Abstract
Key message
PeSNAC1 , a stress-related NAC1 from Phyllostachys edulis , was characterized. Ectopic expression in Arabidopsis indicated that PeSNAC1 together with ped -miR164b participated in the regulation of organ boundaries and stress tolerance.
Abstract
NAC (NAM, ATAF1/2 and CUC2) participates in many different processes regulating plant growth, development, and stress response. A total of 125 NAC genes have been predicted in moso bamboo (Phyllostachys edulis), but their roles are poorly understood. PeSNAC1 targeted by ped-miR164b was focused for further study. The cleavage of PeSNAC1 mRNA guided by ped-miR164b was validated using RLM-5′ RACE. Tissue-specific expression analysis demonstrated that ped-miR164b had a declining trend from root, sheath, leaf, to that of stem, which was opposite to that of PeSNAC1. Transgenic Arabidopsis plants overexpressing either PeSNAC1 (OX-PeSNAC1) or, ped-miR164b (OX-ped-miR164b) driven by the CaMV35S promoter were generated. OX-ped-miR164b plants showed similar phenotype of cuc2 mutants whose growth was seriously suppressed. Compared with Col-0, sense OX-PeSNAC1 plants grew rapidly and flowered earlier, whereas antisense plants grew slowly and exhibited delayed flowering. Sense OX-PeSNAC1 plants had the greatest number of lateral roots, while antisense OX-PeSNAC1 and OX-ped-miR164b plants had fewer lateral roots than Col-0. Under NaCl and PEG6000 stresses, survival rates were higher and F v/F m values declined more slowly in sense OX-PeSNAC1 plants than in Col-0, with lower survival rates and a more rapid decrease in F v/F m values conversely observed in antisense OX-PeSNAC1 and OX-ped-miR164b plants. These findings indicated that ped-miR164b-targeted PeSNAC1 may play key roles in plant development and tolerance to salinity and drought stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant biomass and agricultural productivity are seriously affected by abiotic stress due to the environmental deterioration globally (Ahuja et al. 2010). To ensure survival, plants have developed numerous effective molecular mechanisms over the long course of evolution to protect cellular activities and maintain plant integrity under adverse conditions (Pastori and Foyer 2002). Transcription factors (TFs) play important roles in plant growth and development by regulating the expression of stress-related genes to control plant biological processes (Meshi and Iwabuchi 1995). NAC (NAM, ATAF1/2, CUC2) TFs constitute a large plant-specific gene family involved in the regulation of plant developmental processes and response to environmental stresses (Zhu et al. 2015). The expression of some NAC TFs is negatively regulated by microRNAs (miRNAs) at the post-transcriptional level in a wide variety of eukaryotic organisms (Jeong et al. 2011). In Arabidopsis, miR164 targets five NAC domain-encoding mRNAs: NAC1, CUC1, CUC2, At5g07680, and At5g61430 mRNAs (Mallory et al. 2004). NAC1 plays an important role in the transmission of auxin signals that promote lateral root emergence. AtNAC1 has been characterized as an intermediary in the auxin-signaling pathway that activates genes encoding molecules involved in the specification of lateral root initiation (Xie et al. 2000). In addition, transgenic ZmNAC1-overexpressing Arabidopsis plants have an increased number of lateral roots compared with the wild type (Li et al. 2012), while TaNAC29-overexpressing plants have enhanced tolerance to high salinity and dehydration (Huang et al. 2015).
Bamboos, which are woody grasses with rhizomes and underground roots, are found exclusively in forest habitats. The most economically important bamboo, moso bamboo (Phyllostachys edulis), possesses desirable features such as a fast growth rate, extensive usability, high yield, and strong regeneration capacity. Much attention has been paid to the regulatory mechanism promoting rapid growth in bamboo. One such study involved histological investigations and protein expression analyses using bamboo culm materials from different developmental stages and internodes (Cui et al. 2012). The results of that study revealed that culm development is initially dominated by cell division followed by cell elongation in middle and late stages, with the expressed proteins involved in many physiological and metabolic processes constituting a complex network to regulate culm development. Following the publication of the draft genome of moso bamboo (Peng et al. 2013), a genome-wide analysis of TFs identified 1768 TFs classified into 54 families (http://planttfdb.cbi.pku.edu.cn/). Among them, 125 NAC TFs were clustered into 17 groups and their gene structures and conserved domains were subsequently predicted by Li et al. (2015). In another study, miRNA expression profiling of bamboo was performed using next-generation sequencing (He et al. 2013). In addition, 84 conserved miRNAs belonging to 17 families as well as 81 novel miRNAs have been identified in ma bamboo (Dendrocalamus latiflorus) (Zhao et al. 2013), while 75 miRNAs and 24 miRNA* variants of 22 families have been found in moso bamboo (Xu et al. 2014). Finally, comparative profiling of miRNAs between sympodial and monopodial bamboos has been conducted using two representatives: moso bamboo and ma bamboo, respectively (Zhao et al. 2014b). Despite all these data, little information is available regarding how miRNAs regulate TFs in bamboo.
