Abstract
Key message
CRISPR-Cas9/Cpf1 system with its unique gene targeting efficiency, could be an important tool for functional study of early developmental genes through the generation of successful knockout plants.
Abstract
The introduction and utilization of systems biology approaches have identified several genes that are involved in early development of a plant and with such knowledge a robust tool is required for the functional validation of putative candidate genes thus obtained. The development of the CRISPR-Cas9/Cpf1 genome editing system has provided a convenient tool for creating loss of function mutants for genes of interest. The present study utilized CRISPR/Cas9 and CRISPR-Cpf1 technology to knock out an early developmental gene EPFL9 (Epidermal Patterning Factor like-9, a positive regulator of stomatal development in Arabidopsis) orthologue in rice. Germ-line mutants that were generated showed edits that were carried forward into the T2 generation when Cas9-free homozygous mutants were obtained. The homozygous mutant plants showed more than an eightfold reduction in stomatal density on the abaxial leaf surface of the edited rice plants. Potential off-target analysis showed no significant off-target effects. This study also utilized the CRISPR-LbCpf1 (Lachnospiracae bacterium Cpf1) to target the same OsEPFL9 gene to test the activity of this class-2 CRISPR system in rice and found that Cpf1 is also capable of genome editing and edits get transmitted through generations with similar phenotypic changes seen with CRISPR-Cas9. This study demonstrates the application of CRISPR-Cas9/Cpf1 to precisely target genomic locations and develop transgene-free homozygous heritable gene edits and confirms that the loss of function analysis of the candidate genes emerging from different systems biology based approaches, could be performed, and therefore, this system adds value in the validation of gene function studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The emergence and the amalgamation of systems biology resources, data and findings in experimental research currently, has heralded the identification of hitherto unknown genes involved in the early development of plants. Moreover, it has ensured that not only novel genes are identified but also the interaction of several genes involved in key metabolic and developmental pathways can be elucidated rapidly. The utilization of the different ‘-omics’ approaches has pooled in data from the genomic, transcriptomic levels and has given rise to a picture that enhances our understanding of how the early development in plants occurs, the genes involved with the network and interaction that occur between them. Therefore, a need arises to functionally validate the said genes in vivo to see how do they actually act on the plants and how do their interactions affect its growth. This can be answered by studying the loss of function of said genes by employing various genetic engineering techniques such as precise genome editing, which provides researchers with the ability of clear, precise targeting of desired genes and to create knock outs, thus studying the inevitable loss of function of a gene.
The CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats-/CRISPR associated protein 9) genome editing system provides an efficient way of generating targeted mutations in the plant genome (Jiang et al. 2013). A derivation of the prokaryotic immune system,CRISPR-cas9/cpf1 introduces indels at a desired locus in the genome while the randomly inserted T-DNA containing the Cas9 nuclease can be segregated out through negative selection across several generations. The alteration of a specific DNA locus, without leaving behind heterologous genetic elements, serves as an advantage for this system over other genetic modification (GM) approaches (Xu et al. 2015). The adoption of crops produced from modern genetic engineering has improved and diversified crop agronomical traits, but has been hampered by strict regulatory guidelines that hinder release. However, mutations created through this technology mimic natural spontaneous mutations or induced mutations produced using chemicals (e.g. Eethyl methanesulfonate) or Gamma rays, which have much more relaxed regulations for adoption and delivery to the farmers (Voytas 2013). The genetic variation from these kinds of mutation (natural or induced) has long been harnessed by breeders to understand the genetic basis of desirable agronomical traits and to utilize in their breeding programs. Combining robust CRISPR/Cas9 technology and conventional breeding could speed up the pace in which we can improve the traits of the world’s crops needed to sustain future food demand in our rapidly changing environment.
The CRISPR/Cas9 system used in genome editing requires a single guide RNA (sgRNA) that assists the Cas9 protein in recognizing the targeted locus to induce a DNA double strand break (DSB) (Shen et al. 2013). Mutation at the targeted locus happens through the repair of the DSB, facilitated by the error prone process of the non-homologous end joining (NHEJ) DNA repair pathway (Xie and Yang 2013). The sgRNA is a single stranded RNA molecule that contains a section that is complementary to the target site sequence. This helps the Cas9 enzyme to identify the desired locus. The CRISPR/Cas9 used in this study was derived from Streptococcus pyogenes, which requires a 17–20 bp target site that is directly adjacent to a 5′-NGG PAM (protospacer adjacent motif) sequence to be effectively recognized by the sgRNA (Fu et al. 2013). The abundance of NGG motif sequences provides flexibility in choosing a desired locus to serve as a target site for Cas9 cleavage (Shan et al. 2013). However, the simplicity of this approach poses a risk of causing off-target effects in the genome at sites with sequence similarity to the target site. Several bioinformatics tools are available to help predict regions in the genome that contain similarity to the target region (Doench et al. 2014, 2016; Lei et al. 2014; Xie et al. 2014; Yan et al. 2014). Identified genes/genetic regions can be verified for genome edits (mutation) by PCR amplification and subsequent sequencing.
