flux in atg mutants correlates with their phenotypic severity
To quantify inhibitory effects of various atg mutations on autophagy, we used atg7, atg9, atg11, atg2, and atg18a homozygous mutants expressing autophagy marker GFP-ATG8a, whose transgene expression is driven by UBQ10 promoter (Kim et al. 2013). To ensure equal transgene expression, a single transgenic line proUBQ10::GFP-ATG8a was introduced to atg mutants by genetic crosses.
Uniform fluorescence and antibiotics resistance prior to analysis confirmed homozygosity of GFP-ATG8a transgene in each homozygous mutant population.
Autophagy in Arabidopsis was monitored by the GFP-ATG8 processing assay (Chung et al. 2010; Shin et al. 2014), in which autophagic flux is estimated by the level of free GFP moiety cleaved from GFP-ATG8 by vacuolar proteases. Mutants in ATG8 conjugation system, such as atg7 and atg5, are used as a negative control for the GFP-ATG8 processing assay, because GFP-ATG8 in these mutants cannot be conjugated to PE and, thus, remains mostly as a full-length protein. As we noted previously (Shin et al. 2014), atg7 and atg9 mutants often accumulated partial proteolytic products of GFP-ATG8a (Fig. 2a, b; indicated by brackets). These partial products were prominent also in atg2, atg11, and atg18a and run slightly slower than the free GFP moiety (Fig. 2a, b; indicated by arrowheads), which was most conspicuous in wild type, but barely detected in atg7.
Autophagic flux analysis of atg seedlings by the GFP-ATG8 processing assay. a, b Anti-GFP immunoblot was performed using protein extracts of wild-type (WT) or various atg seedlings expressing ProUBQ10::GFP-ATG8a. 9-day-old seedlings were incubated in nitrogen-sufficient (a) or -deficient (b) medium for 2 days prior to protein extraction. Anti-histone H3 immunoblots are shown as a loading control. c, d Quantification of the protein bands shown in a, b, respectively. Intensity of bands corresponding to free GFP moiety (solid arrowheads), normalized by intensity of full-length GFP-ATG8a bands (open arrowheads), is plotted (mean ± SE; n = 4 [c] or 5 [d]). Columns marked with asterisks represent means of mutants significantly different from WT according to Student’s t test (*P < 0.05; **P < 0.01). NS not significantly different. Protein bands indicated by brackets in a and b were excluded from our analysis, because they represent unstable polypeptides partially processed from GFP-ATG8a, independently of autophagy
The level of free GFP moiety was significantly reduced in all atg mutants that were hydroponically grown in a nitrogen-sufficient condition (Fig. 2a, c). As previously noted (Suttangkakul et al. 2011), nitrogen deprivation led to an increase in the free GFP band in the wild type (Fig. 2b). This increase was also detected in atg9 and atg11 but only partially in atg18a and atg2 (Fig. 2b). Apparently, the ratio of free GFP to GFP-ATG8a band intensity of a given atg mutant in this autophagy-inducing condition correlates with their RSI (compare Fig. 2d with Fig. 1c), despite different nutrient stresses (nitrogen versus carbon starvation).
Endogenous ATG8–PE accumulates in atg2 and atg18a membrane
To understand mechanisms for differential autophagic flux in various atg mutants, we characterized biochemically endogenous ATG8 proteins. Previous studies showed that endogenous ATG8 levels increased in many core atg mutants like atg7 and that conjugation of ATG8 to PE is defective in atg7 but normal in atg11 (Chung et al. 2010; Li et al. 2014). We found that atg2 and atg18a contained a high level of endogenous ATG8, especially ATG8–PE (Fig. 3a, indicated by solid arrowheads; Fig. 3b), which was enriched in a membrane preparation (Fig. 3c). Similarly, atg2 plants expressing GFP-ATG8a also had a significant level of GFP-ATG8a in a membrane fraction, whereas wild-type membrane barely contained GFP-ATG8a (Fig. 3d). Furthermore, all atg mutants tested, except conjugation-incompetent atg7, had a somewhat higher level of ATG8–PE in membrane fractions than the wild type (Fig. 3b, c). These data suggested that ATG8–PE in atg2 and atg18a is incorporated into either the phagophore or aberrant autophagic precursor membrane, whereas unconjugated ATG8 in atg7 cannot be targeted to any membrane.
