Abstract
Key message
Ectopic expression of auxin glycosyltransferase UGT84A2 in Arabidopsis can delay flowering through increased indole-3-butyric acid and suppressed transcription of ARF6, ARF8 and flowering-related genes FT, SOC1, AP1 and LFY.
Abstract
Auxins are critical regulators for plant growth and developmental processes. Auxin homeostasis is thus an important issue for plant biology. Here, we identified an indole-3-butyric acid (IBA)-specific glycosyltransferase, UGT84A2, and characterized its role in Arabidopsis flowering development. UGT84A2 could catalyze the glycosylation of IBA, but not indole-3-acetic acid (IAA). UGT84A2 transcription expression was clearly induced by IBA. When ectopically expressing in Arabidopsis, UGT84A2 caused obvious delay in flowering. Correspondingly, the increase of IBA level, the down-regulation of AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8, and the down-regulation of flowering-related genes such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1(SOC1), APETALA1 (AP1), and LEAFY(LFY) were observed in transgenic plants. When exogenously applying IBA to wild-type plants, the late flowering phenotype, the down-regulation of ARF6, ARF8 and flowering-related genes recurred. We examined the arf6arf8 double mutants and found that the expression of flowering-related genes was also substantially decreased in these mutants. Together, our results suggest that glycosyltransferase UGT84A2 may be involved in flowering regulation through indole-3-butyric acid-mediated transcriptional repression of ARF6, ARF8 and downstream flowering pathway genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants go through a vital physiological change as they transition from vegetative growth to reproductive development. The floral transition is controlled by a complex regulatory network (Srikanth and Schmid 2011; Li et al. 2016; Wang 2014). There are at least four main pathways controlling flowering in the model plant Arabidopsis thaliana, including the vernalization pathway, gibberellin pathway, photoperiod pathway and autonomous pathway (Bastow et al. 2004; Gendall et al. 2001; Murase et al. 2008; Achard et al. 2004; Monte et al. 2003; Corbesier et al. 2007). In Arabidopsis thaliana, many essential molecular candidates for mediating these pathways have recently been uncovered. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) plays an integral role in the flower transition process and in the induction of downstream flowering signal of FLOWERING LOCUS T (FT) (Cho et al. 2016; Abe et al. 2005). The plant-specific transcription factor LEAFY (LFY) and APETALA1 (AP1) are two key regulators controlling flower formation and flower patterning in Arabidopsis. AP1 can act both in parallel with LFY or as a direct target of LFY (Yamaguchi et al. 2012; Weis et al. 2008; Winter et al. 2015). The LFY gene plays an important role in promoting floral transition and flower formation by interaction and coordination with other genes, such as AP1, FT, PI, CO and GA1 (Siriwardana et al. 2012; Campbell et al. 1997).
Plant hormone auxin plays indispensable roles in every aspect of plant life (Vanneste and Friml 2009; Scarpella et al. 2010). Auxin directs both cell elongation and cell division specifically in response to the environment, thus regulating critical aspects of plant growth and development (Ludwig-Müller 2000; Liu et al. 2012; Strader et al. 2011). Indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA) are two main types of auxins naturally occurring in plants. IBA can act as an important auxin precursor and can be converted to IAA through a multiple process similar to fatty acid β-oxidation in Arabidopsis (Strader et al. 2010). However, pieces of evidence indicated that IBA might play an independent role. For instance, IBA could induce Arabidopsis stem adventitious roots at concentrations at which IAA was ineffective (Ludwig-Müller et al. 2005; Zolman et al. 2000). Compared with IAA, it was found that IBA was more effective in promoting lateral root growth in Arabidopsis (Zolman et al. 2000). Moreover, IBA was proved to transport independently the characterized IAA transport machinery (Strader et al. 2010). Thus, IBA can function as auxin depending on its conversion into IAA or just through itself independently.