The recent identification of another class 2 CRISPR effector (Cpf1) broadens the horizon for genome editing. Zetsche et al. (2015) have shown that Cpf1 allows robust genome editing in human cell-lines and can target A/T-rich areas of the genome and thus increases the number of locations that can be edited. Two major orthologues of Cpf1 from Acidaminococcus sp and Lachnospiracae bacterium showed significant genome editing activity. The Cpf1 not only targets adjacent to a T-rich PAM, but also it has certain unique characteristics that differ from Cas9 (Zetsche et al. 2015). The CRISPR-Cpf1 system is simpler, only requiring one crRNA in contrast to Cas9 where one crRNA and a tracrRNA complex are required. Moreover, in contrast to Cas9’s blunt-ended double strand break, the CRISPR-Cpf1 system introduces a staggered DNA double strand break, which can be useful in generating efficient gene insertions in plant systems.
In this study we have used the OsEPFL9 gene as a marker to test the efficiency of the CRISPR-Cas9 and CRISPR-LbCpf1 systems in rice. A developmental gene, OsEPFL9 also known as rice STOMAGEN is part of a family of secretory signal peptides that regulate leaf stomatal density (Kondo et al. 2010; Sugano et al. 2010). In Arabidopsis, AtEPFL9 gene expression positively regulates stomatal development and correlates with increase in stomatal density (Hunt et al. 2010). Knock-down of the AtEPFL9 gene expression produces plants with decreased stomatal density, a phenotype which is good for analyzing the heredity of the mutations even after several generations. OsEPFL9 was targeted in rice to serve as a visible phenotypic screen to support the sequencing results. Segregating mutant lines showed an incomplete dominant phenotype that helps in validating the zygosity of mutant lines. Exon 1 of OsEPFL9 was targeted by CRISPR-Cas9 and CRISPR-LbCpf1 separately. T0 plants that were regenerated showed the presence of the targeted gene edits by both systems. CRISPR-Cas9 mediated knockout lines were taken through several generations to check that the edits were stabilized and that the segregation of the Cas9 had occurred. This study reports, the application of CRISPR-Cpf1 in plants and also reports CRISPR-Cas9 mediated editing of the OsEPFL9 gene in rice, transmission of the edits through generations, segregation of Cas9 and development of a visible phenotype with more than eightfold decrease in stomatal density.
Materials and methods
Construct designing
Generation of pCambia-CRISPR_Cas9 vector
A backbone pCambia-CRISPR_Cas9 binary vector was generated for the establishment of a robust rice genome editing system. The plasmid DNA of pOsU3-sgRNA that contained the transcript of a single guide RNA (sgRNA) and pJIT163-2NLSCas9 CRISPR/Cas9 that expressed rice-codon-optimized Cas9 (Fig. S6) was a kind gift from Caixia Gao’s lab. The two functional cassettes from each vector were cloned into the pCambia vector system for Agrobacterium-mediated rice transformation.
The sgRNA cassette of pOsU3-sgRNA and the Cas9 cassette of pJIT163-2NLSCas9 and the backbone of the modified pCambia1300 were isolated by restriction enzyme digestion and were agarose gel purified respectively, and were ligated to produce the backbone pCambia-CRISPR/Cas9 binary vector.
The backbone pCambia-CRISPR_Cas9 vector was digested with AarI to create two unique sticky ends. Target sequence was formed by annealing a pair of oligos (EPFL9-Cas9-Target-F and EPFL9-Cas9-Target-R) that had compatible sticky ends to the AarI digested backbone pCambia-CRISPR/Cas9 vector. The first exon of the OsEPFL9 gene in rice cv. IR64 (OsIR64_00032g010800.1, Rice SNP-Seek Database, IRIC) was targeted.
Generation of pCambia-LbCpf1 vector
The pCambia-LbCpf1 working backbone vectors were developed from the pCambia-CRISPR_Cas9 vector. The guide RNA scaffold of pCambia-CRISPR_Cas9 was removed by restriction digestion using AarI (ThermoFisher Sceintific) and XbaI (NEB). A pair of oligos (LbCpf1-gRNA-F and LbCpf1-gRNA-R) was annealed to create the compatible sticky end to AarI-XbaI digested pCambia-CRISPR_Cas9, and the annealed oligo carried LbCpf1 specific gRNA scaffold (Zetsche et al. 2015) and the BaeI recognition site to help the insertion of designed target, and a 41-nt long gRNA transcription terminator that was taken from the pCambia-CRISPR_Cas9 (Fig. S8).