In this study, we used a bioinformatics approach to identify the NAC genes targeted by miR164b and isolated sequences of the ped-miR164b precursor, the ped-miR164b promoter, and the target gene PeSNAC1 from moso bamboo. We then validated the ped-miR164b target site in PeSNAC1 using RNA ligase-mediated (RLM) rapid amplification of cDNA ends (RACE). To understand the function of ped-miR164b and PeSNAC1, we further analyzed the expressions of ped-miR164b and PeSNAC1 in moso bamboo and characterized the phenotypes of transgenic Arabidopsis plants expressing either ped-miR164b or PeSNAC1. We found that the expressions of PeSNAC1 and ped-miR164b in different tissues exhibited opposite trends. In addition, PeSNAC1-over-expressing plants grew faster and had more lateral roots than the Columbia-0 (Col-0) wild type, while antisense (PeSNAC1) plants grew more slowly and had fewer lateral roots, some lines in the latter group even had phenotypes similar to those of transgenic plants expressing the ped-miR164b precursor. Further analysis of plants under salinity and drought stresses revealed that sense PeSNAC1-overexpressing plants had higher survival rates and F v/F m values than those of Col-0, while those of antisense and ped-miR164b expressing plants were lower. These findings indicate that PeSNAC1 expression is negatively regulated by ped-miR164b, which in turn may influence tolerance to salinity and drought stresses through regulation of root development. Taken together, our results shed some light on the function of PeSNAC1 targeted by ped-miR164b and can serve as a basis for the study of miRNA target genes in bamboo.
Materials and methods
Plant materials and growth conditions
Seedlings of moso bamboo and wild-type Arabidopsis thaliana (ecotype Col-0) were planted in nutritional bowls (basal diameter/mouth diameter/height = 6.5/10/8 cm) containing vermiculite presoaked with plant nutrient solution (one-third strength B5 medium). The seedlings were grown at 22 ± 1 °C under a 16-h light/8-h dark photoperiod with a light intensity of 200 µmol m−2 s−1.
Sequence retrieval and analysis
Homologous sequences of NAC genes and miR164 in moso bamboo were retrieved from BambooGDB (http://www.bamboogdb.org) (Zhao et al. 2014a). Open reading frames (ORFs) of NAC genes were determined with DNASTAR, BLASTN and BLASTX, and a multiple alignment of NAC proteins was conducted using DNAMAN to find the conserved domains. The GSDS gene structure display server (http://gsds.cbi.pku.edu.cn) was used to uncover the number and the positions of introns (Hu et al. 2015). The secondary stem-loop structure of ped-miR164b was predicted using the RNAfold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (Gruber et al. 2008), cis-elements in the ped-miR164b promoter region were analyzed with PlantCARE (http://bioinformatics.psb.ugent.be/Webtools/plantcare/html/) (Lescot et al. 2002). The target site of ped-miR164b was predicted using psRNATarget (http://plantgrn.noble.org/psRNATarget/?function=3#) (Dai and Zhao 2011).
RNA/DNA extraction and gene isolation
Roots, stems, leaves and sheaths were collected from 1-year-old moso bamboo seedlings and quickly frozen in liquid nitrogen. Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacture’s protocol and treated with RNase-free DNase I (Tiandz, Beijing, China). First-strand cDNA was synthesized from 1 µg total RNA using a Reverse Transcription System (Promega, Beijing, China). cDNA was synthesized from miRNAs using miRNA-specific stem-loop primers according to Unver and Budak (2009). Genomic DNA was extracted from leaves using the CTAB method (Gao et al. 2006).
