Abstract
Mutagenesis continues to play an essential role for understanding plant gene function and, in some instances, provides an opportunity for plant improvement. The development of gene editing technologies such as TALENs and zinc fingers has revolutionised the targeted mutation specificity that can now be achieved. The CRISPR/Cas9 system is the most recent addition to gene editing technologies and arguably the simplest requiring only two components; a small guide RNA molecule (sgRNA) and Cas9 endonuclease protein which complex to recognise and cleave a specific 20 bp target site present in a genome. Target specificity is determined by complementary base pairing between the sgRNA and target site sequence enabling highly specific, targeted mutation to be readily engineered. Upon target site cleavage, error-prone endogenous repair mechanisms produce small insertion/deletions at the target site usually resulting in loss of gene function. CRISPR/Cas9 gene editing has been rapidly adopted in plants and successfully undertaken in numerous species including major crop species. Its applications are not restricted to mutagenesis and target site cleavage can be exploited to promote sequence insertion or replacement by recombination. The multiple applications of this technology in plants are described.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Continued crop plant germplasm improvement is an ongoing requirement to meet the ever increasing demand for food production. Several decades ago mutagenesis and selection was used extensively in the hope of generating new genetic diversity of agronomic value. In this approach, mutations were generated using both radiation and chemical mutagens. Some trait improvements were made (reviewed by Ahloowalia and Maluszynski 2001), however, rational design and tailored production of new alleles were not possible due to a lack of targeted DNA modification tools available at the time. Nevertheless, random mutagenesis coupled with either phenotypic screening or TILLING has been extremely useful for demonstrating gene function and facilitating gene isolation, as has the large-scale production and characterisation of random T-DNA insertion libraries (reviewed by Koornneef 2002; Parry et al. 2009).
A major development in increasing plant germplasm diversity is by transgenesis. Globally, an ever-expanding area of transgenic crop species are being grown (175 million hectares in 2013) that contain an increasing number of transgenes for trait improvement, including insect resistance, herbicide resistance, improved nutritional quality and modified oil (ISAAA 2013). A limitation of current plant transformation technologies is that insertion of transgene sequences into the genome is essentially random. With an expanding number of transgenic traits available this becomes problematic as an increasing number of transgenes located randomly in the genome requires a correspondingly increased breeding effort to keep these unlinked loci together (Que et al. 2010). For example, the Monsanto/Dow AgroSciences maize lines Smartstax contains eight transgenes, six Cry genes and two herbicide resistance genes, located at multiple loci (Que et al. 2010). As future transgene numbers grow, this difficulty will become exacerbated without the development of new technological advances.
The emerging technology of CRISPR/Cas9 gene editing offers the potential to overcome some of the difficulties described above that arise from the imprecise nature of random mutagenesis and the random integration of transgenes into the plant genome. The CRISPR/Cas9 system enables precise targeting and cleavage of a single 20 bp sequence within a large genome. This remarkable specificity can be exploited for highly targeted mutagenesis or targeted gene insertion. There are alternatives to CRIPRS/Cas9 genome editing, including TALENs, zinc finger nucleases and meganucleases, and each system has advantages and disadvantages. CRISPR/Cas9 is the most recent of these technologies and advances in both gene mutation and gene targeting using this system in plants are described herein.
The CRISPR/Cas system
CRISPR/Cas systems (I–III) are prokaryote defense systems, common to many archaea and bacteria, which protect the prokaryotic genome from invading nucleic acids (Makarova et al. 2011). Remnants of invading sequences are co-located in regions of the bacterial genome as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), with invading plasmid or viral sequences (27–72 bp) separated by short direct repeats (21–47 bp). In type II CRISPR/Cas systems, these small remnant sequences are actively transcribed and processed into small CRISPR RNAs (crRNAs) that complex with a trans-activating CRISPR RNA (tracrRNA) and an endonuclease protein, Cas9 (Chylinski et al. 2014). Nucleotides (19–22 bases) at the 5′ end of the crRNA target the Cas9 complex to complementary invading sequences that are subsequently cleaved by the endonuclease (Horvath and Barrangou 2010). A pre-requisite for processing of the target sequence is that a trinucleotide sequence, NGG, is required in juxtaposition to the 3′ end of the target site on the non-complementary strand. This trinucleotide sequence is known as a protospacer adjacent motif (PAM) sequence and is the primary crRNA/Cas recognition site from which strand separation and RNA–DNA heteroduplex formation is initiated (Sternberg et al. 2014). Cas9-mediated cleavage of the complementary and non-complementary DNA strands generally occurs three nucleotides and three to eight nucleotides, respectively, from the PAM sequence within the 19–22 bp region of crRNA and target site identity.