Auxin glycosylation is supposed to be an essential pathway to regulate auxin levels. Auxin glycosyltransferases catalyze the transfer of activated sugars to auxins and thereby regulate their bioactivity, solubility and transport (Ross et al. 2001; Gachon et al. 2005; Chong et al. 2002). Several Arabidopsis genes were identified to encode auxin glycosytransferases. For instance, UGT84B1 was identified to be IAA-preferring glycosyltransferase. Meanwhile, UGT74E2, UGT74D1 and UGT75D1 were IBA-preferring glycosyltransferases (Jackson et al. 2001; Tognetti et al. 2010; Jin et al. 2013; Zhang et al. 2016). The ectopic expression of these genes showed different auxin deficiency phenotypes. Overexpressors of UGT84B1 exhibited shorter stature, curling leaves and compressed rosette. The ectopic expression of UGT74E2 resulted in compressed rosette, shorter stature and enhanced drought tolerance. Meanwhile, UGT74D1 overexpressors only exhibited curling leaves. UGT75D1 overexpression plants only exhibited smaller cotyledons, but with increased abiotic stress adaptation during seed germination (Jackson et al. 2002; Tognetti et al. 2010; Jin et al. 2013; Zhang et al. 2016). Together, these studies indicated that auxin glycosylation exerts important contribution to plant development and environmental adaptation.
UGT84A2 is one member of glycosyltransferase family 1 (GT1) in Arabidopsis. In this study, we identified UGT84A2 to be an IBA-specific glycosyltransferase. Although the UGT84A2 mutants showed similar phenotype to the wild type, the transgenic plants ectopically expressing UGT84A2 showed severe delay in flowering time. Moreover, the overexpression of UGT84A2 led to IBA accumulation and down-regulation of auxin response factors ARF6, ARF8 and key flowering-related genes FT, AP1, LFY and SOC1. Exogenously applied IBA to wild-type plants could mimic the role of UGT84A2 overexpression. Furthermore, we found that arf6arf8 double mutation also delayed the flowering transition and decreased the expression of key flowering genes. These data suggest that glycosyltransferase UGT84A2 may be involved in flowering regulation by indole-3-butyric acid-mediated transcriptional repression of ARF6, ARF8 and those genes of the downstream flowering pathway.
Materials and methods
UGT84A2 glycosyltransferase activity assay
The cDNA sequence of UGT84A2 (At3G21560) was amplified from Arabidopsis thaliana by reverse transcription-PCR (RT-PCR), and then cloned into the prokaryotic expression vector PGEX-3H and mobilized into XL1-Blue. Soluble recombinant protein was induced by isopropyl-beta-d-thiogalactopyranoside (IPTG) and the fusion protein was then extracted and purified by affinity chromatography. The concentration was measured by reacting with Coomassie brilliant blue as described (Hou et al. 2004).
The glycosyltransferase activity assay mix (100 μl) contained 2 μg of purified recombinant UGT84A2 protein, 5 mM UDP-glucose, 1 mM auxins and 100 mM Tris–HCl (pH 7.0). The tested substrates include two naturally occurring auxins (IAA and IBA). The reactions were carried out at 30 °С for 1 h and the reaction mix was analyzed subsequently using reversed-phase HPLC.
The preparation of transgenic plants
The CRISPR/cas9 system was used to mutate the UGT84A2 gene. The gRNA sequence was GAATGAGATGTAAACAACGGAGG and the oligo dimer sequences were 5′-TGATTGGAATGAGATGTAAACAACGGAGG-3′ and 5′-AAACCCTCCGTTGTTTACATCTCATTCCA-3′. The dimer sequences were synthesized and cloned into the CRISPR/cas9 vector. The vector was introduced into Agrobacterium tumefaciens (GV3101 strain) and further used to transform Arabidopsis plants through the floral dip method (Clough and Bent 1998). The 400 bp of PCR fragments containing gRNA sequence amplified from genome DNA of hygromycin-resistant seedlings was sequenced and compared with the wild type. The seeds collected from heterozygous plants were further tested for antibiotic resistance and selected through sequencing to identify homozygous mutant plants. Besides the CRISPR/cas9 mutation allele 84a2-3-13, another deletion allele of UGT84A2, brt1-1 (NASC ID: N66577) was also used in this research. In the brt1-1 allele, the mutation is a deletion of a single nucleotide (T50) by ethylmethane sulfonate mutagenesis that results in a frameshift and a subsequent premature stop codon (Sinlapadech et al. 2007). The UGT84A2 overexpression plants and UGT84A2 promoter::GUS transgenic plants were constructed as described (Zhang et al. 2016).