The intermediate vectors carried LbCpf1 gRNA scaffold, were further digested with HindIII and SalI to remove the Cas9 coding sequence and the terminator. The coding sequence of LbCpf1 was PCR amplified (Cpf1-F and LbCpf1-R) from pcDNA3-huLbCpf1 that was kindly given as a gift by Feng Zhang’s lab (Zetsche et al. 2015) (Fig. S7). The CaMV terminator was PCR amplified using LbCpf1-NLS-F and Cpf1-NLS-R. The two PCR products were used as templates for overlapping PCR (Cpf1-F and Cpf1-NLS-R). The product of the overlap PCR covered a unique HindIII restriction site upstream of the Cpf1 cassette and a unique SalI restriction site downstream of the Cpf1. The overlap PCR product was digested with HindIII and SalI and was then ligated with the HindIII-SalI digested intermediate vectors, to produce the working backbone vector pCambia-LbCpf1.
Guide sequences were produced by oligo annealing that had the compatible sticky ends to the restriction enzyme digested working backbone vectors (AarI for pCambia-CRISPR_Cas9, and BaeI for pCambia-LbCpf1).
All oligo DNA used in this study were ordered from Macrogen. Vector maps were generated using SnapGene.
Rice transformation
The Indica rice cultivar IR64 was used in this transformation. pCambia-CRISPR_Cas9-EPFL9 was transformed to rice via A. tumefaciens mediated transformation using rice immature embryos (IE) (Hiei and Komari 2008) that were collected from immature seeds of rice panicles harvested 12 days after anthesis.
A. tumefaciens transformed with the working vector, was mixed with infection medium. Five microliter of the A. tumefaciens suspension was dropped on top of each IE and the IEs were allowed to co-cultivate for 7 days at 25 °C in the dark. Elongated shoots were removed from IEs after co-cultivation and the IEs were gently blotted on sterile filter paper. Blotted IEs were transferred to the resting medium (Hiei and Komari 2008) and incubated at 30 °C for 5 days under continuous illumination. Each IE was cut into 4 pieces. All cut IEs were incubated on selection medium, containing 30 mg/L hygromycin, for 10 days at 30 °C (Hiei and Komari 2008), and then all were transferred to fresh selection medium for another 10 days. After being incubated twice on the selection medium, hygromycin-resistant calli were selected to transfer to the 3rd time selection for 10 days. After the 3rd time selection, the hygromycin-resistant calli were moved to pre-regeneration medium, containing 50 mg/L hygromycin, to incubate for 10 days at 30 °C (Hiei and Komari 2008). Profilerating calli were then transferred onto regeneration medium (Hiei and Komori 2008), containing 50 mg/L hygromycin and were allowed to grow for 10–15 days until the roots were about 2 mm long. Regenerated plantlets were then transferred and grown in Yoshida Culture Solution (YCS) for 2 weeks (Datta and Datta 2006). Regenerated plants were screened for their transgene (i.e. Cas9 or Cpf1) by PCR using primer Cas9-F and Cas9-R for pCambia-CRISPR_Cas9-EPFL9 and LbCpf1-F and LbCpf1-R for pCambia-LbCpf1-EPFL9. Plants of positive PCR results were maintained and were transferred to soil after 2 weeks growing in YCS.
Surveyor assay
PCR was performed using Phusion® High-Fidelity DNA Polymerase (NEB) with isolated genomic DNA. The product size was 795 bp using primer EPFL9-seq-F and EPFL9-seq-R. For every PCR that was performed for transformed plant, a wild type (WT) control PCR was performed alongside under the same conditions. The total volume of each reaction was 15 µL. After the completion of the PCR, 3.5 µL from each reaction was loaded on agarose gel to confirm the success of PCR reaction.
For the surveyor assay of each sample, 6 µL of the PCR product from the transformed plant and 6 µL of the PCR product from the WT control were well mixed. Each mixed PCR product was hybridized to form DNA heteroduplexes and were then digested with Surveyor Nuclease, following the user guide of Surveyor ® Mutation Detection Kit (IDT, 706020).
Digested DNA was run on 2% agarose gel and samples with visible digested bands (expected size 463 and 332 bp) were selected as surveyor assay positives.
Indel analysis
The PCR products of selected plants were directly sequenced. Each PCR reaction had a volume of 40 µL and was performed using Q5® High-Fidelity DNA Polymerase (NEB). PCR products were purified from agarose gel (QIAquick Gel Extraction Kit, QIAGEN) and were sequenced directly by Sanger sequencing.
When the chromatogram of a sample shows single peaks, sequence is directly aligned with reference sequence (IR64, Rice SNP-Seek Database, IRIC).
When double peaks were observed, the chromatogram was subjected to online indel detection tools poly peak parser (Hill et al. 2014) to detect the alternative sequences and TIDE (Brinkman et al. 2014) to determine the frequency of the alternative sequences. Manual checking was also performed subsequently, due to the minor errors and limitation of each tool. Samples with the ratio of [frequency of alternative sequence]: [frequency of WT sequence] greater than 0.8: 1 were arbitrarily selected. At T0 generation, such plants were considered as heterozygous mutants (having germline mutation) and were brought to further generation.