A minimal version of the type II CRISPR/Cas9 system, generally using the Cas9 gene from Streptococcus pyogenes, has now been used in a variety of eukaryotic organisms, including plants, to target and cleave a specific sequence in the genome. A significant biotechnological simplification for exploitation of this system in vivo has been the design of synthetic small guide RNAs (sgRNAs) that forgo the tracRNA requirement and thereby reduce the CRISPR/Cas9 system to just two genes; one encoding the Cas9 endonuclease and the other a synthetic sgRNA (Jinek et al. 2012), with the latter usually under the regulatory control of a U3 or U6 small nuclear RNA promoter. The requirement for a PAM motif (NGG) adjacent to the target site sequence does constrain the number of potential CRISPR/Cas9 targets in a genome, although in most plant crop species analysed (i.e. rice, sorghum, soybean, tomato) >90 % of genes contain at least one potential CRISPR target site (Li et al. 2013; Xie and Yang 2013; Xie et al. 2014). A curious exception was maize where only 30 % of genes were reported to contain suitable CRISPR/Cas9 targets (Xie et al. 2014). Recently, orthologous type II CRISPR/Cas9 systems from other bacteria with different PAM sequence specificities have been shown to function with similar efficiency in Arabidopsis, which will further expand the number of potential CRISPR target sites available in plant genomes (Steinert et al. 2015).
Following cleavage by Cas9 of the 20 bp target sequence in the plant genome, the target site is generally repaired by nonhomologous end joining (NHEJ) which is an error-prone process that frequently results in small sequence insertions or deletions (indels) (Puchta 2005). The incorporation of indels within gene coding sequences can obviously result in codon insertions or deletions, or more commonly frame shift mutations that result in loss of gene function. CRISPR/Cas9 is, therefore, a remarkably targeted mutation tool when compared with previous random mutagenesis approaches. An interesting point, however, is that although the 20 bp target site is precisely defined with CRISPR/Cas9, the end product modification is still a random event defined by error-prone NHEJ.
Although generally specific CRISPR/Cas9 can occasionally result in off-target site cleavage of related, but non-identical, sequences with base differences tolerated generally only in the 5′ end of the sgRNA (Fu et al. 2013; Endo et al. 2015). Avoiding off-target effects can be achieved by selecting target sequences with minimal similarity to other sequences in the genome. In addition, modifications of the CRISPR/Cas9 system have been designed to reduce off-target site effects including using a mutated Cas9 nuclease that is capable of cleaving only a single DNA strand (Fauser et al. 2014; Schiml et al. 2014). Two sgRNAs with target sites in very close proximity and complementary to opposite strands are, therefore, required for cleavage. This effectively doubles the sequence specificity required for target site cleavage. Nonetheless, when compared with random mutagenesis the frequency of background mutations from CRISPR/Cas9 is trivial. Furthermore, given the relative ease with which many plant species can be backcrossed, the low frequency of potential off-target CRISPR/Cas9 sites is unlikely to be problematic in crop species.
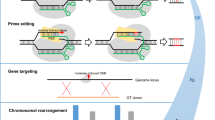
Application of the CRISPR/Cas9 system is not limited to targeted mutation mediated via NHEJ. An interesting feature of DNA sequence integration is that introduced sequences will preferentially insert into a pre-existing break within a genome (Puchta et al. 1996; Hannin et al. 2001; Tzfira et al. 2003; Puchta 2005; Wright et al. 2005; Tzfira et al. 2012). Site-specific CRISPR/Cas9-mediated cleavage can, therefore, facilitate targeted DNA insertion at the cleavage site by nonhomologous recombination. In addition, if the incoming DNA template is flanked by sequences that are homologous to those surrounding the cleavage site (i.e. a repair template) the frequency of homologous recombination mediated by homology-directed repair mechanisms (HDR) can be increased by several orders of magnitude (Puchta et al. 1996; Puchta 2005, Wright et al. 2005). CRISPR/Cas9 cleavage coupled with HDR has the potential to enable engineering of new alleles of endogenous genes or enable sequential insertion of transgenes at the same locus. This targeted cleavage can also be used to remove genes if sequences flanking the gene are simultaneously cleaved.
CRISPR/Cas9 technology has now been used in a variety of plant species (Shan et al. 2013; Li et al. 2013; Nekrasov et al. 2013) where its applications include targeted mutations, gene removal and site-specific transgene integration by HDR (Table 1). Examples of these approaches are discussed below.
Targeted mutations
Numerous genes have now been successfully targeted by CRISPR/Cas9 in different plant species giving rise to loss of function mutant phenotypes that are produced by error-prone NHEJ (Shan et al. 2013; Li et al. 2013; Nekrasov et al. 2013). In diploid transgenic plants containing CRISPR/Cas9 constructs, a mutation can occur in the target sequence of a single allele (monoallelic) or alternatively each allele can be independently mutated to produce a knockout phenotype (biallelic). Homozygous mutations can also occur (e.g. Shan et al. 2013; Zhang et al. 2014), presumably by a gene conversion process. Mutations can occur independently in plant somatic tissue resulting in chimerism with only those mutations present in germ line cells being transmitted to the following generation (Feng et al. 2014; Wang et al. 2015b; Mao et al. 2015; Xu et al. 2015). Progeny derived from CRISPR/Cas9 plants that show mutant phenotypes may, therefore, consist of either homozygous or biallelic mutant genotypes. In subsequent generations, plants that still contain a wild-type allele and CRISPR/Cas9 construct can generate new allelic variants of the target gene sequence (Feng et al. 2014; Xu et al. 2015).