Profiling of IAA
Seedlings of the wild-type Arabidopsis thaliana Col-0, one line of UGT84A2 mutant (84a2-3-13) and one line of UGT84A2 overexpressor (84A2OE8-4) were used as the representatives for profiling of IAA. 10-day-old whole seedlings were collected in five replicates, weighed, immediately frozen in liquid nitrogen and stored at − 80 °С until extraction. Extraction, purification and quantification of IAA were done as described (Sun et al. 2014).
Histochemical GUS assays
Plant tissues were first placed in 90% acetone on ice for 20 min and then washed twice with staining buffer which contains 50 mM sodium phosphate (pH 7.2), 0.2 mM K3Fe(CN)6, 0.2 mM K4Fe(CN)6 and 0.2% Triton X-100. The samples were then incubated overnight at 37 °С with staining buffer containing 2 mM 5-bromo-4-chloro-3-indolyl-b-d-glucuronic acid (X-Gluc). The samples were then washed in 70% ethanol until the materials became transparent before observation under a dissecting microscope.
Quantitative RT-PCR
For quantitative real-time PCR (qRT-PCR), total RNAs were extracted from plant tissues using Trizol reagent (TaKaRa, Japan). For detecting transcription levels of flowering genes, the leaves of 2- to 3-week-old seedlings before flowering were collected for RNA extraction. For detecting UGT84A2 expression under the induction of auxins, 2-week-old seedlings were harvested for RNA extraction. Reverse transcription reactions were performed with the PrimeScript RT reagent kit with gDNA Eraser (Takara, Japan). qRT-PCR reactions were carried out on real-time thermal cycling system (Bio-Rad, USA), and the SYBR-Green Ex Taq II kit (TaKaRa, Japan) was used for detecting gene expression abundances. ACT2 gene was used as a reference to normalize the samples. Primers used in this research are included in Table S1.
Statistical analysis
All experiments were done with three independent biological replicates and three technical repetitions. Data were collected and subjected to Student’s t test. Data represent mean value ± SD. Asterisks indicate significant differences relative to the wild type or control (*P < 0.05, **P < 0.01).
Results
Expression of UGT84A2 was developmentally regulated and induced by IBA
Firstly, we investigated the UGT84A2 promoter::GUS transgenic lines for the tissue-specific expression of UGT84A2. Histochemical staining of GUS activity demonstrated that UGT84A2 had strong expression in germinating seeds, cotyledons and hypocotyls in the germination period. UGT84A2 was also strongly expressed in newly grown leaves and trichomes of early seedlings, but a weak expression was observed in roots. It should be noticed that UGT84A2 was strongly expressed in the meristem of 12- to 14-day-old seedlings. Besides, UGT84A2 was also expressed in the pistil, stigma and embryo (Fig. 1A). These results suggested that UGT84A2 was developmentally regulated and may play a role in modulating plant growth and development.
