Abstract
Key message
The radish WRKY gene family was genome-widely identified and played critical roles in response to multiple abiotic stresses.
Abstract
The WRKY is among the largest transcription factors (TFs) associated with multiple biological activities for plant survival, including control response mechanisms against abiotic stresses such as heat, salinity, and heavy metals. Radish is an important root vegetable crop and therefore characterization and expression pattern investigation of WRKY transcription factors in radish is imperative. In the present study, 126 putative WRKY genes were retrieved from radish genome database. Protein sequence and annotation scrutiny confirmed that RsWRKY proteins possessed highly conserved domains and zinc finger motif. Based on phylogenetic analysis results, RsWRKYs candidate genes were divided into three groups (Group I, II and III) with the number 31, 74, and 20, respectively. Additionally, gene structure analysis revealed that intron–exon patterns of the WRKY genes are highly conserved in radish. Linkage map analysis indicated that RsWRKY genes were distributed with varying densities over nine linkage groups. Further, RT-qPCR analysis illustrated the significant variation of 36 RsWRKY genes under one or more abiotic stress treatments, implicating that they might be stress-responsive genes. In total, 126 WRKY TFs were identified from the R. sativus genome wherein, 35 of them showed abiotic stress-induced expression patterns. These results provide a genome-wide characterization of RsWRKY TFs and baseline for further functional dissection and molecular evolution investigation, specifically for improving abiotic stress resistances with an ultimate goal of increasing yield and quality of radish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Sessile organisms such as plants usually encounter several biotic and abiotic environmental challenges, such as drought, salinity, and heat conditions (Chen et al. 2012). Due to the recent global climatic changes, the optimal productivity of crops has continued to decline due to the increase of abiotic and biotic stresses in plants. Furthermore, the high solubility of toxic metals in water results in uptake by plants, which has deleterious effects on the food chain and human health. To overcome these stresses, higher plants continuously develop more complex strategies for their survival under these conditions (Kulhari et al. 2013). Heavy metals (HMs) could negatively influence the morphology, physiology, and biochemistry of plants (Gautam et al. 2016). HMs have also been shown to significantly decline biomass accumulation (Zhao et al. 2012) in addition to detrimental effects on critical metabolic mechanisms including photosynthesis, inorganic nutrition, and water uptake (Mukhopadhyay and Mondal 2015; Rodriguez et al. 2012; Vernay et al. 2007). Transcription factors (TFs) including WRKY are crucial to the adjustment of gene expression in response to environmental stresses (Zhao et al. 2015). More than 20% of the genes in Arabidopsis thaliana genome seem to translate into proteins involved in gene transcription (Rushton et al. 2010).
Transcription factors (TFs) have sequence-specific DNA-binding domains and perform integral roles in plant development by temporarily and spatially regulating their target genes (Zhang et al. 2011). WRKY family members have been found to play key roles in biotic and abiotic stress responses in plants (Zhao et al. 2015). WRKY protein can interact with the W-box [TGACC (A/T)] in the promoter of their target genes to induce or repress the expression of downstream genes to activate their stress response (Zentgraf et al. 2010). In Arabidopsis, AtWRKY22 initiates submergence tolerance by interacting with the ACS7 promoter and activating downstream ethylene response (Hsu et al. 2013). BhWRKY1 interacts with BhGolS1 promoter involved in promoting dehydration and cold tolerance (Wang et al. 2009). Some WRKY TFs are implicated in the coordination of several biological activities such as salt (Chen et al. 2012), heat (Tang et al. 2013), HM (Wang et al. 2013), and other biotic and abiotic stresses (Tang et al. 2013). Additionally, double mutants, wrky26–wrky25 and wrky33–wrky25; and triple mutants, wrky33wrky26wrky25 in A. thaliana exhibit susceptibility to thermal stress (Li et al. 2011). In Tamarix hispida, ThWRKY7 under CdCl2 treatment improved Cd tolerance by acting as an upstream regulator of ThVHAc1 gene (Yang et al. 2016). Noteworthily, single WRKY gene can regulate multiple abiotic responses by interacting with several VQ proteins (having VQ-linked motif including SIB1 and SIB2 (Sigma Factor-Interacting Protein) to modulate multiple abiotic stresses (Lai et al. 2011). In O. sativa, OsWRKY74 positively regulates phosphate (Pi) homeostasis and signal transduction, Fe deficiency, and cold stress in rice (Dai et al. 2015). Overexpression of GhWRKY41 in tobacco promotes salinity and desiccation tolerance by modulating stomatal conductance and ROS levels (Chu et al. 2016). PsWRKY from Papaver somniferum is implicated to be induced by multiple treatments, including salinity, cold, dehydration, wounding, phytohormones (ABA, MeJA) as well as modulating benzylisoquinoline pathway (Mishra et al. 2013).
Radish (Raphanus sativus L.) production continues to be affected by biotic and abiotic factors. Thus, it is critical to explore the molecular mechanisms regulating multiple biological processes in radish. Recently, drafts of the R. sativus databases were published (Kitashiba et al. 2014; Mitsui et al. 2015), providing a valuable gateway for genome-wide and comparative analysis of putative TFs. Previous transcriptome studies on radish have mentioned the expression of TFs including, NAC, MYB, ERF, and WRKY under stress and normal growth conditions, Pb (Wang et al. 2013), salt (Sun et al. 2016), heat (Wang et al. 2015b), and taproot thickening (Yu et al. 2015). In this study, genome-wide exploration and classification of WRKY TFs were performed. The conserved motif and exon–introns distribution, linkage group localization, and in silico transcriptome expression analysis were conducted. Additionally, the expression of the RsWRKY gene validation was subsequently investigated under different abiotic stresses.
Materials and methods
Sequence data retrieval
The sequences of WRKY genes were retrieved from radish genome database using Hidden Markov Model profile (PF03106). Furthermore, CDD (http://www.ncbi.nlm.nih.gov/Structure) and interproscan (Letunic et al. 2006) online programs were utilized to ascertain the presence of WRKY domain in the non-redundant sequences. The data for the sequences of WRKY TFs from other plants were obtained from (http://planttfdb.cbi.pku). The proportional study was performed using additional 16 previously studied species including plant and fungi (He et al. 2012; Huh et al. 2012; Bencke-Malato et al. 2014) (Fig. 1).