Southern blotting
Southern blotting technique was employed to assess the T-DNA copy number of the T2 transgenic plants. Twelve micrograms of DNA sample was digested using XbaI (New England Biolabs, USA) at 37 °C for 16 h. The digested samples were then separated using 0.8% agarose gel electrophoresis with 1X TAE buffer at 30 volts overnight. DIG-labeled molecular weight marker II (Roche Diagnostics, Germany) was used to determine the apparent molecular size of the bands. Un-transformed rice was used as negative control. Two samples of Cas9 PCR positive were used as positive control. The gel was processed in preparation for neutral transfer. DNA was transferred from the gel into Hybond Nylon+ membrane (GE Healthcare, UK) using capillary method and neutral transfer buffer using 20X SSC (0.3 M tri-sodium citrate acetate dehydrate, pH 7.0, in 3 M NaCl). The blots were hybridized with probe synthesized using PCR DIG Probe Synthesis Kit (Roche Diagnostics, Germany) using primers specific to the Hygromycin gene (HptII-F and HptII-R). Hybridization was done at 42 °C overnight and washed the next day. Anti-DIG Fab Fragment-AP conjugate (Roche Diagnostics, Germany) was used to detect the DIG labeled probe. The blots were detected using CDP-Star Detection Reagent (Roche Diagnostics, Germany) following manufacturers protocols.
Microscopy
Middle portion (about 10 cm long) of the youngest fully expanded leaf of each plant was sampled at maximum tillering stage. The adaxial (upper) epidermis and mesophyll cells were gently scratched and removed with a razor blade. The near transparent abaxial (lower) epidermis was allowed to remain and was transferred to a glass slide with water.
Transmitted bright field images were captured with Olympus BX61 connected to a Hamamatsu ORCA-Flash2.8 camera (0.090 cm2 area image under 10× magnification), Olympus BX63 microscope connected to an Olympus DP71 camera (0.144 cm2 area image under 10× magnification), or Olympus BX51 microscope connected to an Olympus DP71 camera (0.144 cm2 area image under 10 x magnification). Images of 5 areas of each leaf sample were taken for stomata counting.
Off-target analysis
The Cas-OFFinder (Bae et al. 2014) was used to identify the potential off-target sites in rice. Several parameters from higher stringency to lower stringency were employed, as shown below:
-
a.
Mismatch = 1; DNA Bulge Size = 0; RNA Bulge Size = 0
-
b.
Mismatch = 2; DNA Bulge Size = 0; RNA Bulge Size = 0
-
c.
Mismatch = 3; DNA Bulge Size = 0; RNA Bulge Size = 0
-
d.
Mismatch = 0; DNA Bulge Size = 1; RNA Bulge Size = 1
-
e.
Mismatch = 1; DNA Bulge Size = 1; RNA Bulge Size = 1
-
f.
Mismatch = 2; DNA Bulge Size = 1; RNA Bulge Size = 1
-
g.
Mismatch = 3; DNA Bulge Size = 1; RNA Bulge Size = 1
-
h.
Mismatch = 0; DNA Bulge Size = 2; RNA Bulge Size = 2
-
i.
Mismatch = 1; DNA Bulge Size = 2; RNA Bulge Size = 2
All identified sites were mapped in rice IR64 genome. The sites that were successfully mapped in IR64 genome were ranked according to less disturbed to more disturbed at 5–12 positions beyond the tracrRNA:crRNA. The 10 highest ranked sites were selected.
Primers were designed flanking the potential off-target sites (Table S1). PCR products (amplified with Q5® High-Fidelity DNA Polymerase, NEB) were sequenced directly.
Results
CRISPR-Cas9 and CRISPR-Cpf1 mediated genome editing in T0 transgenic rice
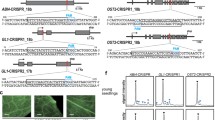
To investigate the editing efficiency of Cas9 and Cpf1, we targeted the Arabidopsis EPFL9 gene orthologue in rice, which is an important positive regulator of stomatal developmental pathway as found in Arabidopsis (Hunt et al. 2010; Rychel et al. 2010; Takata et al. 2013; van Campen et al. 2016). Using both nucleases, Cas9 and Cpf1, we targeted exon-1 of the EPFL9 gene (Fig. S3). The Cpf1 target sequence in this study was chosen very close to the Cas9 target sequence for more accurate comparison (Fig. 1 and Fig. S3). In case of both Cas9 and Cpf1 targets chosen, the cleavage site is after the start codon (underlined in the figure) the result of which would be the disruption of the ORF of the EPFL9 gene. Alignment of the target region of OsEPFL9 (both Cas9 and Cpf1) in this study against other members of the EPF/EPFL gene family showed no significant similarity that would cause off-target effect (Fig. S11).