The primers listed in Table 1 were designed according to the results of the bioinformatics analyses. Leaf cDNA and genomic DNA were used as templates for PCR amplifications performed in 20 μL reaction volumes containing PrimeSTAR DNA polymerase (Takara, Dalian, China). Primer pairs used for the amplifications were as follows: F-SNAC and R-SNAC (for PeSNAC1), F-pre164b and R-pre164b (for the ped-miR164b precursor), and F-pro164b and R-pro164b (for the ped-miR164b promoter) (Table 1). The PCR amplification products were inserted into a pGEM-T easy vector (Promega) and subsequently confirmed by sequencing on an ABI 3730 DNA Analyzer (Applied Biosystems, USA).
Tissue-specific expression analysis
Quantitative real-time PCR (qRT-PCR) and stem-loop qRT-PCR were used in this study to, respectively, detect expression levels of PeSNAC1 and ped-miR164b in different tissues of moso bamboo. Primers specific to individual miRNAs were designed using a method described by Chen et al. (2005). U6 snRNA and PeNTB were, respectively, used as internal controls for miRNA (Ding et al. 2011) and PeSNAC1 (Fan et al. 2013) analyses. Detailed information on primers used in the quantitative analysis is listed in Table 1. qRT-PCR amplifications were carried out using a SYBR Green I Master kit (Roche, Germany) on a QTOWER 2.2 real-time PCR system (Analytick Jena). Each amplification was performed in a final reaction volume of 10 μL containing 5.0 μL 2× SYBR Premix Ex Taq, 0.2 μL of each primer (10 μM), 0.8 μL cDNA and 3.8 μL nuclease-free water. All reactions were repeated three times. Stem-loop qRT-PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, and 62 °C for 10 s. The procedure used for traditional qRT-PCR consisted of 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 10 s. The qRT-PCR experiments were performed as biological triplicates. Expression levels relative to internal reference genes were calculated according to the 2−ΔΔCt method (Livak and Schmittgen 2001).
Rlm-5′ race
Outer and inner primers (listed in Table 1) were designed for RLM-5′ RACE on the basis of miRNA target site prediction. Nested-RACE PCRs were conducted by RLM reverse transcription without CIP and TAP treatments in conjunction with a Full RACE Kit with TAP (Takara). The cDNA for RLM-RACE was synthesized according to the manufacturer’s instructions. Outer PCR was carried out in 20 μL volumes consisting of 2 µL of 10× GC Buffer II, 8 μL 1× cDNA Dilution Buffer II, 2 μL cDNA template, 1 μL miR164b outer primer (10 μM), 1 μL 5′RACE outer primer (10 μM), 0.25 μL LA Taq (5 U μL−1) and 5.75 μL double-distilled water (ddH2O). PCR cycling conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 20 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 10 min. Inner PCR was then performed in 20 μL volume consisting of 4 μL of 5× PrimeSTAR Buffer (Mg2+ plus), 1.6 μL dNTP mix (2.5 mM each), 1 μL outer PCR amplification product, 1 μL miR164b inner primer (10 μM), 1 μL 5′RACE inter primer (10 μM), 0.2 μL PrimeSTAR HS DNA polymerase (2.5 U μL−1) and 11.2 μL ddH2O. The inner PCR protocol involved an initial denaturation of 98 °C for 3 min, followed by 20 cycles of 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 10 min. The generated PCR fragments were cloned into pGEM-T easy vectors and sequenced.
Construction of plant expression vectors and transformation
The ORF sequence of PeSNAC1 was amplified with primers designed to introduce BamH I or Cla I sites. The primers used to generate the sense fragment were F-S and R-S, while those for the antisense fragment were F-A and R-A—all based on the nucleotide sequence of PeSNAC1 (Table 1). PeSNAC1 overexpression vectors were constructed by inserting the fragments into modified pCAMBIA1301 vectors driven by the CaMV35S promoter. The sense and antisense constructions were designated as pC1301-PeSNAC1-S and pC1301-PeSNAC1-A, respectively. The precursor of ped-miR164b was also incorporated into a modified pCAMBIA1301 vector under the control of the CaMV35S promoter by using the precursor fragment with BamH I and Xba I sites at 5′ and 3′ ends, respectively. The promoter fragment was cloned into the Sal I-Xba I site of the binary vector pBI101 to obtain a transcriptional fusion of the ped-miR164b promoter and the GUS coding sequence. The constructs were then separately introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. Transformations of Arabidopsis plants were conducted using the floral dip method (Clough and Bent 1998). Putative transgenic plants harboring PeSNAC1 and ped-miR164b were selected on medium supplemented with 50 mg L−1 hygromycin, while those containing the ped-miR164b promoter were selected using 50 mg L−1 kanamycin. Phenotypes of resistant plants were observed and photographed. Resistant seedlings showing no evidence of segregation were selected for further analysis.