Evolutionary relationships of radish with other species and their number detail of the WRKY family of each species. The left of this figure indicates the categories of the species; the right of this figure shows the number detail of the WRKY family of each species; UG (ungrouped) indicates the subfamily members from a distinct group in a combined phylogenetic tree; GS (Genome size, M)
Protein properties and phylogenetic analysis
The WRKY domains were obtained and utilized for plotting phylogenetic tree using MEGA6 (Hall 2013). The following parameters were adopted: Poisson correction, pairwise deletion, and 1000 bootstrap replicate. To obtain a reliable classification of the different groups, eight highly conserved representative WRKY domains from Arabidopsis were included during tree construction. Furthermore, the online ExPASy proteomics server (http://web.expasy.org/protparam) was used to analyze protein properties of the putative WRKY proteins.
Motif and gene structure analysis of RsWRKY genes
To predict conserved motifs of WRKY proteins were performed using the Multiple EM for motif elicitation software (http://meme-suite.org/tools/meme). Diverse gene structural organizations of RsWRKY genes were identified using GSDS tool (http://gsds.cbi.pku.edu.cn).
GO annotation and prediction of miRNA targeting the RsWRKY genes
Gene ontology annotation of RsWRKY protein sequences was analyzed using an online program (http://www.blast2go.com) as previously described (Conesa et al. 2005). Furthermore, all candidate RsWRKY genes were sought against available radish reference miRNA sequences from our transcriptome sequences (Xu et al. 2013a; Nie et al. 2015; Yu et al. 2015; Sun et al. 2015a; Zhang et al. 2016) using psRNATarget Server (http://plantgrn.noble.org/psRNATarget/). Cytoscape software (Shannon et al. 2003) was applied to visualize the interactions between predicted miRNA and corresponding target RsWRKY genes.
Linkage group localization and distribution of RsWRKY genes
The RsWRKY sequence scaffolds were anchored to the integrated high-density linkage map published by Kitashiba et al. (2014). Gene sequences with similarity ≥98% and length difference ≤5 bp were treated as similar genes between two genomes (Wang et al. 2015c) and subsequently localized to their linkage groups according to their corresponding location parameters using MapInspect software (Liu and Meng 2003). Physical interactions were constructed using Arabidopsis association model in the STRING Protein–Protein Interaction Networks software (Franceschini et al. 2013).
In silico patterns survey of RsWRKY genes
To predict the RsWRKY gene expression profiling, RNA-seq library from NODAI radish genome database was used to retrieve FPKM values of five tissues (cortical, cambium, xylem, root tip and leaf) and six leaf stages (7, 14, 20, 40, 60 and 90 days). Transcriptomic analysis of WRKY-related transcript expression levels in taproot thickening and stresses (heat, salt, Cd, Pb, and Cr) of radish were conducted based on our radish transcriptome data (Wang et al. 2013, 2015b; Xie et al. 2015; Yu et al. 2015; Sun et al. 2016). The MeV version 4.2 software (Saeed et al. 2003) was utilized to plot heat maps for the annotated WRKY genes against their corresponding expression values.
Plant treatments
Seeds of ‘NAU-YH’ radish genotype were grown in the greenhouse for four weeks before further treatments. For heat treatments, seedlings were placed at 42 °C, with 25 °C for the control. For salt treatment, seedlings were treated with 200 mM NaCl and 0 mM NaCl for control. HM treatment involved Cd and Pb, where seedlings were treated with 0 (CK) and 20 mg L−1 CdCl2·2.5H2O and 100 mg L−1 Pb (NO3)2 and 0 (CK), respectively. The samples were harvested at 24 h after each treatment and frozen immediately in liquid nitrogen and stored at −80 °C until further analysis.
Reverse transcription-quantitative polymerase chain reaction
The expression pattern was validated using RT-qPCR, as previously described (Xu et al. 2013b). Beacon Designer 7.7 software was used to design gene-specific primers as listed in Table S1. Total RNAs from triplicate samples were isolated using Trizol (Invitrogen) according to manufacturer’s protocols. PCR was carried out using Start Universal SYBR Green Master mix and performed using iCycler iQ machine (BIO-RAD). The reaction was performed using the following program, 95 °C for 30 s and 45 cycles of 95 °C for 5 s, and 58 °C for 15 s and 72 °C for 20 s. The \(2^{{ - \Delta \Delta C_{\text{T}} }}\) method was used for data quantification (Livak and Schmittgen 2001).
Results
Identification of WRKY genes in radish
In the current study, a sum of 126 potential RsWRKY genes was detectable through genome-wide analysis and designated according to their generic order (Table S2). Additionally, 125 RsWRKY genes confirmed to have complete WRKY domain, and only one (RsWRKY96) had a missing zinc finger motif. Characteristic analysis indicated that the number of aa (amino acids) for these 126 varied from 144 (RsWRKY44) to 1086 (RsWRKY15) aa, with an average protein length of 387 aa. The coding sequence (CDS) length ranged from 435 to 3261 bp with an average length of ~1184 bp (Table S2). The MW (molecular mass) ranged from 1.09 kDa (Kilodalton) (RsWRKY23) to 12.9 kDa (RsWRKY15), and the theoretical isoelectric points varied from 4.96 (RsWRKY103) to 9.89 (RsWRKY54) with 58 members having more than 7 kDa (Table S2).
Relative analysis using pIs (Isoelectric points) and MWs among the three major groups showed that higher proportion of Group II (WRKY GII, 58%) and Group III (WRKY GII, 53%) were acidic, while Group I (WRKY GI) had most of the members (58%) being basic with a wide fluctuation in MWs (Fig. S1). The majority of Group I proteins indicated broader variation in MW’s size and higher pIs compared to Groups II and III members. The detailed information on RsWRKY genes is listed in Table S2.