Designed target sites of the OsEPFL9 gene for the Cas9 and Cpf1 system. a Gene structure of the OsEPFL9 gene. The target sites of Cpf1 and Cas9 are both located on the coding sequence of the first exon. b Target site of Cas9-OsEPFL9 (red boxes) of which the smaller box shows the PAM; c Target site of LbCpf1 (blue boxes) of which the smaller box shows the PAM
One hundred T0 plants of each construct, that were PCR positive for the nuclease (Cas9-F and Cas9-R for pCambia-CRISPR_Cas9-EPFL9 plants, LbCpf1-F and LbCpf1-R for pCambia-LbCpf1-EEFL9 plants), were analyzed by the Surveyor assay. The PCR product of samples that were positive in the surveyor assay were sequenced directly, using the primers flanking the target region (EPFL9-seq-F and EPFL9-seq-R, Fig. 2a). From the 100 pCambia-CRISPR_Cas9-EPFL9 T0 plants that were analyzed, 4 showed double peaks in the sequencing chromatogram (Fig. S1) that indicated heterozygous gene edited plants (4%). For the 100 pCambia-LbCpf1-EPFL9 plants that were analyzed, 10 showed doubled peaks in the sequencing chromatogram (Fig. S2) that indicated heterozygous plants (10%).
Detailed sequence analysis (Fig. 3) of the pCambia-CRISPR_Cas9-EPFL9 plants showed that the maximum deletion size was 37 bp, and the minimum deletion size was 4 bp, averaging about 13 bp. For the pCambia-LbCpf1-EPFL9 plants, the maximum mutation size was 63 bp, and the minimum deletion size was 1 bp, with an average of about 13.5 bp.
The score for the probable gRNA on-target activity, using DESKGEN software (http://www.deskgen.com), showed 69% likelihood for the Cas9-EPFL9, while the Cpf1 showed only 7% likelihood (Fig. 4). Our results show that, even with much lower probable gRNA on-target activity, the Cpf1 system still produced a higher percentage of stably edited mutants.
The sizes of the mutation induced by the two systems were comparable. The induced DNA double strand break is repaired by the NHEJ repair pathway in plants and is affected by local micro-homologous DNA sequences. For this reason, we chose the Cpf1 target site that was close to the target site of the Cas9 mutated site. Despite the fact that low gRNA on-target activity was predicted by DESKGEN in the Cpf1 target site (7% probability for the Cpf1 system, vs. 69% for the Cas9 system), more than double the percentage of edited plants were observed with the LbCpf1 system in T0. Taking into account the high similarity of the vector design of the two systems in this study (Figs. S4, S5), the result indicates that the LbCpf1 system may also be an efficient genome-editing tool for rice and reports application of Cpf1 in a plant system.
Segregation of targeted mutation in the T1 generation
Event 13 and 24 of pCambia-CRISPR_Cas9-EPFL9 were taken to the T1 generation. The Surveyor assay along with the direct sequencing of the PCR product, were performed on the T1 plants. Many T1 samples showed digested bands in the surveyor assay. The sequencing results were seen to have the expected Mendelian segregation pattern (Table 1). As shown in Fig. 5, homozygous (highlighted in green), heterozygous (highlighted in yellow) and azygous (no highlight) were confirmed by analyzing the chromatogram of the sequencing results (data not shown). Furthermore, the mutation seen in T1 plants was the same as in their T0 parent plants. This confirmed that the mutation in the T0 plants was indeed a germline mutation.
PCR of Cas9 and Cpf1 gene and surveyor assay of selected T0 and T1 plants. a All T1 progenies of event 13 that were tested were still positive in Cas9 PCR; clear digested bands were seen in T1 progenies. b Three T1 progenies of event 24 were negative in Cas9 PCR, of which two were confirmed by sequencing to have inherited the mutation at the target site. Homozygous plants are highlighted in green; heterozygous plants are highlighted in yellow. (Color figure online)
PCR analysis of the tested T1 plants of event 13 all still harboured the Cas9 gene. However, with event 24, two progeny (07 and 08) were Cas9 PCR negative and were also homozygous for the targeted mutation. This result shows that Cas9-free homozygous mutants can be obtained as early as the T1 generation in rice.
It is interesting to note that in the case of CRISPR-Cpf1 event 006 T0 and T1 lines there is a difference in the sizes of the band produced after the Surveryor assay. The Surveyor assay, involves the hybridization of a wild-type DNA strand and a transgene DNA strand. The Surveryor enzyme is known to cut the DNA at a location where it encounters a mismatch. The mismatch in case of the event 006 is 63 bp may have contributed to the shifts in band sizes and an increase in band numbers that are observed in the Surveyor assay. Another thing of note is the T1 plant Cpf1-006-01 (Fig. S9), which seems to have developed a new mutation which was not present in the T0 due to the action of the transgene. After sequencing it was discovered that the mutation was different from the rest of the plants of that event, perhaps introduced by the transgene. The reason for it being Surveyor positive is the fact that the enzyme is only capable of displaying if a given sequence has a mutation or not when compared to the wild-type; but not the nature and exact details of the said mutation.