Verification of transgenic plants
Transgenic plants of ped-miR164b were verified by qRT-PCR using F-miR/R-miR primer pair, with the actin gene used as an internal control (Song et al. 2013). In addition, expression analysis of endogenous genes such as CUC1, CUC2 and NAC1 was carried out using primer pairs designed according to their respective sequences (AB049069, AB002560, and NM_104479) (Table 1).
Transgenic plants of the ped-miR164b promoter and PeSNAC1 were validated by RT-PCR using F-GUS/R-GUS, F-S/R-S, and F-A/R-A primer pairs (Table 1). AtUbiquitin (NM180850) was used as an internal control under the same RT-PCR conditions with primers F-AtUbiquitin and R-AtUbiquitin (Yang et al. 2015). Vertical plate culture was used for root growth observations, and the number of lateral root was counted using 20 seedlings of each line. Student’s t test was used for the statistical analysis of the unit lateral root numbers.
Salinity and drought stress treatments
Seeds of transgenic Arabidopsis plants were sown on half-strength Murashige–Skoog (MS) solid medium containing hygromycin, while those of the wild type (Col-0) were cultivated on half-strength MS solid medium alone. Following incubation at 4 °C for 2 days, the seedlings were cultivated under normal conditions. One week after germination, both transgenic and Col-0 plants were transferred to half-strength MS solid medium containing NaCl (200 mM). At the same time, another set of transgenic and wild type plants were transferred to solid medium with incorporated PEG6000 (30 %) to simulate drought stress. Each treatment included 100 seedlings of each line. The survival rate of each NaCl-treated transgenic line was counted after 10 days, while that of each line treated with PEG6000 was counted after 20 days. During this period, chlorophyll fluorescence parameters were measured using an IMAGING-PAM fluorometer (Waltz, Germany) and used to calculate the maximum quantum yield of photosystem II, F v/F m, as (F m − F 0)/F m (Demmig-Adams and Adams 1996), where F 0 is the minimum fluorescence in the dark-adapted state and F m is the dark-adapted maximum fluorescence upon illumination with a pulse (0.6 s) of saturating light. F 0 and F m were determined after 20 min dark adaptation. Parameters were measured for 10 replicates per treatment per line. To determine significant differences (p < 0.05), all data were statistically analyzed by one-way analysis of variance in SPSS.
GUS activity assay
GUS activity was examined in T3 seedlings harboring the ped-miR164b promoter fused with the GUS reporter gene. The transgenic plants were stained in a 0.1 % X-Gluc solution according to the method of Jefferson (1987). After incubation at 37 °C for a period of 3 h to overnight, tissues were destained in 70 % ethanol until chlorophyll pigments were completely bleached. Finally, photos were taken using a camera (SONY DSC-HX50).
Results
Identification and isolation of PeSNAC1 and ped-miR164b
Six of the 125 NAC domain-encoding genes in the bamboo genome database (Zhao et al. 2014a) were identified as miR164b-targeted genes (S1): PH01001309G0120, PH01000110G0680, PH01000183G1320, PH01000093G0340, PH01000483G1000, and PH01000501G0450. These genes belonged to the stress-related NAC (SNAC) subgroup (Zhu et al. 2015) and were accordingly named PeSNAC1–6. PeSNAC1 was isolated from moso bamboo leaves using RT-PCR with primers N-F and N-R. The open reading frame (ORF) was 867 bp, and the genomic sequence corresponding to ORF was 3675 bp including three exons and two introns. A multiple alignment revealed that PeSNAC1 shared a highly conserved N terminal DNA binding domain (>94.52 % homology) and a highly variable C-terminal transcriptional regulatory domain compared with NACs of other plants such as Oryza sativa, Zea mays, Triticum aestivum, and Aarbidopsis thaliana (S2), this finding indicates that NACs have not been conserved during evolution. In addition, a putative miR164b-binding region was located downstream of the NAC domain in the PeSNAC1 coding region (626–646 bp).