The mutations that are produced by NHEJ repair of Cas9 cleaved sites are often small indels. In a study of over 800 CRISPR/Cas9-induced mutations at 12 target sites in seven genes in Arabidopsis T2 plants, Feng et al. (2014) demonstrated that 90 % of mutations were either small insertions or deletions at the target site sequence. Similarly, in rice small indels were most frequently observed with insertions and deletions occurring at similar frequencies (Zhang et al. 2014). However, Xu et al. (2015) report differences between different target loci in rice with some targets sites showing a higher frequency of larger deletions. They suggest that regions of micro-homology at these latter loci may promote deletions and that the sequence structure of the target locus may influence the final repair product (Xu et al. 2015). It is also of interest that a tenfold higher frequency of small insertions rather than deletions was observed in the Arabidopsis regulator of telomere length (RTEL1) gene (Fauser et al. 2014).
In some species, a high frequency of mutations in T0 plants can be obtained. In poplar, more than 90 % of mutations produced in a targeted phytoene desaturase gene were biallelic or homozygous in T0 regenerants resulting in albino phenotypes, with 50 % of all regenerated plants successfully mutated (Fan et al. 2015). In maize, high frequencies (77–100 %) of biallelic mutations were observed in T0 plants (Svitashev et al. 2015), while in soybean monoallelic and biallelic mutations were detected at a target site in 28 and 31 % of transgenic events, respectively (Li et al. 2015). In rice, biallelic mutations were detected in 31–41 % of T0 plants (Xu et al. 2015). However, this latter study also demonstrated that many of these mutations did not show expected inheritance patterns and in some instances were not inherited in T1 progeny, consistent with T0 plant chimerism. Similar chimerism in rice was reported by Zhang et al. (2014). The reported frequency of chimerism differs between studies but, nonetheless, it appears that at least one generation of inheritance is necessary to confirm the heritability of new CRISPR/Cas9-derived alleles.
In Arabidopsis, due to the floral dip in planta transformation system, chimerism occurs in T1 plants with somatic mutations often detected in the T1 generation (Feng et al. 2014; Jiang et al. 2014; Wang et al. 2015b and references therein). Chimerism was suggested to arise from poor expression of the Cas9 gene (usually 35S driven) in the egg cell. Using an egg cell-specific promoter, a higher frequency of bialleic or homozygous mutations was detected in the T1 (Wang et al. 2015b) which simplified mutant phenotype identification (Wang et al. 2015b). Germ line-specific promoters have also been used in Arabidopsis to improve the frequency of heritable, as opposed to somatic, mutations (Mao et al. 2015).
Once a CRISPR/Cas9 mutation has been transmitted through the germ line it remains stably transmitted thereafter, even in the presence of the original CRISPR/Cas9 transgenes. Presumably additional modifications of this allele are not possible as the original 20 bp target sequence has been altered beyond further CRISPR/Cas9 recognition. Inheritance of these mutant alleles has been shown in numerous plant species including Arabidopsis (Feng et al. 2014; Jiang et al. 2014; Fauser et al. 2014; Wang 2015a, b), rice (Zhou et al. 2014; Zhang et al. 2014; Xu et al. 2015), maize (Svitashev et al. 2015), lettuce (Woo et al. 2015) and potato (Butler et al. 2015). As expected, in some progeny these mutations have segregated away from CRISPR/Cas9 constructs making these modified plants transgene free, which becomes relevant when considering the transgenic status of these plants as discussed below.
Additional studies have demonstrated in plants, as shown previously in animal systems, that multiple genes can be simultaneously modified with CRISPR/Cas9. In maize, three loci, lig1, ms26 and ms45 were successfully targeted in the same cells with no apparent reduction in editing efficiency arising from multiplexing. Stable transgenics were successfully generated with mutations in all three genes (Svitashev et al. 2015). In rice, two genes were targeted with separate sgRNAs and the double mutation frequency in T0 plants (5–28 %) was approximately equal to the multiplication product of mutation frequencies at each target site (Zhang et al. 2014). This was demonstrated for four different gene pairs (Zhang et al. 2014). Also in rice, multiplex editing of a cyclin-dependent kinase gene family was achieved in three gene members in an approach which interestingly exploited the ability of CRISPR/Cas9 to tolerate target site mismatches (Endo et al. 2015). In Arabidopsis, three genes involved in trichome development were targeted by two sgRNAs and mutant phenotypes were observed in the T1 generation consistent with biallelic double and triple mutations that were inherited and confirmed in the T2 generation (Xing et al. 2014). Also in Arabidopsis, six genes were simultaneously targeted with six sgRNAs and plants produced that contained mutations at multiple loci with one plant containing modifications all six target loci (Zhang et al. 2015). Other examples of multiplex editing have been reported in maize (Xing et al. 2014), tobacco (Gao et al. 2015) and tomato (Brooks et al. 2014).