Screening of the Cas9-free homozygous mutant at T2 generation
Table 2 shows the T1 lines that were selected and brought forward to the T2 generation. Three homozygous lines of each event were selected, and for event 24, the two Cas9 PCR negative lines were included. One heterozygous line of each event was also advanced as a backup if the homozygous lines gave no real Cas9-free progenies.
Thirty seeds of each T1 line were germinated. All germinated plants were immediately screened by PCR using the Cas9-F and Cas9-R primer pair. Plants with negative PCR results were selected for sequencing of the target region.
One T2 progeny of Cas9-013-01, one of Cas9-013-02 and one of Cas9-013-11 was Cas9 PCR negative. Five T2 progenies of Cas9-013-13 were Cas9 PCR negative and were sequenced. The sequencing results of all five lines showed a homozygous targeted mutation, which was consistent with their T0 and T1 parent plants (data not shown).
Eight T2 progenies of Cas9-024-01 and seven of Cas9-024-04 were Cas9 PCR negative, while all T2 progenies of Cas9-024-07 and Cas9-024-08 were Cas9 PCR negative (data not shown).
We selected five T2 progenies of Cas9-024-07 and all five PCR negative T2 progenies of Cas9-013-13, as well as two Cas9 PCR positive as positive controls (from Cas9-024-01) to perform Southern Blot to confirm the Cas9 PCR negative plants were Cas9-free (Fig. 6).
Southern Blot of selected Cas9 PCR negative plants. a Cas9 PCR using primer Cas9-F and Cas9-R, the green boxes indicate Cas9 free plants, the red boxes indicate the positive PCR controls; b Southern Blot, the green boxes indicate transfene free plants, the red boxes are the positive control for the transgene. (Color figure online)
Analysis of altered stomatal phenotype in the edited homozygous transgene-free T2 plants
The average stomatal density of the abaxial epidermis in the middle portion of the 6th leaf was measured at the maximum tillering stage. T2 Cas9-EPFL9 plants with homozygous mutations at the target site were measured.
Figure 7 shows the average stomatal density of event 13 and event 24 of T2 homozygous Cas9-EPFL9 plants compared to the wild-type control. Both events were seen to have more than an eightfold decrease in the average stomatal density. This clearly demonstrates the importance of OsEPFL9 expression for stomatal production in rice and strongly supports the role of the stomagen peptide orthologue in rice epidermal development. Furthermore, it identifies OsEPFL9 as a useful target gene in planta for tractable testing of gene targeting techniques.
Analysis of potential off-target effects of CRISPR-Cas9 modified plants
Cas-OFFinder (Bae et al. 2014) was used to identify potential off-target loci in the rice genome. The 10 loci with the highest ranking off-target potential were aligned to the IR64 genome and primers were designed to amplify each region (Table 3). Two T2 Cas9-free homozygous mutant plants were analyzed, Cas9-013-13-29 and Cas9-024-07-03. The same regions were also amplified using wild-type genomic DNA as comparison. Sequencing chromatograms are shown in Fig. S10. The primers that were used for amplifying these regions are listed in Table S1.
No secondary off target mutations were detected near to the potential off-target loci in Cas9-EPFL9 plants. A previous study reported that 5–12 positions beyond the tracrRNA:crRNA base-pairing interaction are important for efficient Cas9 binding and target recognition (Jinek et al. 2012). The target sequence identified in this study was selected to avoid possible off-target sequences that have overall similarity to the desired target.
All Cas9-EPFL9 plants with homozygous target mutations were seen to have drastic reductions in stomatal density, including Cas9-free plants. No off-target effects were found in the Cas9-free homozygous mutant plants from potential off-target analysis. The results indicate that the phenotype was due to the dysfunction of the stomatal development related gene EPFL9 as reported previously (Hunt et al. 2010).
Discussion
In the current scenario of research, systems biology plays an important role in the identification of genes that are involved in the regulation of the multitude of pathways that enable a plant to grow and survive. The approach used by systems biology of collecting and integrating data generated by studying the plant under different ‘-omics’ viz. genomics, phenomics, transcriptomics, metabolomics, ionomics, etc. have generated data to help and understand the various genes that are involved in plant development. Varied systems biology approaches have been used to assess understand and fill the gaps in between the genes and metabolites and the underlying interaction between the genes and the regulators and their expression, involving both BOTTOM-UP and TOP-DOWN approach (Saito and Matsuda 2010; Yoshida et al. 2010). But as new data are being generated, functional validation of said genes also becomes extremely important to actually visualize the effects of said genes on a plant. The most favored method for this kind of validation is the loss of function of genes and to see how does the loss of the genes affect the growth of a plant. While in the past several methods have been used to generate loss of function mutants in plants, none have been precise and accurate resulting in knock out of undesired genes and genomic locations. With the advent of genome editing it has now become possible to precisely target particular locations in the genome of our interest thus helping in generation loss of function mutants in the genes of interest and help in functionally validate the genes new or known and visualize the interaction amongst, completing the small gaps in the picture that was created by systems biology approach.