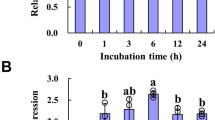
The expression of UGT84A2 was developmentally regulated and induced by IBA. A Histochemical staining for GUS activity of the UGT84A2 promoter::GUS transgenic lines. a, 1-day-old seedling; b, 2-day-old seedling; c, 3-day-old seedling; d, 5-day-old seedling; e and f, 7-day-old seedling; g, 10-day-old seedling; h and i, 12-day-old seedling; j, 14-day-old seedling; k and l, flower and silique. B Expression level of UGT84A2 gene induced by IBA. C Expression level of UGT84A2 gene induced by IAA. Values are means ± SD of three biological replicates. Asterisks indicate significant differences relative to 0 h (Student’s t test: *P < 0.05, **P < 0.01)
To know whether or not UGT84A2 was involved in plant hormone regulation, we further investigated the induced expression patterns under hormone treatments. Experimental results indicated that UGT84A2 gene was clearly induced by IBA (Fig. 1B). In contrast, UGT84A2 was only faintly induced by IAA (Fig. 1C). Other hormones cannot induce UGT84A2 expression. This observation suggested that UGT84A2 might play a role in auxin regulation of plant growth and development.
UGT84A2 catalyzed IBA glycosylation to form IBA–glucose conjugates
IAA and IBA are two naturally occurring auxins. Considering that UGT84A2 is a putative glycosyltransferase, we tested its enzymatic activity towards auxins. We found that UGT84A2 had significant catalytic activity toward IBA, but not IAA (Fig. 2A, B). To verify the glycosylated products, reaction products were subjected to LC–MS analysis. The molecular weight of IBA–glucose ester is 365.00. As shown in Fig. 2C, putative IBA–glucose conjugation gave dominant ions m/z 366.14 (M + H+); m/z 383.17 (M + NH4+) and m/z 388.12 (M + Na+) in the positive ionization mode. The molecular weight of IBA was 203 and the molecular weight of the product was increased by 162 (one glucose molecule mass minus one H2O molecule mass), which corresponds to the addition of one glucose molecule to IBA. These results provide evidence that the UGT84A2 can catalyze the glycosylation of IBA to form IBA–Glc.
Enzyme activity analysis of glycosyltransferase UGT84A2. A HPLC analysis of reaction products from IBA catalyzed by UGT84A2. GST was used as negative control. B HPLC analysis of reaction products from IAA catalyzed by UGT84A2. GST was used as negative control. C LC–MS analysis of reaction products from IBA catalyzed by UGT84A2. D The glycosyltransferase activities of the crude protein extracts from overexpression lines (84A2OE8-4,84A2OE2-2), mutants (84a2-3-13, brt1-1) and wild type (WT)
To deepen the understanding of the UGT84A2 physiological role, the UGT84A2-overexpressing plants and knockout mutants were generated. Two independent homozygous lines (named 84A2OE2-2 and 84A2OE8-4) with high UGT84A2 transcript levels, and two null mutation alleles of UGT84A2 (a CRISPR/cas9 mutation allele 84a2-3-13 and an ordered mutation allel brt1-1 from the European Arabidopsis Stock Centre) were used in this study (Fig. 3). At first, we examined the enzyme activity of crude proteins extracted from the overexpression and mutant plants toward IBA. As shown in Fig. 2D, the UGT84A2 overexpression plants with higher expression levels displayed stronger enzyme activity toward IBA, while the mutants displayed weaker enzyme activity compared to wild type. These results provided further evidence that UGT84A2 was an active glycosyltransferase toward IBA both in vitro and in vivo.
The preparation of UGT84A2 overexpression lines and knockout lines. A UGT84A2 transcript level in different overexpression lines. Actin2 was used as the reference gene. B The UGT84A2 knockout line (84a2-3-13) was prepared by CRISPR/cas9 technology. An adenine base was deleted in the UGT84A2 coding region of the mutant 84a2-3-13 compared to the wild type (indicated by arrow)
The delayed flowering and altered IBA level in UGT84A2 overexpression lines
As mentioned above, two independent overexpression lines (84A2OE2-2 and 84A2OE8-4) and two null mutants (84a2-3-13 and brt1-1) were obtained and used in this study to investigate the physiological role of UGT84A2. Although the flowering time of two ugt84a2 mutants was similar to the wild type, the UG84A2 overexpression plants exhibited a delay of up to 1 week in flowering (Fig. 4A). To investigate the molecular mechanism underlying the later flowering phenotype of UGT84A2OE, we compared the expression level of several key genes specifically involved in major flowering pathways. It was found that the expression levels of FT,SOC1, AP1 and LFY, which could promote flowering, were down-regulated dramatically in the overexpression lines (Fig. 4B). These data suggest that UGT84A2 plays an important role in Arabidopsis flowering.