Phylogenetic classification of RsWRKY genes
Twenty-five RsWRKY proteins from the 126 RsWRKY identified proteins in the current study, were found to have two WRKY domains, while the rest had only one. When RsWRKY protein had two domains, the domain was designated RsWRKY together with either letter N for N-terminal or C for the C-terminal domain (Fig. 2). To predict the degree to which the WRKY and zinc finger motif were conserved in each group, the sequence logos were produced using weblogo online software (Crooks et al. 2004) (Fig. 2; Fig. S2). The phylogenetic relationship of the RsWRKY proteins was constructed by multiple sequence alignment of their WRKY domains (Fig. 3) which clustered into three main groups (Group I, Group II and Group III). For each group, at least one representative was randomly selected from Arabidopsis AtWRKY genes (AtWRKY1, 58, 61, 14, 40, 56, 11 and 64).
Multiple sequence alignment of the WRKY domain among R. sativus WRKY genes. Red indicates conserved WRKY amino acid domains; green indicates zinc finger motifs; dashes indicate gaps. ‘N’ and ‘C’ indicate the N-terminal and C-terminal WRKY domain of a specific in Group I WRKY gene. The conserved domains were identified using the Pfam program. The overall height of each stack on the logo illustrates the sequence conservation at each position. Each residue letter height symbolizes the relative frequency of the corresponding residue (color figure online)
Unrooted phylogenetic tree representing relationships among WRKY domains of radish and selected Arabidopsis domains. The domain sequences of all RsWRKY and Arabidopsis AtWRKY representatives were aligned using ClustalW and the phylogenetic tree constructed with the neighbour joining method in MEGA 6.0. Group I proteins with the suffix ‘N’ or ‘C’ indicates the N-terminal WRKY domains or the C-terminal WRKY domains. The different colors highlight different groups or subgroups of WRKY domains (color figure online)
Although RsWRKY98 had one WRKY domain similar to Group II and III, it was considered as an orphan gene termed as ‘ungrouped’ due to its low homology and divergence in WRKY domain and therefore not clustering in any group. It was also noted that group one had 31 WRKY proteins, out of which 25 had two WRKY domains. Nonetheless, some others (RsWRKY28, 117, 122, 115, 103, and 121) contained only one WRKY domain and clustered with C-terminal WRKY domains, indicating that they might have either undergone domain loss or acquisition events during evolution (Ross et al. 2007).
Despite high conservation of the WRKYGQK in WRKY TFs, four variants in the signature WRKY domain were identified in six RsWRKYs (Table 1): WRKYGQR (RsWRKY105); WRKYGKK (RsWRKY79, RsWRKY121); WRKYGYA (RsWRKY109), and WRKYGYK (RsWRKY113, RsWRKY120). In addition, one zinc finger form variation, C2H was identified in RsWRKY96 gene (Table 1).
Linkage group localization and distribution of RsWRKY genes
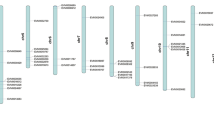
Sixty-seven RsWRKY genes (53%) of total RsWRKY genes were anchored onto the approximate locations of the nine linkage groups (LGs) R1–R9 of the radish genome (Fig. 4; Table S3). Among the nine LGs, LG R2, R3, and R4 constitute the highest number of WRKY TFs, with 9, 8, and 22 genes, respectively, whereas the least number of TFs was located on LG R1 (one gene). The highest WRKY gene number in LG R4 was ascribed to the increased number of Group I, Group IIc and Group III with 6, 6, and 4 genes, respectively. Interestingly, Group II was responsible for 56, 55, and 38% of the total TF family in LG R2, R4, and R3, respectively. Furthermore, tandem duplications of RsWRKY genes were explored along the nine LGs. Tandem duplication was considered when a pair or more homologous WRKY genes confined within the same range distance. Nine RsWRKY gene clusters containing 22 tandemly duplicated genes were located on LGs R2, R3, R4, R5, R6, and R8 (Fig. 4). Most of the highest tandem duplication was found on LG R4, which had four gene clusters containing nine genes.
(I) Linkage map of group localization and distribution of WRKY genes in radish, Tandem-duplicated genes are indicated with vertical gray lines. The size of a linkage group is indicated by its relative length. (II) The percentages of RsWRKY genes on each linkage group are demonstrated by the pie accordingly
Motif composition and gene structure of RsWRKY proteins
MEME motif analysis revealed that different RsWRKY proteins had distinct conserved motifs (Table 2). Motif 1 and 2 are closely related to WRKY domain and are found in all radish WRKY amino acid sequences other than RsWRKY49 without motif 2 (Fig. 5). Motif 1, 2, and 9 jointly contain the C-terminal WRKY domain, while, motif 4, 8, and 10 made up the N-terminal WRKY domain in radish, these six motifs were highly conserved in all RsWRKY proteins. Motif 4, 3, and 10 were highly conserved in Group I. Motif 7 was prominently conserved in GIIb. Group IId has four unique motifs 1, 2, 6, and 9. Notably, RsWRKY96 was not clustered in any group due to the lack of the complete WRKY domain, but contained two motifs (motif 1 and 2), which are prominent in the C-terminal of Group II-b. Therefore, it was speculated that RsWRKY96 is a member of Group IId. The width of the motifs was diverse with motif 8 being the longest while motif 6 had the shortest motif length.
Unrooted phylogenetic tree based on the full-length sequences, conserved motifs composition and gene structure of 126 RsWRKY proteins. The phylogenetic tree was constructed by MEGA6.0. Different colors represented various groups, MEME was used to predict motifs and these motifs represented with boxes, while exon–intron structure of radish WRKY genes; blue indicates untranslated 5′- and 3′-regions; yellow indicates exons; black lines indicate introns (color figure online)
Intron–exon structure analysis revealed the presence of intron–exon in all RsWRKY gene coding sequences. Remarkably, closely related genes in the phylogenetic tree shared similar structure compositions, indicating functional similarities within a subfamily. Moreover, the number of exons ranged from 3 to 12 (RsWRKY3). Normally, most RsWRKY genes in a common pair or triplets indicated similar intron–exon distribution, for example, RsWRKY28 and RsWRKY98.