Polyploid plants represent an obviously increased challenge for CRISPR/Cas9 gene editing given the need to mutate all homoealleles to generate a mutant phenotype. In wheat, TALENS rather than CRISPR/Cas9 have been used to successfully simultaneously edit all three homoealleles of the MLO gene albeit with monoalleic modifications occurring at each locus (Wang et al. 2014). However, the MLO gene located on the wheat A genome was also successfully modified using CRISPR/Cas9 (Wang et al. 2014). In tetraploid potato, CRISPR/Cas9 modification was reported at each ALS1 locus and surprisingly involved the same 4 bp deletion suggesting a preference for this mutation type at this locus (Butler et al. 2015). While simultaneous modifications of homeoloci can occur in polyploids, the low likelihood of biallelic/homozygous mutations occurring at multiple loci suggests that progeny analysis or crossing will be required in many cases to effectively produce mutant phenotypes in polyploids using these technologies.
In some instances loss of function alleles produced by CRISPR/Cas9 can generate potentially useful agronomic traits. In one instance, targeted CRISPR/Cas9 modification of an endogenous plant gene produced virus-resistant plants. Plant viruses require host proteins to complete their lifecycle such as the eukaryotic translation initiation factor eIF4E and loss of this protein can provide virus resistance (Chandrasekaran et al. 2016). The cucumber elF4E gene was targeted by CRISPR/Cas9 and deletion alleles generated. T3 plants homozygous for deletion alleles and which no longer contained CRISPR/Cas9 transgenes were selected. These plants showed resistance to viruses from the Potyviridae family, i.e. cucumber vein yellowing virus, zucchini yellow mosaic virus and papaya ring spot mosaic virus-W (Chandrasekaran et al. 2016).
Potentially useful CRISPR/Cas9 mutations are not restricted to ORF modifications
This system can also be valuable in defining critical promoter regulatory elements in plants where the modified gene exists in its native genomic location and chromatin state. A conventional promoter deletion analysis was undertaken on the rice OsRAV2 gene promoter using a combination of stable rice transgenics and transient Agrobacterium assays in tobacco to identify a candidate GT-1 (GAAAAA) promoter element responsible for transcriptional induction upon salt stress (Duan et al. 2016). This sequence was subsequently edited in the endogenous rice gene using CRISPR/Cas9, whereupon its modification abolished salt inducibility of this endogenous gene (Duan et al. 2016). In a similar approach, also in rice, a region of the OsSWEET14 gene was identified that serves as a binding site for a secreted transcription factor produced by the bacterial rice pathogen Xanthomonas oryzae pv. oryzae. This pathogen transcription factor upregulates this host gene which promotes bacterial infection (Li et al. 2012). Abolition of this region of the OsSWEET14 promoter using TALENs prevented pathogen induction of this plant gene and increased rice resistance to this bacterial disease (Li et al. 2012). The OsSWEET14 gene promoter has also subsequently been modified using CRISPR/Cas9 (Jiang et al. 2013). Gene editing technologies, therefore, also offer possibilities for modifying endogenous gene expression by targeting critical regulatory elements.
Targeted deletions
Successful multiplex CRISPR/Cas9 editing in plant cells makes the possibility of producing targeted deletions an obvious proposition. Small deletions have been produced in several plant species. In tomato, heritable deletions of expected size were produced in T0 plants at the ARGONAUTE7 locus using two sgRNAS with target sites separated by 90 bp (Brooks et al. 2014). One T0 plant out of 29 contained a single homozygous deletion of expected size. Two additional plants were chimeric with an allele of the expected size and other alleles of smaller size. In maize, two sgRNAs were produced that targeted the ZmHKT gene with these sites separated by approximately 35 bp. Analysis of 20 transgenic T0 lines showed that 12 plants had a mutation efficiency of approximately 100 % at each locus and modified deletion alleles with the intervening sequence between each CRISPR/Cas9 site removed were present in two lines (Xing et al. 2014). Similar CRISPR/Cas9-induced deletions have been made in Arabidopsis (Li et al. 2013; Feng et al. 2014; Johnson et al. 2015) and tobacco (Gao et al. 2015).
Larger deletions have also been produced by CRISPR/Cas9. In rice, production of 170 kb and 245 kb deletions was achieved in T0 plants using multiple sgRNAs with target sites at either end of the deleted sequence (Zhou et al. 2014). Each intervening sequence encoded five and ten genes, respectively. In both examples, plants were monoallelic for these deletions but also had evidence of modification of each individual target site on the corresponding allele (Zhou et al. 2014). Inheritance of these deletions was not shown in rice; however, heritability experiments of large CRIPSR/Cas9-induced deletions would be best undertaken in a polyploid species such as wheat which can readily accommodate large deletions and chromosome loss. The ability to target and delete large segments of chromosomes has the potential to rapidly reduce the size of chromatin segments introgressed into plant genomes from wild relatives for crop trait improvement. These segments frequently also encode deleterious phenotypes linked to the trait of interest that can be difficult to remove by conventional breeding approaches. CRISPR/Cas9 may be a useful tool for the rapid reduction of these frequently nonrecombinogenic segments.