Two candidates belonging to the miR164 family (ped-miR164a and ped-miR164b) were identified in the bamboo genome database. The mature sequence of ped-miR164b, which was 21-bp long (5′-UGGAGAAGCAGGGCACGUGCU-3′), had a different nucleotide at its 3′ end compared with the standard mature sequence of miR164 (5′-UGGAGAAGCAGGGCACGUGCA-3′). This result indicates that the miR164 family sequence is indeed highly conserved (Koyama et al. 2010). The precursor of ped-miR164b was isolated by PCR using specific primers (F-pre164b and R-pre164b) designed based on the PH01000543 sequence. The 82-bp precursor sequence could form a stable stem-loop structure with the generated mature sequence on the 5′ end of the arm, consistent with miR164a and miR164b in Arabidopsis (Mallory et al. 2004). The promoter region sequence upstream of the ped-miR164b precursor, obtained using primers F-pro164b and F-pro164b, comprised 1397 bp containing essential promoter elements, such as the TATA-box and the CAAT-box. Some putative cis-acting regulatory elements, such as ABRE, MBS, and ARE (S3), were also identified, thus indicating that ped-miR164b may be involved in abiotic stress regulation.
Identification of the ped-miR164b-mediated cleavage site
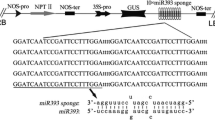
According to PsRNATarget prediction, a strongly matching target site for miR164b was identified in the PeSNAC1 ORF at 626–646 bp. To verify that PeSNAC1 is the direct target of ped-miR164b, RLM-5′RACE was used to confirm the ped-miR164b-mediated cleavage site. A cleavage product generated by the processing of the PeSNAC1 mRNA fragment by ped-miR164b was successfully detected (Fig. 1a); as confirmed by sequencing, this product was 119 bp long. Sequencing also revealed that 9 of 10 cDNA clones possessed this cleavage site, which was located in the middle of the ped-miR164b-PeSNAC1 base-pairing interaction region and corresponded to the 11th nucleotide position of the mature ped-miR164b sequence (Fig. 1b). This result confirmed that transcripts of PeSNAC1 are targeted for degradation by ped-miR164b at miR164-directed cleavage positions identical to those previously identified in the target mRNA in Arabidopsis (Guo et al. 2005) and Z. mays (Li et al. 2012).
Analysis of target site for ped-miR164b in PeSNAC1 transcript. a The resulting agarose gel showed the nested PCR product that we cloned and sequenced, and the product was expected to be approximately 120 bp. 1 DNA ladder; 2 PCR product. b The frequency of 5′ RACE clones corresponding to the cleavage site (indicated by arrow) was shown as a fraction, with the number of clones matching the target message in the denominator
Tissue-specific expression analysis of ped-miR164b and PeSNAC1
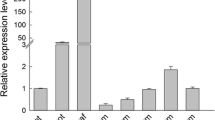
Because the expression patterns of miRNAs can provide clues to their functions (Yao et al. 2007), qRT-PCR was used to detect expression levels of ped-miR164b and PeSNAC1 in different tissues of moso bamboo. According to the qRT-PCR analysis, both PeSNAC1 and ped-miR164b were expressed in the examined tissues. The expression of ped-miR164b exhibited a declining trend from roots, sheaths, leaves, and stems, with the highest level in roots, the lowest in stems, and similar levels in leaves and sheaths (Fig. 2a). The expression of PeSNAC1 showed an opposite trend to that of ped-miR164b, with the lowest level in roots and the highest in stems (Fig. 2b). This result suggests that ped-miR164b has a regulatory effect on the instant amount mRNA of PeSNAC1.