Phenotype of UGT84A2 overexpression and mutant lines. A UGT84A2 overexpression lines (84A2OE2-2, 84A2OE8-4) delayed flowering, while mutants (brt1-1, 84a2-3-13) had the similar phenotype to the wild type. B Expression levels of flowering-related genes FT, SOC1, AP1 and LFY in UGT84A2 overexpression lines grown in soil for 3 weeks. Values are means ± SD of three biological replicates. Asterisks indicate significant differences relative to WT (Student’s t test: *P < 0.05, **P < 0.01)
UGT84A2 can catalyze the glycosylation of IBA. We supposed that UGT84A2 could affect the IBA homeostasis in planta. To gain insights into the relationship between UGT84A2 and auxin, further investigations of IBA contents of UGT84A2 transgenic lines were taken. Because IBA measurement is very difficult and often technically challenging, we used only one representative line from each of the mutants and overexpressors. To increase the reliability of the data, we used two different internal references, [13C6] IAA and [13C6] IBA, in the same measurement. When [13C6] IAA was used as an internal reference, the IBA content of 84a2-3-13(0.019 pg/mg) was almost the same as that of the the wild type (0.017 pg/mg). However, the IBA content of 84A2OE 8-4 (0.042 pg/mg) was about twofold that of the wild type. Similarly, when [13C6] IBA was used as internal reference, although the IBA contents of both 84a2-3-13 and WT were below the detection threshold, the IBA content of 84A2OE 8-4 showed a much higher value of 0.15 pg/mg. These data indicated that the overexpression of UGT84A2 led to the perturbation of IBA homeostasis (Table 1).
Exogenously applied IBA affected flowering transition
Given that UGT84A2 was identified to be an IBA glucosyltransferase, and the IBA content of UGT84A2 overexpression lines was increased, whether or not the IBA is responsible for the flowering transition was investigated next. Interestingly, upon growth on MS medium containing IBA, the flowering of wild type was delayed up to 1 week (Fig. 5A, B). We also detected the expression levels of key flowering genes in wild type with or without IBA treatment. It was found that when growing on MS medium supplied with 5 or 10 μM IBA, the expression levels of key genes such as FT, AP1, LFY and SOC1 were also down-regulated compared with plants growing on MS medium without IBA (Fig. 5C), which corresponded to UGT84A2 overexpression lines. These data imply that IBA homeostasis was involved in flowering time probably through down-regulating FT, AP1, LFY and SOC1.
Effects of IBA on flowering transition and flowering-related genes. A, B Phenotype and flowering rates of wild-type plants grown on MS medium supplied with 0, 5 or 10 µM IBA for 3 weeks. C Expression levels of FT, AP1, LFY and SOC1 genes in wild-type plants grown on MS medium supplied with 0 µM (control), 5 or 10 µM IBA for 2 weeks. Values are means ± SD of three biological replicates. Asterisks indicate significant differences relative to control (Student’s t test: *P < 0.05, **P < 0.01)
IBA suppressed the transcription of ARF6 and ARF8
If IBA was involved in flowering time, then the new question arose: how does IBA play a role in flowering induction? We further investigated the expression levels of auxin response factor genes (ARFs) in transgenic lines and under IBA treatment. It was found that the expression levels of both ARF6 and ARF8 were down-regulated in UGT84A2 overexpression plants, but there was no change with other ARF genes (Fig. 6A). This result indicated that UGT84A2 might suppress the expression of flowering-related genes through down-regulating the expression levels of ARF6 and ARF8. Since IBA can also affect flowering transition and suppress the expression of flowering-related genes, we supposed that the bridge between IBA and flowering genes might also be ARF6 and ARF8. So, we further tested the role of IBA on ARF expression. Interestingly, ARF6 and ARF8 transcriptions were indeed suppressed by exogenously applied IBA (Fig. 6B). Together, it was indicated that UGT84A2 may affect the flowering time through modulating IBA homeostasis and influencing the expression of flowering-related genes, in which ARF6 and ARF8 were most likely the bridges between IBA and flowering-related genes.