Several genome editing tools have been discovered prior to CRISPR/Cas9 but none of them surpassed its ease of use and efficiency (Hsu et al. 2014; Kumar and Jain 2015; Liang et al. 2014). All of these genome-editing tools rely on a system that has a DNA sequence-recognizing element and a DNA-cleaving element to induce genome modifications. Unlike the Zinc finger nucleases or TALEN (transcription activator-like effector nucleases) which use protein complexes to serve as DNA recognizing elements, CRISPR/Cas9 makes use of short RNA molecules (sgRNA) that can be modified easily and cheaply (Voytas 2013; Chen and Gao 2013; Mali et al. 2013; Reyon et al. 2012). The attractiveness of this genome engineering tool has been proven by its wide adoption in animals, particularly in medicine, and in plants (Jiang et al. 2013; Fu et al. 2013; Brooks et al. 2014; Chang et al. 2013; Cho et al. 2013; Deriano and Roth 2013; Friedland et al. 2013). Recent development of the technology expanded its capacity, by altering the nuclease domain of the enzyme and adding a nucleotide-modifying enzyme. With this, researchers were able to induce specific single nucleotide changes to human genes responsible for genetic diseases (Komor et al. 2016; Plosky 2016). Such modifications would be useful for plant systems if introduced in the correct way and in the correct context. Recently, genome editing by single strand oligonucleotide coupled with CRISPR associated protein was demonstrated (Sauer et al. 2016). This provides a promising tool for future application of SNP modifications for crop plants. Furthermore, intron mediated site-specific gene replacement and insertion opens up new doors for generating mutations/allele replacements by NHEJ (Li et al. 2016). Successful application of transiently expressed CRISPR/Cas9 or in vitro transcripts of Cas9 coding sequence and guide RNA in wheat callus cells showed efficient genome editing in hexaploid bread wheat, as well as tetraploid durum wheat (Zhang et al. 2016). This system shows promise for application across a range of crop species. It will also be important to identify further unique nucleases that target more efficiently than Cas9. Cas9 targeting is generally restricted to the G/C rich area of the genome. Inclusion of Cpf1 with its capability of targeting T/A rich areas of the genome would increase the available editing tools and permit broader coverage of the genome that can be edited. Recent studies have shown the use of Cpf1 to successfully edit the rice genome (Xu et al. 2016; Endo et al. 2016). We also demonstrate here that CRISPR/Cpf1 mediated successful editing of rice gene.
As it was seen in the surveyor assay the presence of the transgene in subsequent generations could give rise to new mutations indicating that the transgene is still active. Thus, it is important to obtain transgene free, stable mutants in the subsequent generations. In agreement with other reports we show successful transmission of genome edits through subsequent generations and the production of clean homozygous lines.
Classical transgenic development happens by introducing the GOI (gene of interest) randomly into the genome, which results in a large amount of downstream work to characterize the offspring in search of a suitable event where the GOI has landed in a safe locus devoid of unwanted effects. CRISPR-mediated gene editing can aid targeted gene insertion to a particular locus by its unique gRNA-nuclease-aided site-specific targeting. Moreover, the CRISPR system (Cas9/Cpf1-gRNA) is not required to remain in the genome and can be segregated out after the editing is complete. The final product is, therefore, transgene free and thus may require no/less legislative legal regulation, which could reduce the financial burden of premarket approval and also increase the social acceptance of genome-edited crops. In this work, we demonstrate that the role of the epidermal patterning factor EPFL9/stomagen is a positive regulator of stomatal development in rice; that transgene-free homozygous gene editing is possible as early as the T1 generation; and furthermore we report the application of CRISPR/Cpf1 in plants. This proves that this new method of genome editing can be applied to create loss of function mutants and thus help in validating putative candidate genes for early development in plant. The integration of both the ability to find novel genes and to visualize the interaction between different genes, and functionally validate them in vivo will lead to a better understanding of how early development in plant proceeds ahead.