Comprehensive analysis of microRNA targeting RsWRKY genes
A total of 15 known miRNAs plus 9 candidate novel miRNAs belonging to 25 miRNA families were identified as putative miRNAs targeting 32 RsWRKYs transcripts in this study (Table S4). The relationship network between candidate miRNAs and their targets was constructed (Fig. 6). RsWRKY12 and RsWRKY82 were the most targeted transcripts with each being successfully targeted by five radish miRNAs. They were closely followed by RsWRKY 7 and 55 which were successfully targeted by 4 putative miRNAs. A microRNA rsa-miRn36 with nine RsWRKY target transcripts was proven to be the most abundant identified miRNAs. It is noteworthy that rsa-miRn36 was identified as salt-responsive novel miRNA corresponding to Unigene1698 (Sun et al. 2015b).
Gene ontology annotation and interactions among specific RsWRKY proteins
GO analysis showed that these 126 RsWRKY genes were assigned into 35 biological processes, 4 cellular components, and 3 molecular functions (Fig. S3A). In biological process category, a group of 7 metabolic processes had the highest representation (144 sequences, 90.5%) (Fig. S3A), followed by ‘response to stress’ (50 sequences, 39.7%) and ‘response to chemical’ (49 sequences, 38.9%). In cellular classification, there was an equal number of sequence representation among the four designated categories with each having 102 (81%) sequences (Fig. S3B). In molecular components, ‘sequence-specific DNA binding” had the highest representation of 115 (91.3%) sequences, followed by ‘transcription factor activity’ with (90.5%) sequences. ‘Calmodulin binding’ was the least represented in this group annotation with only 13 (10.32%) members (Fig. S3C).
Subsequently, the 50 RsWRKY candidates potentially involved in plant stress responses were utilized for protein–protein interaction using STRING 9.1 software with the confidence parameter set at a threshold of 0.15. The 50 proteins belonged to diverse groups, (12 proteins (Group I), 3 (Group IIa), 6 (Group IIb), 12 (Group IIc), 7 (Group IId), 3 (Group IIe), 6 (Group III) and 1 (Ungrouped) (Table S5). Subsequently, functional and physical interactions were examined using their database to retrieve the protein interactions. Four proteins that showed high sequence similarity with WRKY33 (RsWRKY18, 19, 22) were involved in the stronger (thicker lines) interaction network (Fig. 7). MKS1 and MPK3 genes formed a strong interaction with WRKY33 and WRKY25 (RsWRKY8, 11, 12), respectively.
Reverse transcription-quantitative polymerase chain reaction
Gene profiling indicated that the 36 RsWRKYs representative genes exhibited distinct patterns under stress and normal growth conditions (Fig. 8). In detail, 24 and 20 RsWRKY transcripts were significantly elevated under Cd and Pb treatments, respectively. Moreover, 30 and 24 genes were found to be potentially responsive to salt and heat stresses, respectively. Co-expression of several RsWRKYs plays vital roles in abiotic stress adjustments. For instance, the expression levels of 13 RsWRKY genes (RsWRKY6, RsWRKY10, RsWRKY11, RsWRKY13, RsWRKY15, RsWRKY17, RsWRKY20, RsWRKY23, RsWRKY24, RsWRKY28, RsWRKY29, RsWRKY30, and RsWRKY98) indicated significant higher expression with a cross-responsive approach to Pb, Cd, salt, and heat. RsWRKY31 and RsWRKY114 had the highest expression levels (>22 folds) under all abiotic stresses. Additionally, stress-specific responses were also observed, RsWRKY28 expression was preferentially expressed under heat stress with no significant response to other stresses. RsWRKY31 and RsWRKY114 had 25- and 22-fold changes, respectively, under different stress treatments as compared to normal growth condition. It was noted that the expression level of 29 RsWRKY genes was relatively higher under heat treatment, among which RsWRKY31 was the most upregulated under all stress treatments. The average fold change value was 1.36 and 0.96 in Cd and Pb stresses, respectively. Additionally, the average fold change in heat and salt was 4.23- and 1.96-folds, respectively (Fig. 8; Table S8).
RT-qPCR was used to analyze the expression profiles of 36 RsWRKY genes under cadmium, lead, heat, and salt stress. CK is untreated seedling. The Actin gene was used as an internal control for RT-qPCR. Error bars were obtained from three technical replicates. Asterisks reveal the gene significantly upregulated or downregulated under abiotic stresses by t test (*P < 0.05, **P < 0.01)
Discussion
Identification and characterization of RsWRKY genes
In higher plants, WRKY proteins regulate multiple biological processes specifically in stress response (Chen et al. 2015), and have been found throughout protozoa, fungi, and other plant species (Wu et al. 2015). In the present study, a total of 126 WRKY putative genes were identified from radish genome database; additional 16 species were incorporated for comparative analysis. It was found that WRKY genes in O. lucimarinus were relatively advanced compared to C. reinhardtii about their number of WRKY genes (Fig.1). Surprisingly, O. lucimarinus had higher WRKY protein coverage in the genome compared to most of the higher plant species. Such extensive coverage illustrated the basic and complex importance of WRKY proteins through the evolution of this gene superfamily in the green lineage. The coverage of WRKY proteins in Brassica rapa and Arabidopsis thaliana was the highest in comparison to all other species used in this study, indicating that WRKY proteins play critical and diverse functions in Brassicaceae family. With the highly advanced evolutionary level, higher species normally acquire additional genes for their complex biological mechanisms. Conversely, the genome WRKY gene proportion in complex plants do not necessarily tally with their evolutional stage (Wu et al. 2015). Stress normally curtails optimal biological processes in plants, WRKY genes in conjunction with other stress responsive genes may synergistically promote tolerance in plants by adjusting their expression patterns.