Homology-directed repair (HDR) to create new alleles
Site-specific cleavage coupled with the co-addition of a homologous repair sequence can create new alleles via homology-directed repair. This approach can be used to rapidly introduce new alleles without linkage drag or to introduce allelic variants that do not exist naturally. For example, in Nicotiana benthamiana protoplast assays, the phytoene desaturase (PDS) gene was successfully modified using CRISPR/Cas9 and a repair template (Li et al. 2013). A unique AvrII restriction site was encoded on a 600 bp PDS sequence and this sequence change was incorporated into the endogenous PDS sequence by gene replacement at 9 % efficiency. Similar experiments in Arabidopsis, however, were unsuccessful (Li et al. 2013).
In contrast, a GUS reporter gene with an internal duplication (GUUS) was transformed into Arabidopsis along with a sgRNA targeting this gene and Cas9 gene (Feng et al. 2014). 11 % of T1 plants showed chimeric GUS expression indicating that restoration of the GUS ORF had occurred in somatic tissue by homologous recombination. Analysis of T2 families from 16 T1 plants identified two families (13 %) in which a corrected GUS gene was stably inherited. Staining in GUS-positive T2 plants was uniform, consistent with germ line inheritance of the corrected reporter gene in contrast to the GUS chimerism observed in T1 plants (Feng et al. 2014). Molecular analysis also detected NHEJ events present in T2 progeny that were twice as frequent as homologous recombination events (Feng et al. 2014). A CRIPSR/Cas9 system derived from Staphylococcus aureus could also reconstitute GUS activity in Arabidopsis by homologous recombination with similar efficiencies to that observed using the Streptomyces pyogenes-based CRISPR/Cas9 system (Steinert et al. 2015).
In an experimentally similar study in Arabidopsis, it was demonstrated that a Cas9 nuclease modified to produce nickase activity (i.e. capable of cleaving only a single DNA strand at the target site) could also reconstitute GUS activity by HDR (Fauser et al. 2014). Two versions of the GUS gene were engineered that were each inactivated by a sequence encoding an I-SceI homing endonuclease site. Each reporter gene also contained repeated regions of the GUS gene that could serve as repair templates for homologous recombination. A direct comparison between Cas9, Cas9 nickase and I-SceI homing endonuclease cleavage in promoting homology-directed reactivation of these GUS reporter genes was undertaken. Interestingly, the single-strand nickase protein was as efficient, if not better, than the two nucleases that produced double-strand DNA breaks in reactivating GUS expression (Fauser et al. 2014).
In maize, a single specific amino acid substitution in the acetolactate synthase (ALS) gene provides resistance to sulfonylurea herbicides. Maize embryos were bombarded with a Cas9 gene and sgRNA gene targeting the ALS2 gene in close proximity to the critical amino acid. DNA repair templates as either 127-mer oligonucleotides or a 794 bp DNA fragment were also co-introduced, with each encoding an appropriate change at the critical amino acid in addition to several additional changes to confirm that herbicide-resistant plants were derived by template repair. During tissue culture callus was cultured on media containing the sulfonylurea chlorosulfuron and 2–4 repair events were detected per thousand embryos processed (Svitashev et al. 2015). T1 plants were confirmed as showing chlorosulfuron resistance. A very similar experiment was also undertaken in soybean using a 1 kb repair sequence of the ALS1 gene encoding chlorosulfuron resistance and again an error-free sequence exchange event was recovered after ALS1 was disrupted using Cas9 and a sgRNA (Li et al. 2015). HDR has also been achieved in the rice ALS gene (Sun et al. 2016). These experiments demonstrate how powerful this approach can be in those instances where a specific sequence change in a repair template is known to confer a beneficial phenotype.
HDR gene insertion
Targeted genome integration of plant transgenes potentially enables the sequential addition of transgenes at the same locus. This “cis gene stacking” would greatly simplify subsequent breeding efforts with all transgenes inherited as a single locus. Several studies have made advances in targeted gene integration by flanking transgenes of interest with additional sequences that are homologous to a desired insertion site. When coupled with CRISPR/Cas9 cleavage of the target site the transgene can be incorporated into this locus by homology-directed repair that is facilitated by flanking sequence homology.
HDR was used to insert an NPTII selectable marker gene into the alcohol dehydrogenase (ADH1) gene of Arabidopsis (Schiml et al. 2014). This selectable marker gene was flanked by approximately 670 bp of ADH1 sequence on either side (i.e. ADH5′/NPTII/ADH3′) and encoded on a T-DNA with Cas9 and a sgRNA gene that targets a site present in ADH1. The same sgRNA target site was placed on either side of the ADH5′/NPTII/ADH3′ sequence such that CRISPR/Cas9 cleavage would not only cleave ADH1 but also release ADH5′/NPTII/ADH3′ as a double-stranded DNA fragment. Stable Arabidopsis transgenics were produced and two plants out of 1400 T2 seedlings examined contained heritable gene targeting events whereby ADH5′/NPTII/ADH3′ had integrated into the ADH1 gene by HDR and this modified gene was transmitted to T3 progeny. However, further molecular analysis indicated that error-free HDR had occurred in only one of these lines (Schiml et al. 2014).