Endogenous ATG8–PE is associated with membrane in wild type (WT) and atg2, atg9, atg11, and atg18a mutants. a Anti-ATG8 immunoblot showing unconjugated ATG8 (open arrowheads) and PE-conjugated ATG8 (black arrowheads). Each lane contains protein extract obtained from 15 mg FW seedlings grown in solid MS medium for 9 days. b Quantification of endogenous ATG8–PE level in WT and atg mutants, using anti-ATG8 immunoblot analysis in a. Gray columns represent the intensity of ATG8–PE bands normalized by UGPase band intensity (mean ± SE; n = 3 [for atg18a] or 5 [for others] biological replicates). Means of atg2 and atg18a mutants were significantly different from WT, according to Student’s t test (*P < 0.05; **P < 0.01). NS not significantly different. ND not determined. The white column for atg7 represents the normalized intensity of unstable ATG8 protein band co-migrating with ATG8–PE bands in WT and other mutants. c, d Anti-ATG8 (c) and anti-GFP (d) immunoblot analysis of total extract (T) and soluble (S) and 100K membrane (M) fractions prepared from seedlings of indicated genotypes. Anti-UGPase (cytosolic) and anti-VDAC1 (mitochondrial) immunoblots are shown as controls for soluble and membrane fractions, respectively
PI3K activity is required for the accumulation of autophagic vesicles in atg2 and atg18a
To gain insight into the relation of the PI3K complex with other groups of core ATG proteins, we investigated the effects of the PI3K inhibitor Wm on autophagic flux in wild type and various atg backgrounds (Fig. 4a). The Wm effect is most obvious when GFP-ATG8a transgenic seedlings are treated with ConA for 16 h (Shin et al. 2014). As expected, Wm effect on the ratio of free GFP to GFP-ATG8a was evident in the transgenic seedlings with wild-type background (Fig. 4a, b). In contrast, none of the atg mutants showed any significant Wm effect, although weaker mutants like atg9 and atg11 may have a residual sensitivity to Wm (Fig. 4a, b).
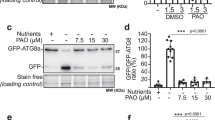
PI3K is required for efficient autophagy in the wild type and for over-accumulation of GFP-ATG8 puncta in atg2 and atg18a. a, b GFP-ATG8a processing assay to show the effect Wm on autophagic flux of wild type (WT) and various atg mutants. Anti-GFP immunoblot analysis (a) was performed using protein extracts from 9-day-old seedlings that were N-starved for 2 days and incubated with 0.5 µM ConA ± 30 µM Wm 16 h. Anti-histone H3 immunoblots are shown as a loading control. b The graph shows the ratios of free GFP (solid arrowheads) to full-length GFP-ATG8a (open arrowheads) band intensities (mean ± SE; n = 4 [atg18a] or 5 [other mutants and WT]). Protein bands indicated by brackets in a were excluded from analysis because they represent unstable, partially processed polypeptides from GFP-ATG8a, independently of autophagy. c–f Microscopic observation of GFP-ATG8a puncta in wild-type (WT), atg7, atg2, atg9, atg11, and atg18a hypocotyl cells. c Confocal images of hypocotyl cells of indicated genotypes expressing ProUBQ10::GFP-ATG8a. Seedlings were incubated in MS liquid medium for 9 days and treated with either DMSO (upper row) or 30 µM Wm (lower row) for 1 h prior to observation. Scale bar 5 µm. Graphs show intensity of GFP-ATG8a fluorescence (d), punctum number per 50,625 µm2 (e), and punctum area (f), calculated from confocal images like ones shown in c (mean ± SE; n = 17 images or more for DMSO controls and 10 images or more for Wm-treated samples). b, d–f Columns marked with asterisks at the bottom represent means significantly different from WT (genetic effect) or DMSO (drug effect) controls according to two-way (genotype × drug) ANOVA followed by Tukey’s test (*P < 0.05; **P < 0.01; N not significant)
We compared the effect of atg mutations on autophagic vesicles by observing wild-type and atg seedlings expressing GFP-ATG8a by confocal microscopy. Using images obtained from roots (Supplementary Fig. S1) and hypocotyls (Fig. 4c–f), we quantified overall intensity of GFP-ATG8a fluorescence, which appeared to correlate with autophagy inhibition; wild-type cells showed a weaker GFP-ATG8a signal than any atg mutant did (Fig. 4d and Supplementary Fig. S1d; white columns). Similarly, autophagic inhibition was obvious when the atg mutants expressing GFP-ATG8a were incubated with ConA to stabilize autophagic bodies in the vacuole (Supplementary Fig. S1b). Autophagic bodies were most abundant in wild-type root cells, whereas they were absent in atg7, very rare in atg2 and atg18a, and occasionally observed in atg11, but easily detected in atg9. Although quantification of autophagic bodies was not feasible due to a variable effect of ConA, these data suggested that the increased intensity of GFP-ATG8a in atg mutants resulted from inefficient targeting of GFP-ATG8a into the vacuole for degradation.