Structural conservation and divergence of radish WRKY gene family
The phylogenetic tree indicated the classification of 126 RsWRKY genes into three broad groups (Group I, Group II, and Group III), Group II was expanded further to five subgroups (IIa to IIe) according to Arabidopsis. Group I members had a pair of WRKY domains with six members (RsWRKY28, 117, 122, 115,103, and 121) having a single domain. The same phenomenon was consistent with other species such as Arabidopsis (Wei et al. 2012). The evolutionary loss of WRKY domain seems lower in dicotyledons than in monocotyledons. Therefore, Group I can be inferred from the origin of other groups and appeared early in the evolution.
The size of WRKY family size is diverse among different species and there were 72, 182,128, and 149 members in Arabidopsis, Glycine max, Malus domestica, and Zea mays (Wei et al. 2012), respectively. Compared with Arabidopsis (genome size 125 Mb) (Rushton et al. 2010), G. max (975 Mb) (Bencke-Malato et al. 2014), M. domestica (881.3 Mb) (Meng et al. 2016) and Z. mays (Wei et al. 2012) (2400 Mb) in R. sativus (520 Mb) (Wei et al. 2012), the number of the RsWRKY family is relatively smaller. Group I took nearly 20% of the total WRKY genes, which was also similar with Arabidopsis, B. rapa, and A. indica among others (Fig. 3). It was found that the Group II is made up of the largest clade. Comparing the three main groups (Group I, II, and III,) Group II has undergone significant expansion by having majority members, Populus trichocarpa, Camellia sinensis, A. indica, and Cucumis sativus and had 90.4%, 88%, 91.3%, and 86%, respectively. Moreover, it is considered that differences in the proportion of WRKY genes in Group II are the main reason for the diversity in WRKY family size of different species. Noteworthily, the RsWRKY98 clustered solely on the evolutionary tree, this was consistent with Arabidopsis classification where several WRKY member including ATWRKY10, ATWRKY38, and ATWRKY52, could not fit neatly into any group, This might partially be attributed to the loss of the N-terminal WRKY domain. Additionally, AtWRKY38 and AtWRKY52 could either belong to Group III or represent members of a novel group of genes (Eulgem et al. 2000). This observation suggests that RsWRKY98 could belong to either GII or GIII, or be considered as a member of a novel group of genes according to its position on the phylogenetic tree. Additionally, the independent positioning of the RsWRKY98 could also suggest that it evolved from N-terminal of WRKY gene after sub-functionalization and retaining ancestral gene function. Continuous duplication event could positively contribute to the plant survival under various abiotic stresses (Paterson et al. 2009).
Multiple sequence alignment analysis indicated that six RsWRKY had a variant WRKY domain and one had an alteration in its zinc finger motif. In WRKY TF, WRKYGQK-converged sequence plays a fundamental role as a binding site located beside TTGACY motif in AtWRKY TFs (Ciolkowski et al. 2008). If the genes are targeted by a variant WRKY domain, the motif may result to an alternative function from that of the usual WRKYGQK motif (Eulgem et al. 2000), therefore, WRKY TFs with altered WRKY domain may end up recognizing a DNA-binding sequence other than a W-box sequence [(C/T) TGAC(C/T)] (Maeo et al. 2001). It can be deduced that the variations of the WRKY domain are likely to impact on expression patterns of stress-responsive genes targeted by RsWRKY TFs.
Gene structure of all the 126 RsWRKY indicated variation in exon–intron pattern. Members located on the same subgroup showed closely related conserved patterns; this is an indication of functional similarities among members in the same evolutionary group (Wang et al. 2014). Furthermore, exon–intron reorganization by addition or losing may result from and combinations of various linkage groups (Guo et al. 2014). The present study shows an example of such diversification on WRKY gene (RsWRKY61) which has a single exon, while other RsWRKY members within the same phylogenetic group (Group III) possess two to five exons in their genomic sequences. Moreover, RsWRKY3 was found to bear 12 exons. Notably, the members with the highest number of exons categorically clustered in Group I followed by Group II. Therefore, it could be deduced that Group I and II constitute the ancestral genes, from which other groups originate.
The evolutionary relationship and protein interaction networks of RsWRKY genes
A large number of gene duplication occurrences in radish facilitate gene function prediction and evolution analysis (Fig. 4). In many species, whole-genome duplication events mostly result in the enlargement of a gene family (Cannon et al. 2004). This phenomenon also occurred in R. sativus (Shen et al. 2013). In WRKY TFs, tandem gene duplication has also occurred in many other plant species including rice and cucumber (Ling et al. 2011). In cucumber, duplication events in CsWRKY family generated additional Group-III WRKY genes (Ling et al. 2011). In this study, 28.5% (20/70) RsWRKY genes evolved from tandem gene duplication (Fig. 4). It was observed that 40% (9 out of 22) RsWRKY genes on LG R4 were involved in duplication as indicated by the higher number of RsWRKY genes on LG R4. Consequently, tandem duplication probably took an integral role in the enlargement of RsWRKY gene family in radish.
The current study deduced that gene expansion mainly happened in Group II which constituted 50% (11/22) of the total genes involved in the duplication. Among the Group II subgroups Group IIc was leading in the number (5) of WRKY sequences involved in duplication event, these findings were contrary to Arabidopsis and rice, where most of the tandem duplication occurred in Group III. Additionally, the conserved WRKY genes and physical proximity of TFs shared a common group and were located on the same tandem cluster, R1 (RsWRKY10, 11 and 28), R4 (RsWRKY16, 21 and 23) in Group I, R3 (RsWRKY29 and 77), and R8 (RsWRKY69 and 70) in Group II. This phenomenon has also been observed in poplar (Jiang et al. 2014) and grape (Guo et al. 2014) genomes, suggesting paralogous segments resulting from ancestral polyploidization occurrence (Song et al. 2013).