In maize, a DNA repair template encoding a phosphinothricin acetyltransferase selectable marker gene flanked by 1 kb of DNA sequence on either side with homology to the LIG1 gene was introduced into maize along with Cas9 and a sgRNA targeting the LIG1 sequence. Callus was screened by PCR for site-specific insertion and 2.5–4 % of calli showed evidence of correct target sequence integration when biolistic transformation was used (Svitashev et al. 2015). Interestingly, no case of targeted integration was observed when 192 embryos transformed by Agrobacterium transformation were screened. Seven independent biolistic regenerant plants were characterised by DNA blot hybridisation and two had restriction patterns consistent with homology-directed gene integration that was inherited in T1 plants. The remaining plants had extra, rearranged and randomly integrated copies of the selectable marker gene (Svitashev et al. 2015).
In a similar study in soybean, two target sites in close proximity on the distal end of chromosome 4 were targeted for HDR using a hygromycin phosphotransferase selectable marker gene flanked on each side by 1 kb of target site sequence homology (Li et al. 2015). Transgenic events were again produced by biolistic transformation and flanking PCR analysis detected potential HDR integration events in 4 % of T0 callus at each target site. Subsequent analysis, however, indicated that many HDR events were not present in regenerated T0 plants presumably due to callus chimerism. A number of plants regenerated also contained additional or imprecise insertions. However, an error-free HDR event with no additional transgene sequences was generated and homozygous T1 progeny produced.
An interesting modification of a gene targeting experiments described above was undertaken in tomato using CRISPR/Cas9 (or TALEN constructs) that cleaved a site in the 3′ end of the promoter of the ANT1 gene (Cermak et al. 2015). ANT1 encodes an MYB transcription factor which when overexpressed results in anthocyanin accumulation and intense purple-coloured tissue. Nuclease constructs were encoded on a modified bean yellow dwarf virus genome that was capable of undertaking replication within the plant cell. Also encoded on this replicon was a 35S promoter sequence and a selectable marker gene flanked on each side by approximately 900 bp of flanking sequence homology to the ANT1 cleavage target site. Exact HDR targeting of this sequence results in the ANT1 gene being under the regulatory control of the 35S promoter and an obvious visible accumulation of purple anthocyanin pigment. This replicon sequence was introduced into tomato cells on an Agrobacterium T-DNA where it was predicted to initiate rolling circle replication and generate hundreds to thousands of copies. Gene targeting frequencies of approximately 10 % were observed using this replicon which was an order of magnitude greater than that observed for a nonreplicating T-DNA vector sequence (Cermak et al. 2015). Subsequent molecular analyses indicated that more than two-thirds of the insertions were precise with no unanticipated sequence modifications and targeted modifications were transmitted to progeny. Interestingly, no evidence of off-target T-DNA or replicon sequence insertion was observed meaning that separation of nuclease sequences and the target sequence by segregation was not required (Cermak et al. 2015). This study, coupled with previous results from the same laboratory (Baltes et al. 2014) demonstrate that high repair sequence copy numbers coupled with site-specific DNA cleavage more efficiently promotes gene targeting via homology-directed repair processes.
Other CRISPR/Cas9 applications
Inactivation of the nuclease domains of Cas9 can generate an sgRNA/Cas9 complex that lacks catalytic activity but can bind to specific DNA target sites specified by sgRNA sequence complementarity. When targeted to appropriate genic regions this complex can inhibit transcription initiation and transcription elongation by RNA polymerase or binding of transcription factors. This has been demonstrated in E. coli and mammalian cells and co-expression of dual sgRNAs enabled simultaneous co-regulation of two genes in E. coli (Qi et al. 2013). Conversely, fusion of a transcription activation domain to a catalytic deficient Cas9 protein can enable it to behave as a transcriptional activator when targeted to an appropriate promoter site using sgRNA complementarity in human cells, although greater success was observed when multiple sites were targeted in the same promoter sequence (Perez-Pinera et al. 2013; Maeder et al. 2013). Transient expression assays have shown similar results in Nicotiana benthamiana where endogenous genes and transgenes could be activated or repressed by a catalytically inactive Cas9 protein fused with either transcriptional activation or repression domains. These modified Cas9 proteins were targeted to specific regions in plant promoters using sgRNA complementarity (Piatek et al. 2015). Other applications using the site-specific targeting abilities of the CRISPR/Cas9 complex are also being explored including epigenomic modifications (Puchta 2015).
An alternative CRISPR/Cas9 application in plants is to produce virus resistance in a process akin to the endogenous role of CRISPR/Cas systems in bacteria and archaea. Targeting the D/S DNA replicative form of gemini viruses using CRISPR/Cas9 was successful in generating indels in essential regions of viral genomes and generating virus-resistant transgenic plants. This was shown for beet severe curly top virus in Arabidopsis and Nicotiana benthamiana (Ji et al. 2015), bean yellow dwarf virus in Nicotiana benthamiana (Baltes et al. 2015) and tomato yellow leaf curl virus in Nicotiana benthamiana (Ali et al. 2015).