In addition to overall fluorescence, we determined the abundance and dimension of GFP-ATG8a puncta, which presumably represent phagophores and autophagosomes in the wild type. GFP-ATG8a puncta were detected in hypocotyl and root cells of wild type, atg9, and atg11, but significantly more were observed in other atg mutants, especially in atg2 (Fig. 4e and Supplemental Fig. S1e; white columns). The nature of the puncta in atg7, atg2, and atg18a is not clear, but the GFP-ATG8a puncta in atg7 are unlikely functional autophagosomes, considering the lack of autophagic bodies in atg7 (Supplementary Fig. S1b) and their much larger size than wild-type puncta (Fig. 4f and Supplemental Fig. S1f; white columns). It is noteworthy that previous studies suggested that atg4a atg4b double mutant, which, similarly, cannot conjugate ATG8 to PE (Chung et al. 2010), accumulate GFP-ATG8 aggregates (Yoshimoto et al. 2004). The puncta in atg2 and atg18a were smaller than those in atg7 were (Fig. 4f), suggesting that the nature of GFP-ATG8a puncta in atg2 differs from that of the atg7 puncta.
Treatment with Wm for 5 h reduced the number of cytoplasmic GFP-ATG8a puncta in nutrient-deprived wild-type roots (Merkulova et al. 2014) and completely inhibited accumulation of autophagic bodies in ConA-treated roots (Zhuang et al. 2013). Remarkably, accumulation of GFP-ATG8a puncta in atg2 and atg18a mutants was suppressed by treatment with Wm for 1 h (Fig. 4e; Supplementary Fig. S1e; white and gray columns), indicating that PI3K is required for the formation of GFP-ATG8a puncta in atg2 and atg18a backgrounds. In contrast, Wm did not affect the abundance of GFP-ATG8a puncta in atg7, again supporting the notion that GFP-ATG8a puncta in atg2 and atg7 have different properties.
Taken together, our microscopic and biochemical analysis indicated that in atg2 and atg18a mutants, ATG8-containing autophagic vesicles accumulated in the cytoplasm, because the vesicles cannot be delivered to the vacuole. Our data also suggested that PI3K is required for the formation of GFP-ATG8a puncta in atg2 and atg18a.
ATG11 and ATG9 are required for the accumulation of autophagic vesicles in atg2
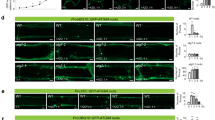
To identify genetic interactions between core ATG genes, we carried out an epistasis analysis using atg single and double mutants. By genotyping progenies from genetic crosses between atg2, atg7, atg9, and atg11 expressing proUBQ10::GFP-ATG8a transgene, we isolated six combinations of double homozygous mutants: atg2 atg7, atg2 atg9, atg2 atg11, atg7 atg9, atg7 atg11, and atg9 atg11. Confocal microscopy of hypocotyl (Fig. 5) and root (Supplementary Fig. S2) revealed that double mutants of atg2 atg7, atg7 atg9, and atg7 atg11 mostly looked like atg7 single mutant. This is not surprising, because GFP-ATG8a should not be conjugated to PE (and thus either remains in the cytosol or forms aggregates) in these genotypes. Interestingly, abundance of GFP-ATG8a puncta in atg2 atg11 and atg2 atg9 double mutants was reduced to ~ 20 to 25 and ~ 40 to 60%, respectively, when compared to atg2 (Fig. 5b and Supplementary Fig. S2b), indicating that ATG11 and, to a lesser extent, ATG9 are required for the over-accumulation of GFP-ATG8a puncta in atg2 single mutants. GFP-ATG8a puncta were significantly larger in these double mutants than in atg2 single mutants (Fig. 5c and Supplementary Fig. S2c).