The genomic comparison or protein interaction is a technique for transferring available genomic information from a model plant to those currently studied. Six proteins, which exhibited increased sequence similarity with Arabidopsis WRKY18 (RsWRKY93, 94 and 102), WRKY33 (RsWRKY18, 19), and WRKY53 (RsWRKY63 and 74) were located at the center of the network node, are involved in stronger interaction network illustrated by thicker lines (Fig. 7). MPK3 and MKS1 are also essential phosphorylation partner genes in the function of WRKY genes (Li et al. 2012). According to previous studies, AtWRKY8 via its C-terminal domain, interacts with target genes to regulate multiple abiotic stresses including salt and heat (Wang et al. 2015a). AtWRKY 33, 25, and 26 showing high homology to 7 RsWRKY genes (Fig. 6), were found to coordinate induction of plant thermotolerance (Li et al. 2011). Intriguingly, the soloist gene was featured in the protein interaction network which indicated homology with AtWRKY29. The results indicated multiple interactions among WRKY genes; this was commensurate to the RT-qPCR result indicating co-expression of RsWRKY genes under multiple stress responses.
WRKY proteins play roles in abiotic stress response
Accumulating reports propose that the abiotic stresses deter the optimal physiological and biochemical mechanism in plants (Joshi et al. 2016). In the dynamic growing conditions, a critical factor underlying stress resistance in sessile plants is their ability to conceptualize, integrate, and adjust to biotic and abiotic environmental cues, which could consequently modulate molecular and physiological metabolism for their survival. These deleterious conditions sometimes occur simultaneously and negatively affect plant growth causing significant losses in agricultural production. Such abiotic stresses also activate the WRKY TFs and trigger a network of signal transduction to enhance the stress tolerance in plants (Schluttenhofer and Yuan 2015). Recent advances suggest that WRKY TF family is crucial in numerous plant processes such as plant development and patterns to abiotic stresses (Zhai et al. 2016). In Arabidopsis, rice, and Chinese cabbage, at least 26, 54, and 28 WRKY genes were identified in response to abiotic stress, respectively (Jiang and Deyholos 2006; Zhou et al. 2008; Goel et al. 2016). The majority of RsWRKY genes showed higher expression under abiotic stress treatments than control. We also found involvement of 13 RsWRKY genes in all the abiotic stresses and almost all (35/36) to more than one stressor. Similarly, AtWRKY25 and AtWRKY33 has been known to respond to both heat and salt treatments (Li et al. 2011), while TcWRKY53 is simultaneously induced by cold, salt, and PEG treatments (Wei et al. 2008). These findings support that WRKY genes can co-regulate more than one stressor by a synergistic or antagonistic mechanism (Shaik and Ramakrishna 2013). In addition, co-expression of WRKYs creates a complex network, which resists stress by multiple expression. In rice, overexpression of two WRKY TFs (OsWRKY11 and OsWRKY45) promotes water stress tolerance under drought condition (Qiu and Yu 2009). The result from this study also indicated that 29 RsWRKY genes responded significantly to salt stress, and confirms cross regulation among WRKY genes in response to abiotic stress. Overexpression of ZmWRKY33 in Arabidopsis improved salt stress tolerance of the transgenic plants (Li et al. 2013a). Consistently, RsWRKY90 was upregulated in salt stress similar to its orthologue ATWRKY70, which highly expresses under osmotic stress (Li et al. 2013b).
Surprisingly, the RsWRKY98 showed preferential expression in responses to HMs as compared to the control, while in the salt condition it was downregulated. Comparatively, its orthologous gene (AtWRKY29) has been implicated to be involved in mitogen-activated protein kinase (MAPK) pathway during stress responses (Göhre et al. 2012), implying that the RsWRKY98 could be among the few TFs phosphorylated and activated by kinases. In particular, MEKK1–MAPK kinase 2(MKK2)–MPK4/MPK6 cascade has been shown to function as part of cold and salt stress signaling (Teige et al. 2004). It could provide underpin for the unique positioning of RsWRKY98 gene on the phylogenetic tree. This affirms that the WRKY family continuously evolve as the plants adjust to the ever-changing environmental cue from selective pressure, resulting in new RsWRKY members by either duplication or polyploidization. In Sorghum bicolor, tolerance to desiccation could be associated with the gene duplication events (Paterson et al. 2009). In Brassica napus, evolutionary expansion of BnaWRKY gene family results in new members with conserved or different roles in multiple stress responses (He et al. 2016).
Notably, RsWRKY78 and RsWRKY59 were found to be upregulated under salt treatments, while their orthologous genes, AtWRKY7 and AtWRKY59 were downregulated (Scarpeci et al. 2013). Conversely, the RsWRKY023, RsWRKY32, and RsWRKY80 were downregulated under salt conditions, indicating that the complex responses among WRKY genes in response to abiotic stresses. Additionally, overexpression of ZmWRKY33 in Arabidopsis plants showed enhanced osmotic stress tolerance compared to wild-type (Liu et al. 2013). Furthermore, current study reported that most RsWRKY genes were negatively regulated during HM treatments (Pb and Cd), In Musa species, most of the WRKY genes were downregulated Pb and Cd, Arsenic, and chromium HM stresses (Goel et al. 2016). In Arabidopsis, double and triple mutations of WRKY18, WRKY40, and WRKY60 were highly resistant to Cd stress compared to single mutant or wild-type (Chen et al. 2010). Additionally, overexpression of ThWRKY7 exhibited increased tolerance to Cd stress in T. hispida by binding to the W-box in the promoter region of ThVHAc1 genes (Yang et al. 2016). These results indicate the critical roles of WRKY TFs in modulations of stress responsive genes in abiotic stress tolerances and provide the possibility of engineering for multiple stress tolerances in radish and other root vegetable crops.
Conclusion
In this study, 126 RsWRKY genes were identified from the R. sativus genome sequence. Classification and expression analyses revealed that some RsWRKY genes broadly participate in the regulation of plant response to abiotic stress. Meanwhile, 36 candidate RsWRKY genes were identified to be involved in abiotic stress responses and their expression profiles were confirmed by RT-qPCR. In particular, a total of 13 genes were confirmed to have multiple stress responses in Cd, Pb, heat, and salt stress. Thus, it could be concluded that WRKY genes may have multifunctional roles under various abiotic stresses, and could contribute in controlling signaling processes linked to transcriptional adjustments in plants under harsh environmental conditions. These WRKYs may potentially be utilized for breeding new radish cultivars resistant to multiple stresses. This study facilitates further identification of critical stress-related WRKYs for their transgenic application in radish.