References
Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30(10):1473–1475. doi:10.1093/bioinformatics/btu048
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42(22):e168–e168
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system1. Plant Physiol 166:1292–1297
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ (2013) Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23:465–472
Chen K, Gao C (2013) TALENs: customizable molecular DNA scissors for genome engineering of plants. J Genet Genomics 40:271–279
Cho SW, Kim S, Kim JM, Kim J-S (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232
Datta K, Datta SK (2006) Indica rice (Oryza sativa, BR29 and IR64). Methods Mol Biol 343:201–212
Deriano L, Roth DB (2013) Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 47:433–455
Deskgen. http://www.deskgen.com
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE (2014) Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol 32:1262–1267
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. doi:10.1038/nbt.3437
Endo A, Masafumi M, Kaya H, Toki S (2016) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6:38169. doi:10.1038/srep38169
Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3:824–834
Hill JT, Demarest BL, Bisgrove BW, Su YC, Smith M, Yost HJ (2014) Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn 243(12):1632–1636
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Hunt L, Bailey KJ, Gray JE (2010) The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186:609–614
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:1–12
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier EA (2012) Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–21
Komor AC, Kim YB, Packer MS, Zuris J, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 61:5985–5991
Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, Kakimoto T (2010) Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol 51:1–8
Kumar V, Jain M (2015) The CRISPR-Cas system for plant genome editing: advances and opportunities. J Exp Bot 66:47–57
Lei Y, Lu L, Liu H-Y, Li S, Xing F, Chen L-L (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C (2016) Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat Plants 2:16139
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013) Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838
Polsky BS (2016) CRISPR-mediated base editing without DNA double-strand breaks. Mol Cell 62:477–478
Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30:460–465
Rychel AL, Peterson KM, Torii KU (2010) Plant twitter: ligands under 140 amino acids enforcing stomatal patterning. J Plant Res 123(3):275–280
Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61:463–489
Sauer NJ, Narváez-Vásquez J, Mozoruk J, Miller RB, Warburg ZJ, Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL, Walker KA (2016) Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol 170(4):1917–1928
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X (2013) Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23:720–723
Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463:241–244
Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M (2013) Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8(6):e65183. doi:10.1371/journal.pone.0065183
van Campen J, Yaapar MN, Narawatthana S, Lehmeier C, Wanchana S, Thakur V, Chater C, Kelly S, Rolfe SA, Quick WP, Fleming AJ (2016) Combined chlorophyll fluorescence and transcriptomic analysis identifies the P3/P4 transition as a key stage in rice leaf photosynthetic development. Plant Physiol. doi:10.1104/pp.15.01624
Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64:327–350
Xie K and Yang Y (2013) RNA-Guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983
Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for the CRISPR–Cas9-Mediated genome editing in model plants and major crops. Mol Plant 1:1–10
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:11491
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J (2016) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. doi:10.1111/pbi.12669
Yan M, Zhou SR, Xue HW (2015) CRISPR Primer Designer: Design primers for knockout and chromosome imaging CRISPR‐Cas system. J Integr Plant Biol 57(7):613–617
Yoshida R, Saito MM, Nagao H, Higuchi T (2010) Bayesian experts in exploring reaction kinetics of transcription circuits. Bioinformatics 26:i589–i595
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163.3: 759–771
Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu JL, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun, vol 7. doi:10.1038/ncomms12617
Acknowledgements
We thank Prof. Caixia Gao of Chinese Academy of Science, China for providing us Cas9 and gRNA scaffold constructs. Also we would like to thank Dr. Feng Zhang of Broad Institute MIT for providing us pcDNA3-huLbCpf1. We also thank Florencia Montecillo and Juvy Reyes for helping in rice transformation and molecular work respectively.
Author contribution statement
XY, AKB and JD contributed equally. AB, PQ, JG and XY designed the experiments and wrote the manuscript. Vector designing and construction was done by XY, AKB, AB and TK (CRISPR-508 Cas9). XY,SM, CPB, CC, AB (CRISPR-Cpf1). Surveyor assay and sequencing was performed 509 by KP, XY, SM, CPB (Cas9 plants and Cpf1 plants) and AKB (Cas9 plants). Rice 510 transformation was supervised by XY. Microscopy was performed by JD, HL and RC. Data 511 analysis was performed by AB and XY. Southern blotting was performed by CPB.
Author information
Authors and Affiliations
Contributions
AB, PQ, JG and XY designed the experiments and wrote the manuscript. Vector designing and construction was done by XY, AKB, AB and TK (CRISPR-Cas9). XY,SM, CPB, CC, AB (CRISPR-Cpf1). Surveyor assay and sequencing was performed by KP, XY, SM, CPB (Cas9 plants and Cpf1 plants) and AKB (Cas9 plants). Rice transformation was supervised by XY. Microscopy was performed by JD, HL and RC. Data analysis was performed by AB and XY. Southern blotting was performed by CPB.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Communicated by Nese Sreenivasulu.
Xiaojia Yin, Akshaya K. Biswal and Jacqueline Dionora have Contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yin, X., Biswal, A.K., Dionora, J. et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep 36, 745–757 (2017). https://doi.org/10.1007/s00299-017-2118-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2118-z