Microscopic observation of GFP-ATG8a puncta in hypocotyl cells of wild type (WT) and atg single and double mutants. a Confocal images of hypocotyl cells of indicated genotypes expressing ProUBQ10::GFP-ATG8a. Seedlings were incubated on MS solid medium for 9 days prior to observation. Scale bar = 5 µm. b, c Graphs showing GFP-ATG8a punctum abundance per 50,625 µm2 (b) and dimension (c), calculated from confocal images like ones shown in panel a (mean ± SE; n = 9 images or more). Column pairs marked with asterisks represent significantly different means between single and double mutants, according to two-way (genotype 1 × genotype 2) ANOVA followed by Tukey’s test (*P < 0.05; **P < 0.01)
To see if PI3K action depends on ATG11 and ATG9, we tested Wm effect on atg2 atg9 and atg2 atg11 double mutants. GFP-ATG8a-expressing atg2 single mutants and atg2 atg9 and atg2 atg11 double mutants were treated with Wm for 1 h, before observation of their hypocotyl cells by confocal microscopy (Fig. 6a). These double mutants were still sensitive to Wm (Fig. 6b), indicating that PI3K activity is required for the accumulation of GFP-ATG8a puncta in atg2 atg11 and atg2 atg9 double mutants. Wm did not significantly change the dimension of GFP-ATG8a puncta in hypocotyl cells of atg2 single and double mutants (Fig. 6c), as in wild type (Fig. 4f).
The requirement of PI3K for autophagic vesicle accumulation in atg2 is independent of ATG9 and ATG11. a Confocal images of DMSO-or Wm-treated hypocotyl cells of atg2, atg2 atg9, and atg2 atg11 seedlings expressing GFP-ATG8a. The seedlings were incubated in MS liquid medium for 9 days and treated with either DMSO or 30 µM Wm for 1 h prior to observation. Scale bar = 5 µm. b, c Graphs showing abundance (b) and dimension (c) of GFP-ATG8a puncta (mean ± SE; n = 9 images or more). Column pairs marked with asterisks represent significantly different means between single and double mutants, according to two-way (genotype 1 × genotype 2) ANOVA followed by Tukey’s test (*P < 0.05; **P < 0.01). d A model of autophagy in Arabidopsis. Early (autophagy initiation) events during autophagosome formation involve ATG1 and VPS34 complexes and ATG9. Block arrows indicate genetic interactions inferred from this study. The dashed arrows represent possible regulation among core ATG proteins, as suggested by studies using yeast and mammalian homologs
To summarize, our double mutant analysis detected genetic interactions between atg2 and atg11, and between atg2 and atg9, showing that ATG11 (and by inference, ATG1 complex function) and ATG9 are necessary for the formation of excess GFP-ATG8a puncta in atg2. PI3K appears to act upstream of ATG2, independently of ATG11 and ATG9.
Discussion
Technical challenges have delayed a clear understanding of how plant ATG genes are organized to control and execute autophagy. Unlike the case for yeast, a complete set of core atg mutants is not available for Arabidopsis. In addition, autophagy markers for plant cells are scarce and we have not yet developed enough quantitative methods for assessing autophagy induction and inhibition. Here we attempted to overcome these challenges by using a PI3K inhibitor and representative atg mutants, including recently identified atg11 (Li et al. 2014) and by developing quantitative methods employing GFP-ATG8, the most widely used marker for plant autophagy. Our finding that atg2 and atg18a mutants accumulate numerous GFP-ATG8a puncta in the cytoplasm was fundamental to double mutant analysis and PI3K inhibitor study. We present a model of autophagy hierarchy in Arabidopsis (Fig. 6d), based on data from the genetic analysis we conducted. We propose that PI3K, ATG11, and ATG9 are involved in autophagy initiation, whereas ATG2 is necessary for a later step during autophagosome biogenesis.
ATG2–ATG18 for phagophore expansion and/or autophagosome maturation
Several lines of evidence support the hypothesis that initial formation of the phagophore is normal in Arabidopsis atg2 and atg18a mutants, but autophagosome formation is hampered at either the expansion or the maturation stage (Fig. 6d). Our immunoblot analysis showed that atg2 and atg18a contain higher levels of full-length GFP-ATG8a (Fig. 2) and endogenous ATG8–PE (Fig. 3a) than the wild type. ATG8–PE is enriched in atg2 membrane (Fig. 3c), implying that ATG8–PE in atg2 is efficiently targeted to membrane, similar to the wild type. A fraction of GFP-ATG8a was also detected in atg2 membrane (Fig. 3d). A ~ 50-fold increase in the abundance of GFP-ATG8a puncta in atg2 compared to wild type (Fig. 4e and Supplemental Fig. S1e) is also consistent with our hypothesis. atg2 and atg18a contained few autophagic bodies (Supplemental Fig. S1b), indicating that most of the cytoplasmic autophagic vesicles in them did not reach the vacuolar lumen. As a result, autophagic flux (Fig. 2), adaptation to C starvation (Fig. 1c), and pexophagy (Fig. 1d) (Farmer et al. 2013; Shibata et al. 2013) were severely compromised in atg2. Our proposed role of ATG2 and ATG18A at the late stage of autophagosome biogenesis is consistent with a previous observation that atg2 leaf cells accumulated ATG8-positive, phagophore-like structures near the ER and aggregate-containing peroxisomes (Yoshimoto et al. 2014).