Author contribution statement
BKK designed the experiments and wrote the manuscript. BKK, FL, TM, and ZF performed validation experiments. WY and WR contributed powerful analytical tools. ZX and EMM contributed to proofreading of this manuscript. LL and XL conceived the study and managed the experiments. All authors read and approved the final manuscript.
Abbreviations
- aa:
-
Amino acids
- BLAST:
-
Basic local alignment search tool
- bp:
-
Base pair
- Cd:
-
Cadmium
- CDS:
-
Coding sequence
- GO:
-
Gene ontology
- HM:
-
Heavy metal
- LG:
-
Linkage group
- MW:
-
Molecular weight
- Pb:
-
Lead
- pI:
-
Isoelectric point
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
- TF:
-
Transcription factor
References
Bencke-Malato M, Cabreira C, Wiebke-Strohm B, Bücker-Neto L, Mancini E, Osorio MB, Homrich MS, Turchetto-Zolet AC, De Carvalho MC, Stolf R (2014) Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol 14:236
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:1–21
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:281
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Chen L, Yang Y, Liu C, Zheng Y, Xu M, Wu N, Sheng J, Shen L (2015) Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem Biophys Res Commun 464:962–968
Chu X, Wang C, Chen X, Lu W, Li H, Wang X, Hao L, Guo X (2016) Correction: the cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 11:e0157026
Ciolkowski I, Wanke D, Birkenbihl R, Somssich I (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol Rep 68:81–92
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Crooks GE, Hon G, Chandonia J-M, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Dai X, Wang Y, Zhang W-H (2015) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J Exp Bot 67:947–960
Eulgem T, Rushton P, Robatzek S (2000) Somssich I (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Von Mering C (2013) STRING v9. 1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815
Gautam S, Anjani K, Srivastava N (2016) In vitro evaluation of excess copper affecting seedlings and their biochemical characteristics in Carthamus tinctorius L. (variety PBNS-12). Physiol Mol Biol Plants 22:121–129
Goel R, Pandey A, Trivedi PK, Asif MH (2016) Genome-wide analysis of the Musa WRKY gene family: evolution and differential expression during development and stress. Front Plant Sci 7:209
Göhre V, Jones AM, Sklenář J, Robatzek S, Weber AP (2012) Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol Plant Microbe Interact 25:1083–1092
Guo C, Guo R, Xu X, Gao M, Li X, Song J, Zheng Y, Wang X (2014) Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot 65:1513–1528
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012) Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep 31:1199–1217
He Y, Mao S, Gao Y, Zhu L, Wu D, Cui Y, Li J, Qian W (2016) Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS One 11:e0157558
Hsu F-C, Chou M-Y, Chou S-J, Li Y-R, Peng H-P, Shih M-C (2013) Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 25:2699–2713
Huh SU, Choi LM, Lee GJ, Kim YJ, Paek KH (2012) Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon Tobacco mosaic virus infection. Plant Sci 197:50–58
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:1
Jiang Y, Duan Y, Yin J, Ye S, Zhu J, Zhang F, Lu W, Fan D, Luo K (2014) Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot 65(22):6629–6644
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029
Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21:481–490
Kulhari A, Sheorayan A, Bajar S, Sarkar S, Chaudhury A, Kalia RK (2013) Investigation of heavy metals in frequently utilized medicinal plants collected from environmentally diverse locations of north western India. SpringerPlus 2:676
Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu J-Q, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23:3824–3841
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34:D257–D260
Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233:1237–1252
Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8:e1002767
Li H, Gao Y, Xu H, Dai Y, Deng D, Chen J (2013a) ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. J Plant Growth Regul 70:207–216
Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET (2013b) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 200:457–472
Ling J, Jiang W, Zhang Y, Yu H, Mao Z, Gu X, Huang S, Xie B (2011) Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom 12:454
Liu RH, Meng J (2003) MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 25:317–321
Liu S, Wang X, Wang H, Xin H, Yang X, Yan J, Li J, Tran L-SP, Shinozaki K, Yamaguchi-Shinozaki K (2013) Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9:e1003790
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem 65:2428–2436
Meng D, Li Y, Bai Y, Li M, Cheng L (2016) Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol Biochem 103:71–83
Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta M, Shanker K, Shukla RK (2013) Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS One 8:e52784
Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, Katayose Y, Mukai Y, Kanamori H, Kurita K (2015) The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci Rep 5:10835
Mukhopadhyay M, Mondal TK (2015) Effect of zinc and boron on growth and water relations of Camellia sinensis. Natl Acad Sci Lett 38:283–286
Nie S, Xu L, Wang Y, Huang D, Muleke EM, Sun X, Wang R, Xie Y, Gong Y, Liu L (2015) Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.). Sci Rep 5:14034
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65:35–47
Rodriguez E, Santos C, Azevedo R, Moutinho-Pereira J, Correia C, Dias MC (2012) Chromium (VI) induces toxicity at different photosynthetic levels in pea. Plant Physiol Biochem 53:94–100
Ross C, Liu Y, Shen Q (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49:827–842
Rushton P, Somssich I, Ringler P, Shen Q (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Scarpeci TE, Zanor MI, Mueller-Roeber B, Valle EM (2013) Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol 83:265–277
Schluttenhofer C, Yuan L (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 167:295–306
Shaik R, Ramakrishna W (2013) Genes and co-expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS One 8:e77261