Ectopic expression of ped-miR164b in Arabidopsis
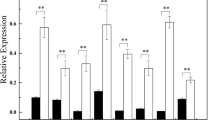
Six ped-miR164b overexpressing (OX-ped-miR164b) lines were obtained through resistance screening. These OX-ped-miR164b lines exhibited seriously suppressed growth and varying degrees of phenotypic changes. For instance, roots were shorter and lateral roots were fewer in the overexpressing lines than in Col-0 (S4), and some cotyledons and petioles were fused to different degrees, such as cup-shaped, small or dissymmetric leaves and fused petioles, resulting in failed seed production and even death (Fig. 3A). These phenotypes are similar to those of Arabidopsis cuc2 enhancer mutants (Hibara et al. 2006), whose CUC2 transcript is targeted for degradation by miR164 (Raman et al. 2008). The only two OX-ped-miR164b lines (miR-2 and miR-6) that were able to produce seeds without resistance segregation in the T3 generation were further selected for gene expression analysis. According to the qRT-PCR results, ped-miR164b was much more highly expressed in the two transgenic lines than in Col-0 (p < 0.01) (Fig. 3B). In addition, expression analysis of endogenous genes such as CUC1, CUC2, and NAC1 in miR-6 showed that they were all much more weakly expressed than in Col-0 (p < 0.01) (Fig. 3C). This result indicates that ped-miR164b can negatively regulate the expression of CUC1, CUC2, and NAC1.
Ectopic expression of ped-miR164b in A. thaliana. A Phenotype of ped-miR164b transgenic plants. a Wild type (Col-0); b–h ped-miR164b transgenic plants (b dissymmetric leaf; c, d fused petiole; e, f fused cotyledon; g, h cup shaped cotyledon). Bar 0.5 cm. B Expression analysis of miR164b in transgenic plants using qRT-PCR. C Expression analysis of CUC1, CUC2 and NAC1 in miR-6 and Col-0 using qRT-PCR. Vertical bars represent standard deviation of three independent biological replicates. Asterisks indicate the strongly significant difference between the transgenic lines and Col-0 (p < 0.01)
We also generated transgenic Arabidopsis plants harboring the ped-miR164b promoter fused with GUS. GUS activity was detected in young roots, leaves, hypocotyls, and the shoot apical meristem of these plants (Fig. 4). The higher promoter activity of the gene in roots might indicate that ped-miR164b participates in the regulation of root development.
Ectopic expression of PeSNAC1 in Arabidopsis
Nine sense PeSNAC1-overexpressing (OX-PeSNAC1) lines and nine antisense OX-PeSNAC1 lines were acquired by hygromycin selection. Morphological observation revealed that the seedlings of sense OX-PeSNAC1 lines grew more rapidly and flowered earlier than Col-0, with growth and flowering of antisense OX-PeSNAC1 lines showing the opposite trend compared with Col-0. Although a significant difference in aboveground phenotypes was not observed between sense OX-PeSNAC1 lines and Col-0, OX-ped-miR164b lines and antisense OX-PeSNAC1 line A-1 had similar phenotypes, including small, dissymmetric or absent leaves that may have attributed to their failure to produce seeds (S5). Four sense OX-PeSNAC1 T3 transgenic lines (S-1, S-6, S-8 and S-9) and four antisense OX-PeSNAC1 lines (A-1, A-2, A-8 and A-9) showing no evidence of segregation were the focus of further analysis. The results of semi-quantitative PCR analysis indicated that PeSNAC1 was transcribed in both sense (Fig. 5a) and antisense (Fig. 5b) OX-PeSNAC1 transgenic plants but not in Col-0.
Root morphology of transgenic plants
To investigate the effects of OX-PeSNAC1 and OX-ped-miR164b on Arabidopsis roots, seedlings of S-1, A-2 and miR-6 lines were randomly selected for observation and measurement of lateral roots on vertical plates. In 7-day-old seedlings, no significant difference was observed in taproot lengths of transgenics and Col-0. At this time, no lateral root initials were apparent in Col-0, A-2 or miR-6, whereas short lateral roots and two to four lateral root initials were found in some S-1 transgenic lines (data not shown).
At 14 days after germination, taproot lengths of S-1 and A-1 were, respectively, slightly longer or slightly shorter than those of Col-0; however, these differences were not significant. In contrast, miR-2 taproot lengths were significantly shorter than those of Col-0 (n = 20, p < 0.01). The average number of lateral roots per centimeter of taproots of S-1 (3.52 ± 0.37) was higher than that of Col-0 (2.3 ± 0.55) (p < 0.01); this observation is consistent with findings in ZmNAC1-overexpressing plants (Li et al. 2012). On the other hand, average numbers of lateral roots per centimeter in primary roots of A-2 and miR-6 were 0.98 ± 0.28 and 1.83 ± 0.47, respectively, significantly less (p < 0.01) than the number in Col-0 (Fig. 6). These results indicated that both ped-miR164b and PeSNAC1 are involved in root development, especially with respect to the number of lateral roots.