ATG1 and PI3K complexes and ATG9 direct autophagy initiation
Abundant GFP-ATG8a puncta in atg2 background were helpful to map ATG1 and PI3K complexes and ATG9 at early steps of autophagosome biogenesis. atg2 atg11 double mutants had much less GFP-ATG8a puncta than atg2 single mutants (Fig. 5b and Supplemental Fig. S2b), although atg11 single mutant did not show a significant difference from the wild type (Fig. 4e and Supplemental Fig. S1e). As we discussed earlier, phagophore initiation may not be greatly affected in atg2 single mutants, but inefficient expansion/closure of phagophore membrane may lead to accumulation of small GFP-ATG8 puncta. In atg2 atg11, the initiation step may be defective due to atg11, which masks the effect of atg2 mutation. Similarly, PI3K inhibitor Wm also suppressed the accumulation of GFP-ATG8 puncta in atg2 (Fig. 4e and Supplemental Fig. S1e). Thus, our observation is in line with a model in which, the ATG1–ATG11–ATG13 and the PI3K complexes, direct phagophore initiation at a proper site (Fig. 6d). A similar model was proposed for the mammalian ULK1/ATG1 and autophagic PI3K complexes, which synergistically act to promote phagophore initiation (Carlsson and Simonsen 2015).
Our double-mutant analysis also indicated that ATG9 acts upstream of ATG2 (Fig. 5b and Supplemental Fig. S2b). Although we do not have direct evidence revealing the relation among ATG1 and PI3K complexes and ATG9, our genetic data favor independent actions by these 3 groups of ATG genes. Double mutants of atg2 atg11 and atg2 atg9 still showed sensitivity to Wm, although they were less sensitive than atg2 single mutant was (Fig. 6b). Consistent with our result, Zhuang et al. (2017) recently reported that Wm and atg9 differentially affected appearance of YFP-ATG18a puncta. Since a mammalian Atg9 homolog is phosphorylated by ULK1 (Zhou et al. 2017), it will be interesting to test whether plant ATG1 also regulates ATG9 by direct phosphorylation (Fig. 6d).
Single mutations in ATG11 and ATG9 did not show a pronounced effect on autophagy, except when they were assessed by the GFP-ATG8 processing assay in a non-inducing condition (Fig. 2c). Notably, Zhuang et al. (2017) also reported that abnormal ER tubules labeled with YFP-ATG8e were abundant in atg9 roots when autophagy was induced by drugs like benzothiadiazole and dithiothreitol. Wm and atg5 inhibited the formation of the ER tubules in atg9. However, it is unknown whether abnormal ER tubules can be observed when atg9 seedlings are grown under fed or starving conditions. We detected a tubule-like GFP-ATG8a signal in atg9 seedlings grown hydroponically (Supplementary Fig. S1g), although they were too rare to be quantified.
We do not know the nature of enlarged GFP-ATG8a puncta in atg2 atg9 and atg2 atg11 double mutants (Fig. 5c and Supplementary Fig. S2c). However, because autophagy is severely inhibited in these mutants, it is tempting to speculate that the enlarged puncta are not functional autophagosomes, but possibly represent ectopic protein aggregates that would normally be degraded by autophagy, like GFP-ATG8a puncta in atg4a atg4b double mutants (Yoshimoto et al. 2004).
ATG8 functions remain unknown
Unfortunately, our study hardly provided further insight as to where ATG7 acts in the plant genetic pathway for autophagy. As we used GFP-ATG8 as an autophagic marker, it is not surprising that atg7 mutation, which blocks ATG8 conjugation to PE, overrides effects of the other mutations. Clearly, future experiments with autophagic markers other than ATG8 will help determine the functions of ATG8.