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shen D, Sun H, Huang M, Zheng Y, Qiu Y, Li X, Fei ZJ (2013) Comprehensive analysis of expressed sequence tags from cultivated and wild radish (Raphanus spp.). BMC Genom 14:721
Song X, Li Y, Hou X (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom 14:1
Sun X, Xu L, Wang Y, Yu R, Zhu X, Luo X, Gong Y, Wang R, Limera C, Zhang K, Liu L (2015a) Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom 16:197
Sun Y, Qiu Y, Zhang X, Chen X, Shen D, Wang H, Li X (2015b) Genome-wide identification of microRNAs associated with taproot development in radish (Raphanus sativus L.). Gene 569:118–126
Sun X, Xu L, Wang Y, Luo X, Zhu X, Kinuthia KB, Nie S, Feng H, Li C, Liu L (2016) Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep 35:329–346
Tang J, Wang F, Wang Z, Huang Z, Xiong A, Hou X (2013) Characterization and co-expression analysis of WRKY orthologs involved in responses to multiple abiotic stresses in Pak-choi (Brassica campestris ssp. chinensis). BMC Plant Biol 13:1
Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15:141–152
Vernay P, Gauthier-Moussard C, Hitmi A (2007) Interaction of bioaccumulation of heavy metal chromium with water relation, mineral nutrition and photosynthesis in developed leaves of Lolium perenne L. Chemosphere 68:1563–1575
Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X (2009) A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230:1155
Wang Y, Xu L, Chen Y, Shen H, Gong Y, Limera C, Liu L (2013) Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to Lead (Pb) stress with next generation sequencing. PLoS One 8:e66539
Wang L, Zhu W, Fang L, Sun X, Su L, Liang Z, Wang N, Londo JP, Li S, Xin H (2014) Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol 14:1
Wang M, Vannozzi A, Wang G, Zhong Y, Corso M, Cavallini E, Cheng Z-MM (2015a) A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front Plant Sci 6:417
Wang R, Xu L, Zhu X, Zhai L, Wang Y, Yu R, Gong Y, Limera C, Liu L (2015b) Transcriptome-wide characterization of novel and heat-stress-responsive microRNAs in radish (Raphanus sativus L.) using next-generation sequencing. Plant Mol Biol Rep 33:867–880
Wang Z, Tang J, Hu R, Wu P, Hou XL, Song XM, Xiong AS (2015c) Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genom 16:17
Wei W, Zhang Y, Han L, Guan Z, Chai T (2008) A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep 27:795–803
Wei KF, Chen J, Chen YF, Wu LJ, Xie DX (2012) Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res 19:153–164
Wu Z-J, Li X-H, Liu Z-W, Li H, Wang Y-X, Zhuang J (2015) Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Funct Integr Genom 15:741–752
Xie Y, Ye S, Wang Y, Xu L, Zhu X, Yang J, Feng H, Yu R, Karanja B, Gong Y (2015) Transcriptome-based gene profiling provides novel insights into the characteristics of radish root response to Cr stress with next-generation sequencing. Front Plant Sci 6:202
Xu L, Wang Y, Xu Y, Wang L, Zhai L, Zhu X, Gong Y, Ye S, Liu L (2013a) Identification and characterization of novel and conserved microRNAs in radish (Raphanus sativus L.) using high-throughput sequencing. Plant Sci 201–202:108–114
Xu L, Wang Y, Zhai LL, Xu Y, Wang LJ, Zhu XW, Gong YQ, Yu RG, Limera C, Liu LW (2013b) Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J Exp Bot 64:4271–4287
Yang G, Wang C, Wang Y, Guo Y, Zhao Y, Yang C, Gao C (2016) Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of cadmium stress. Sci Rep 6:18752
Yu R, Wang Y, Xu L, Zhu X, Zhang W, Wang R, Gong Y, Limera C, Liu L (2015) Transcriptome profiling of root microRNAs reveals novel insights into taproot thickening in radish (Raphanus sativus L.). BMC Plant Biol 15:30
Zentgraf U, Laun T, Miao Y (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89:133–137
Zhai L, Xu L, Wang Y, Zhu X, Feng H, Li C, Luo X, Everlyne MM, Liu L (2016) Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.). Sci Rep 6:21652
Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39:D1114–D1117
Zhang W, Xie Y, Xu L, Wang Y, Zhu X, Wang R, Zhang Y, Muleke E, Liu L (2016) Identification of microRNAs and their target genes explores miRNA-mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus L.). Front. Plant Sci 7:1054
Zhao H, Wu L, Chai T, Zhang Y, Tan J, Ma S (2012) The effects of copper, manganese and zinc on plant growth and elemental accumulation in the manganese-hyperaccumulator Phytolacca americana. J Plant Physiol 169:1243–1252
Zhao H, Wang S, Chen S, Jiang J, Liu G (2015) Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii × Populus nigra. Gene 565:130–139
Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503
Acknowledgements
The current work was partly funded by Grants from the Natural Science Foundation of China (31372064, 31501759, 31601766), National Key Technology Research and Development Program of China (2016YFD0100204-25), Key Technology R&D Program of Jiangsu Province (BE2016379), and Jiangsu Agricultural Science and Technology Innovation Fund (CX(16)1012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Qiaochun Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2017_2190_MOESM1_ESM.tif
Fig. S1 Putative isoelectric points and molecular weights of WRKY proteins Group I, II and III in R. sativus (TIFF 248 kb)
299_2017_2190_MOESM2_ESM.tif
Fig. S2 The schematic diagram of the logo diagrams of ten motifs analyzed in all the 126 WRKY protein sequence in radish (TIFF 497 kb)
299_2017_2190_MOESM3_ESM.tif
Fig. S3 Gene ontology (GO) analysis and distribution of 126 RsWRKY genes into biological process, cellular component, and molecular functions (TIFF 410 kb)
299_2017_2190_MOESM4_ESM.tif
Fig. S4 Heat map showing WRKY genes expression pattern in radish expressed at various developmental stages in leaves, root, root tips, and cambium of radish. The heat map was created using Multi-Experiment Viewer (MeV 4.8) program (TIFF 2064 kb)
299_2017_2190_MOESM5_ESM.tif
Fig. S5 Heat map illustrating WRKY gene transcripts pattern from our radish lab data at different growth stages and biotic stresses. The heat map was created using Multi-Experiment Viewer (MeV 4.8) program (TIFF 1381 kb)
Rights and permissions
About this article
Cite this article
Karanja, B.K., Fan, L., Xu, L. et al. Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Rep 36, 1757–1773 (2017). https://doi.org/10.1007/s00299-017-2190-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2190-4