Expression levels of ARF6 and ARF8 in different genotypes and in wild type treated with IBA. A Expression levels of ARF6 and ARF8 in different UGT84A2 genotypes. B Expression levels of ARF6 and ARF8 in wild type after treatment with IBA. The values are means ± SD of three biological replicates. Asterisks indicate significant differences relative to WT or control (Student’s t test: *P < 0.05, **P < 0.01)
Effects of ARF6 and ARF8 on flowering time
To further explore the direct effects of ARF6 and ARF8 on flowering time, arf6arf8 double mutant was used in this research. We observed that arf6arf8 also showed delayed flowering induction, which was consistent with the phenotype of the UGT84A2 overexpression plants (Fig. 7A, B). Further detection demonstrated that the key genes involved in flowering pathways, FT, SOC1, LFY and AP1, were significantly suppressed in arf6arf8 double mutants (Fig. 7C). These results further indicated that ectopic expression of UGT84A2 delayed flowering most likely through indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes,and then leading to the down-regulation of flowering genes in Arabidopsis.
arf6arf8 double mutant was delayed in flowering transition. A Seedling phenotype of wild type and arf6arf8 double mutants grown in soil for 21 days. B Late flowering phenotype of arf6arf8 double mutant grown in soil for 35 days. C Expression levels of FT, AP1, LFY and SOC1 in arf6arf8 double mutant. Values are means ± SD of three biological replicates. Asterisks indicate significant differences relative to WT (Student’s t test: *P < 0.05, **P < 0.01)
Discussion
Auxins are critical regulators for plant growth and developmental processes, such as apical dominance, tropisms, postembryonic organogenesis, vascular development and responses to environmental stresses (Vanneste and Friml 2009; Scarpella et al. 2010). Despite the identification of several endogenous auxins, most research has been focused on the primary free auxin IAA, and the research on the in vivo function of the other naturally occurring auxin IBA has been rather limited. However, IBA may play crucial roles in modulating plant development. IBA makes up more than 25% of the total auxins present in Arabidopsis seedlings (Ludwig-Müller et al. 1993). It was proved that IBA could induce auxin-responsive reporter genes, as well as affect adventitious root initiation, induce lateral roots and promote shoot and hypocotyl elongation (Zolman 2000; Ludwig-Müller et al. 1993). IBA is usually believed to be a precursor for IAA biosynthesis and is converted into IAA through β-oxidation. Hence, IBA could act as an auxin, depending on its conversion to IAA. However, increasing experimental evidence indicated that IBA itself could also act as an auxin. For instance, IBA could induce adventitious roots of Arabidopsis stem at concentrations at which IAA was out of action (Ludwig-Müller et al. 2005). Moreover, several IBA-resistant mutants were sensitive to IAA, but without defects in β-oxidation, suggesting that IBA has direct auxin effects independently of IBA-to-IAA conversion (Strader et al. 2010, 2011). Recently, the identification of the specific IBA efflux carriers indicated that IBA can be transported under the situation lacking IAA transport machinery (Strader et al. 2009). These data suggested that IBA can function as auxin either depending on its conversion into IAA or just by itself independently of IAA. It is known that auxin glycosylation is important for modulating auxin homeostasis. At present, four Arabidopsis glycosyltransferases have been found to glucosylate auxins to form their glucose conjugates. UGT84B1 was identified to be IAA-preferring glycosytransferase, whereas UGT74E2, UGT74D1 and UGT75D1 were believed to be IBA-preferring glycosyltransferases (Jackson et al. 2001; Tognetti et al. 2010; Jin et al. 2013; Zhang et al. 2016). Here, we demonstrated that UGT84A2 only has high activity toward IBA, but not IAA. These findings suggest that there are multiple glycosyltransferases with functional redundancy toward the same kind of phytohormones by virtue of plant evolution, which may be beneficial for plants to make fine adaptation responses to environmental changes.