Are the final products considered genetically modified organisms?
An immediate question gene editing technologies like CRIPSR/Cas9 raise is whether the final products are considered as genetically modified organisms or not (Abbott 2015). In those examples where NHEJ results in indels at the target site and the original CRISPR/Cas9 transgenes are removed by segregation, the resultant plants are essentially indistinguishable from those that could have theoretically arisen by spontaneous mutation. Should these plants be considered as transgenic organisms given that they no longer contain any transgenes? The regulatory frameworks for genetically modified organisms that were developed several decades ago have arguably not evolved with current technological advances in gene modifications and will require re-evaluation to deal with these new products (Camacho et al. 2014). More vexing for regulators is the study of Woo et al. (2015) where pre-assembled Cas9/sgRNA protein/RNA complexes were introduced into protoplasts of Arabidopsis, rice, lettuce and tobacco and targeted mutagenesis frequencies of up to 45 % observed in regenerated plants. Mutations in these plants were generated by transient expression without a transgene, as such, ever being present. A similar experiment was also undertaken in petunia protoplasts (Subburaj et al. 2016). Given the success of CRISPR/Cas9 modifications in many eukaryotic organisms, these regulatory questions are not confined to plants and need to be addressed for numerous species including human cell therapies. Another important issue to be resolved is the legal ownership of CRISPR/Cas9 technology. Two competing patents have been lodged, one by Feng Zhang from the BROAD Institute and MIT and the second by Jennifer Doudna and Emmanuelle Charpentier from the University of California, Berkeley and Helmholtz Center for Infection Research, Germany, respectively (Rood 2015; Akst Akst 2016).
In summary, the CRISPR/Cas9 system is another technology that offers targeted gene mutation and opportunities for homologous recombination and targeted gene insertion. Arguably it is a simpler platform than other gene editing technologies such as TALENs and zinc finger nucleases which require the production of complex engineered proteins that contain multimeric domains and it is far more readily adaptable than meganucleases (Straub and LaHaye 2013; Belhaj et al. 2013). While targeted mutations using this system have been demonstrated in numerous plant species, HDR and targeted gene insertion are less routine. Nonetheless, CRISPR/Cas9 is likely to serve as a vital tool in progressing these latter approaches which are becoming more essential due to an expanding number of valuable transgenic traits.
Author contribution statement
ML, BG and MA wrote and approved this manuscript.
References
Abbott A (2015) Europe’s genetically edited plants stuck in legal limbo. Nature 528:319–320
Ahloowalia BS, Maluszynski M (2001) Induced mutations—a new paradigm in plant breeding. Euphytica 118:167–173
Akst (2016) Who owns CRISPR cont’d. Scientist, Article 45072
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, Voytas DF (2015) Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat Plant (Article 15145)
Belhaj K, Chaoparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39–49
Brooks C, Nekrasov V, Lippman ZB, van Eck J (2014) Efficient editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 system. Plant Physiol 166:1292–1297
Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 14:e0144591
Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W (2015) CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One 10(8):e0136064. doi:10.1371/journal.pone.013606
Camacho A, van Deynze A, Chi-Ham C, Bennett AB (2014) Genetically engineered crops that fly under the US regulatory radar. Nat Biotechnol 32:1087–1091
Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232–246
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. doi:10.1111/mpp.12375
Chylinski K, Makarova KS, Charpentier E, Koonin EV (2014) Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105
Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnology 271:90–97
Duan Y-B, Li J, Qin R-Y, Xu R-F, Li H, Yang Y-C, Ma H, Li L, Wei P-C, Yang J-B (2016) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol Biol 90:49–62
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56:41–47
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Nat Sci Rep 5 (Article 12217)
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Feng Z, Zhang B, Ding W, Liu X, Yang D-L, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu J-K (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Feng Z, Mao Y, Xu N, Zhang B, Wej P, Yang D-L, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu J-K (2014) Multigenerational analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111:4632–4637
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Keith Young J, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110
Hannin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J (2001) Gene targeting in Arabidopsis. Plant J 28:671–677
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170
Hyun Y, Kim J, Cho SW, Choi Y, Kim J-S, Coupland G (2015) Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241:271–284
ISAAA (2013) Brief 46. Executive summary. Global status of commercialized biotech/GM crops. http://www.isaaa.org/resources/publications/briefs/46/executivesummary/
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467:76–82
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16–26
Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1:15144
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jiang W, Yang B, Weeks DP (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One 9:e99225
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Johnson RA, Gurevich V, Filler S, Samach A, Levy AA (2015) Comparative assessments of CRISPR-Cas nuclease’ cleavge efficiency in planta. Plant Mol Biol 87:143–156
Koornneef M (2002) Classical mutagenesis in higher plants. In: Gilmartin PM, Bowler C (eds) Molecular plant biology. A practical approach, vol 1. Oxford University Press, Oxford, pp 1–11
Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA guided Cas9 nuclease. Genome Biol 16:258
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li Z, Liu Z-B, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169:960–970