Indicators of autophagy inhibition and induction
In this study, we tested several parameters to quantify autophagy inhibition in core atg mutants. The amount of free GFP processed from GFP-ATG8 has been used to estimate autophagic flux in Arabidopsis (Shin et al. 2014), but here we found that a ratio of free GFP to GFP-ATG8 is also a useful and more sensitive parameter to detect moderate inhibition of autophagy, as in atg9 and atg11 mutants (Fig. 2).
We found that overall intensity of GFP-ATG8 fluorescence is a convenient indicator of autophagy inhibition (Fig. 4d and Supplementary Fig. S1d). We speculate that impaired autophagic flux in atg mutants results in the accumulation of cytosolic GFP-ATG8a, which is responsible for the increased fluorescence in the mutants and is consistent with our anti-GFP immunoblot data (Fig. 2c).
In contrast, the abundance of GFP-ATG8 puncta, correlates poorly with the extent of autophagy inhibition in atg mutations. Although atg7 showed slightly more severe phenotypes and more defective autophagy than atg2 (Figs. 1, 2), punctum number in atg7 was smaller than it was in atg2, and marginally increased compared to wild type. Autophagic flux is significantly decreased in atg9 and atg11, but there was no striking or consistent difference in the abundance and dimension of GFP-ATG8a puncta between wild type and single atg9 and atg11 mutants (Figs. 4, 5 and Supplementary Figs. S1, S2).
The abundance of GFP-ATG8a puncta is not a reliable indicator of autophagy induction either. We incubated wild-type seedlings under a nitrogen-starvation condition for 2 days to increase autophagic flux (compare Fig. 2c with 2d), but GFP-ATG8a puncta remained rather scarce (Supplementary Fig. S1e). From a technical prospect, our data again raise concern about using the abundance of GFP-ATG8 puncta as a sole criterion for autophagy induction.
Author contribution statement
TC and KDS conceived and designed research. SK, JHK, and KDS conducted experiments. SK and TC analyzed data. TC and SK wrote the manuscript. All authors read and approved the manuscript.
References
Carlsson SR, Simonsen A (2015) Membrane dynamics in autophagosome biogenesis. J Cell Sci 128:193–205
Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62:483–493
Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell 59:285–297
Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B (2013) Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25:4085–4100
Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143:1132–1139
Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F (2008) In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 283:1921–1928
Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129:1181–1193
Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47:1641–1652
Itakura E, Mizushima N (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776
Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25:4956–4966
Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, Molina A, Escudero V, Shindo T, van der Hoorn RA, Gust AA, Nurnberger T (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66:818–830
Li F, Chung T, Vierstra RD (2014) AUTOPHAGY-RELATED (ATG)11 plays a critical role in general autophagy and senescence-induced mitophagy in Arabidopsis. Plant Cell 26:788–807
Merkulova EA, Guiboileau A, Naya L, Masclaux-Daubresse C, Yoshimoto K (2014) Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol 55:715–726
Michaeli S, Galili G, Genschik P, Fernie AR, Avin-Wittenberg T (2016) Autophagy in plants—what’s new on the menu? Trends Plant Sci 21:134–144
Oxley D, Ktistakis N, Farmaki T (2013) Differential isolation and identification of PI(3)P and PI(3,5)P2 binding proteins from Arabidopsis thaliana using an agarose-phosphatidylinositol-phosphate affinity chromatography. J Proteom 91:580–594
Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C (2014) Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 53:471–483
Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750
Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25:4967–4983
Shibutani ST, Yoshimori T (2014) A current perspective of autophagosome biogenesis. Cell Res 24:58–68
Shin KD, Lee HN, Chung T (2014) A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 37:399–405
Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23:3761–3779
Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12:209–218
Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138:2097–2110
Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA Jr, Klionsky DJ (2001) Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem 276:30442–30451
Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42:535–546
Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16:2967–2983
Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y (2014) Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 127:1161–1168
Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, Luo Q, Feng D, Zhu Y, Chen Q (2017) Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res 27:184–201
Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25:4596–4615
Zhuang X, Chung KP, Cui Y, Lin W, Gao C, Kang BH, Jiang L (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA 114:E426-E435
Acknowledgements
We thank Dr. Richard Vierstra for providing ATG8 antiserum. This work is supported by grants NRF-2014R1A1A1A05003740 and “Cooperative Research Program for Agriculture Science & Technology Development (PJ01110801), Rural Development Administration, Korea” to T.C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Communicated by Youn-Il Park.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kang, S., Shin, K.D., Kim, J.H. et al. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep 37, 653–664 (2018). https://doi.org/10.1007/s00299-018-2258-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2258-9