Previous researches reported that the overexpression of these auxin glycosyltransferase genes resulted in perturbation of auxin homeostasis and related phenotypes. For example, UGT84B1 overexpression plants had higher concentration of free IAA (Jackson et al. 2002), and overexpression of UGT74E2 caused the increased IBA level while IAA level was unchanged (Tognetti et al. 2010). Overexpressors of UGT84B1 and UGT74E2 showed same phenotypes in rosette leaves, branches and stature. Also, ectopic expression of UGT84B1 also resulted in wrinkled leaves and reduced gravitropism, while UGT74E2 overexpressors showed the delayed flowering phenotype compared with wild type (Jackson et al. 2002; Tognetti et al. 2010). Different from the above, overexpressors of UGT74D1 only showed curling leaves compared to wild type, and UGT75D1 overexpression plants only exhibited smaller cotyledons (Jin et al. 2013; Zhang et al. 2016). In this study, ectopic expression of UGT84A2 resulted in IBA accumulation and late flowering transition, but no other clear phenotypes. These findings suggested the important roles of auxin glycosyltransferases in modulating auxin homeostasis and in regulating plant development, depending on different glycosyltransferase members and from different aspects.
Flowering is a key transition point in plant growth and development. It was found that a complex regulatory network and multiple components were involved in this transition process (Bastow et al. 2004; Gendall et al. 2001; Murase et al. 2008). At least six distinct pathways controlling flowering in Arabidopsis thaliana were reported. In this study, although the flowering time of two ugt84a2 mutants was similar to the wild type, the UG84A2 overexpression plants exhibited late flowering phenotype and our further analysis showed that the expression levels of flowering key genes FT,SOC1, AP1 and LFY, which promote flowering, were down-regulated dramatically. These data suggest that UGT84A2 plays an important role in Arabidopsis flowering transition. Moreover, the ectopic expression of UGT84A2 perturbed auxin homeostasis. The IBA content was increased in UGT84A2 overexpression plants. Given that UGT84A2 was identified to be an IBA glucosyltransferase, the question arises whether or not IBA is responsible for the flowering transition in overexpression lines. When grown in MS medium containing IBA, we found that the flowering time of wild type was delayed, and the gene transcription of FT, AP1, LFY and SOC1 was also down-regulated. These data imply that IBA homeostasis was indeed involved in flowering time. Then, how does IBA play a role in flower induction? Our further investigation indicated that the expression of both ARF6 and ARF8 was down-regulated in UGT84A2 overexpression plants and was also specifically suppressed by exogenously applied IBA. These findings prompt us to analyze the relationship between ARF6, ARF8 and flowering transition. Consistent with the observation mentioned above, arf6arf8 double mutant also delayed flowering transition and the expression levels of FT,SOC1, AP1 and LFY were also down-regulated in this double mutant. Therefore, we concluded that UGT84A2 may affect flowering time through modulating IBA homeostasis and influencing the expression of flowering-related genes. In addition, ARF6 and ARF8 were the most likely components mediating IBA and the flowering process.
Author contribution statement
BKH and GZZ conceived and designed the research. GZZ, SHJ and PL conducted the experiments. GZZ, XYJ and YJL contributed analytical tools and analyzed data. GZZ and BKH wrote the manuscript. All authors read and approved the manuscript.