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genom 41:63–68
Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung KJ (2013) CRISPR-RNA-guided activation of endogenous human genes. Nat Methods 10:977–979
Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9:467–477
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J-K (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Mao YY, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu J-K (2015) Development of germ-line-specific CRISPR–Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532
Mercx S, Tollet J, Magy B, Navarre C, Boutry M (2016) Gene inactivation by CRISPR–Cas9 in Nicotiana tabacum BY-2 suspension cells. Front Plant Sci. doi:10.3389/fpls.2016.00040
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu L-J (2013) Targeted mutagenesis in rice using CRISPR–Cas system. Cell Res 23:1233–1236
Michno J-M, Wang X, Liu J, Curtin SJ, Kono TJY, Stupar RM (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6:243–252
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plant Nicoatina benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Parry MAJ, Madgwick PJ, Bayon C, Tearall K, Hernandez-Lopez A, Baudo M, Rakszegi M, Hamada W, Al-Yassin A, Ouabbou H, Labhilili M, Phillips AL (2009) Mutation discovery for crop improvement. J Exp Bot 60:2817–2825
Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA (2013) RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat Biotech 10:973–976
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aoudia M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13:578–589
Puchta H (2005) The repair of double stranded DNA breaks in plants. J Exp Bot 56:1–14
Puchta H (2015) Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J. doi:10.1111/tpi.13100
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 93:5055–5060
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Que Q, Chilton M-DM, de Fontes CM, He C, Nuccio M, Zhu T, Wu Y, Chen JS, Chi L (2010) Trait stacking in transgenic crops—challenges and opportunities. GM Crops 1:220–229
Rood J (2015) Who owns CRISPR? Scientist (Article number 42595)
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as a paired nickase for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80:1139–1150
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, Gao C (2013) Targeted genome modification of crop plants using a CRISPR–Cas system. Nat Biotech 31:686–688
Shan Q, Yang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologous from Streptococcus thermophiles and Staphylococcus aureus. Plant J 84:1295–1305
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67
Straub A, LaHaye T (2013) Zinc fingers, TAL effectors or Cas9-based DNA binding proteins; whats best for targeting desired genome loci? Mol Plant 6:1384–1387
Subburaj S, Chung SJ, Lee C, Ryu SM, Kim DH, Kim JS, Bae S, Lee GJ (2016) Site-directed mutagenesis in Petunia × hybrid protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep (In press)
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y (2015) Targeted mutagenesis in soybean using the CRISPR–Cas9 system. Sci Rep 5:10342. doi:10.1038/srep10342
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L (2016) Engineering herbicide resistant rice plants through CRISPR/Cas9-mediated homologous recombination of the acetolactate synthase. Mol Plant. doi:10.1016/j.molp.2016.01.001
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945
Tang F, Yang S, Liu J, Zhu H (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-Like protein but not the one previously reported. Plant Phys 170:26–32
Tzfira T, Frankmen L, Vaidya M, Citovsky V (2003) Site-specific integration of Agrobacterium tumefacians T-DNA via double-stranded intermediates. Plant Physiol 133:1011–1023
Tzfira T, Weinthal D, Marton I, Zeevi V, Zuker A, Vainstein A (2012) Genome modification in plant cells by custom-made restriction enzymes. Plant Biotechnol J 10:373–389
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat G3(3):2233–2238
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–952
Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X (2015a) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep 34:1473–1476
Wang Z-P, Ling H-L, Dong L, Zhang H-Y, Han C-Y, Wang X-C, Chen Q-J (2015b) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target gene in Arabidopsis in a single generation. Genome Biol 16:144–156
Woo JE, Kim J, Kwon SI, Corvalan C, Choo SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR–Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Wright DA, Townsend JA, Winfrey RJ, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44:693–705
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6:1975–1983
Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol Plant 7:923–926
Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Ban C-Y, Liu B, Wang X-C, Chen Q-J (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J (2014) Gene targeting using the Agrobacterium-mediated CRISPR–Cas system in rice. Rice 7:5–8
Xu R-F, Li H, Qin R-Y, Li J, Qiu C-H, Yang Y-C, Ma H, Li L, Wei P-C, Yang J-B (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Nat Sci Rep 5:11491
Yin K, Han T, Liu G, Chen, Wang Y, Yunzi A, Yu L, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5 (Article 14926)
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu J-K (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu J-K (2015) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. doi:10.1007/s00299-015-1900-z
Zhang B, Yang X, Yang C, Li M, Guo Y (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep 6 (Article 20315)
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Zhou X, Jacobs TB, Xue L-J, Harding SA, Tsai C-J (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate: coA ligase specificity and redundancy. New Phytol 208:298–301
Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J (2016) Efficiency and inheritance of targeted mutagenesis in maize using CRISPR–Cas9. J Genet Genom 43:25–36
Acknowledgments
We wish to thank the Two Blades Foundation for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare they have no conflict of interest.
Additional information
Communicated by T. Cardi.
Rights and permissions
About this article
Cite this article
Luo, M., Gilbert, B. & Ayliffe, M. Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep 35, 1439–1450 (2016). https://doi.org/10.1007/s00299-016-1989-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-1989-8