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427:164–167
Campbell JA, Davies GJ, Bulone V, Henrissat B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326:929–939
Cho LH, Yoon J, An G (2016) The control of flowering time by environmental factors. Plant J. doi: 10.1111/tpj.13461
Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14:1093–1107
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Corbesier L, Vincent C, Jang S, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549
Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107:525–535
Hou BK, Lim EK, Higgins G, Bowles DJ (2004) N-Glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279:47822–47832
Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276:4350–4356
Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ (2002) Overexpression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterization of transgenic lines. Plant J 32:573–583
Jin SH, Ma XM, Han P, Wang B, Sun YG, Zhang GZ, Li YJ, Hou BK (2013) UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS One 8:e61705
Li L, Li X, Liu Y, Liu H (2016) Flowering responses to light and temperature. Sci China Life Sci 59:403–408
Liu X, Barkawi L, Gardner G, Cohen JD (2012) Transport of indole-3-butyric acid and indole-3-acetic acid in Arabidopsis thaliana hypocotyls using stable isotope labeling. Plant Physiol 158:1988–2000
Ludwig-Müller J (2000) Indole-3-butyric acid in plant growth and development. Plant Growth Regul 32:219–230
Ludwig-Müller J, Sass S, Sutter EG, Wodner M, Epstein E (1993) Indole-3-butyric acid in Arabidopsis thaliana. Identification and quantification. Plant Growth Regul 13:179–187
Ludwig-Müller J, Walz A, Slovin JP, Epstein E, Cohen JD, Dong WQ, Town CD (2005) Overexpression of Maize IAGLU in Arabidopsis thaliana alters plant growth and sensitivity to IAA but not IBA and 2,4-D. Plant Growth Regul 24:127–141
Monte E, Alonso JM, Ecker JR, Zhang YL, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15:1962–1980
Murase K, Hirano Y, Sun TP, Hakoshima T (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456:459–463
Ross J, Li Y, Lim E, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2:REVIEWS3004
Scarpella E, Barkoulas M, Tsiantis M (2010) Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol 2:a001511
Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C (2007) The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid:UDPG glucosyltransferase. Plant J 49:655–668
Siriwardana NS, Lamb RS (2012) The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. Int J Dev Biol 56:207–221
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Strader LC, BarteL B (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21:1992–2007
Strader LC, Culler AH, Cohen JD, Bartel B (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153:1577–1586
Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23:984–999
Sun XH, Ouyang Y, Chu JF, Yan J, Yu Y, Li XQ, Yang J, Yan CY (2014) An in-advance stable isotope labeling strategy for relative analysis of multiple acidic plant hormones in sub-milligram Arabidopsis thaliana seedling and a single seed. J Chromatogr A 1338: 67–76
Tognetti VB, Aken O, Morreel K, Vandenbroucke K, Cotte B, Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, Genty B, Stubbs KA, Inze D, Breusegem F (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22:2660–2679
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136:1005–1016
Wang JW (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65:4723–4730
Weis M, Lim EK, Bruce NC, Bowles DJ (2008) Engineering and kineticcharacterisation of two glucosyltransferases from Arabidopsis thaliana. Biochimie 90:830–834
Winter CM, Yamaguchi N, Wu MF, Wagner D (2015) Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Plant Physiol 155:55–73
Yamaguchi A, Abe M (2012) Regulation of reproductive development by non-coding RNA in Arabidopsis: to flower or not to flower. J Plant Res 125:693–704
Zhang GZ, Jin SH, Jiang XY, Dong RR, Li P, Li YJ, Hou BK (2016) Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol Biol 90:77–93
Zolman B, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid response in Arabidopsis thaliana reveals four mutant classes. Genetics 156:1323–1327
Acknowledgements
This research was supported by the National Natural Science Foundation of China (nos. 91217301 and 31570299 to BKH; no. 31500230 to SHJ) and by Shandong Provincial Natural Science Foundation of China (no. ZR2017PC007 to GZZ). We thank Dr. Jason W. Reed (Department of Biology, University of North Carolina) for kindly providing us with the arf6arf8 double mutant seeds. brt1-1 mutant was ordered from the European Arabidopsis Stock Centre (NASC). We thank Dr. Jinfang Chu (The Plant Hormone Platform, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with the measurement of IBA level.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Prakash P. Kumar.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, GZ., Jin, SH., Li, P. et al. Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep 36, 1995–2006 (2017). https://doi.org/10.1007/s00299-017-2225-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2225-x