Effects of overexpression of PeSNAC1 and ped-miR164b on lateral root development. Col-0 and transgenic seedlings of S-1, A-2, and miR-6 were grown on MS medium with 2 % sucrose. Seedlings were photographed 14 days after germination. Lateral root numbers were counted with a binocular microscope directly. Numbers of lateral roots per centimeter of primary root of Col-0, S-1, A-2, and miR-6 were presented at bottom (n = 20). Bar 1 cm
Analysis of transgenic plant resistance to stresses
To examine whether the transgenic plants exhibited tolerance to salinity and drought stresses, a comparative analysis was conducted using transgenic lines S-1, A-2 and miR-6 treated with NaCl and PEG6000. After 7 days NaCl treatment, A-2 and miR-6 seedlings exhibited severely inhibited phenotypes compared with Col-0, with leaf rolling and even death observed. In contrast, S-1 seedlings showed more tolerance than those of Col-0 (S6). The measurement of chlorophyll fluorescence kinetics parameters can be used as a quick, nonintrusive probe in studies of plants under stress. For example, the maximal photochemical efficiency (F v/F m) is an index to estimate the degree of photoinhibition (Maxwell and Johnson 2000). F v/F m values of Col-0 and all the transgenic lines displayed a downward trend during NaCl treatment. The only obvious difference among lines was observed after 4 days NaCl treatment. Compared with Col-0, F v/F m values of A-2 and miR-6 decreased rapidly, and that of S-1 declined slowly (S7). After 7 days treatment, F v/F m was maintained at a relatively high level in S-1; in contrast, the F v/F m value of A-2 was similar to that of miR-6, which remained at a lower level than that of Col-0 (Fig. 7a). No significant difference in F v/F m was observed between transgenic plants and Col-0, but a significant difference (p < 0.05) was found between S-1 and miR-6. According to calculated statistics, the survival rate of S-1 (~90 %) was higher and those of miR-6 (~20 %) and A-2 (~30 %) were lower than that of Col-0 (~50 %) after 10 days NaCl treatment (Fig. 7b).
Response of Col-0 and transgenic plants to salinity stress. a Comparison analysis of F v/F m between transgenic plants and Col-0 after 7 days NaCl treatment. Vertical bars represent standard deviation of ten replicates. Asterisks indicated significant differences between S-1 plants and miR6 (p < 0.05). b Survival rate analysis of transgenic plants and Col-0 after 10 days NaCl treatment. Cotyledon greening were determined as survival. Vertical bars represent standard deviation of the mean (n = 3, treatments with 60 seedlings). Asterisks indicate the significant difference between the transgenic lines and Col-0 (p < 0.05)
During drought stress, A-2 and miR-6 seedlings showed similar responses—yellowing and even dying. Compared with Col-0 (S8), seedlings of S-1 were the last to turn into yellow. Similar to the trend observed under salinity stress, F v/F m values in Col-0 and transgenic lines declined over the course of the drought stress treatment. During the first 6 days of drought treatment, there was no obvious difference in F v/F m values between transgenic plants and Col-0. At 9 days of treatment, F v/F m was obviously decreased, especially in miR-6 and A-2, with S-1 (S9) exhibiting the smallest decline. After 14 days treatment, F v/F m of miR-6 was significantly lower and that of S-1 was significantly higher than that of Col-0 (p < 0.05). Although the value of A-2 was lower than that of Col-0, the difference was not significant (Fig. 8a). After 20 days treatment with PEG6000, the survival rate of S-1 (~85 %) was higher and survival rates of miR-6 (~5 %) and A-2 (~2 %) were lower than that of Col-0 (~50 %) (Fig. 8b). These results indicate that PeSNAC1 can confer abiotic stress tolerance on transgenic Arabidopsis.
Response of Col-0 and transgenic plants to drought stress. a Comparison analysis of F v/F m between transgenic plants and Col-0 after 14 days PEG6000 treatment. Vertical bars represent standard deviation of ten replicates. Asterisks indicated significant differences between transgenic plants and Col-0 (p < 0.05). b Survival rate analysis of transgenic plants and Col-0 after 20 days PEG6000 treatment. Cotyledon greening were determined as survival. Vertical bars represent standard deviation of the mean (n = 3, treatments with 60 seedlings). Asterisks indicate the significant difference between the transgenic lines and Col-0 (p < 0.05)
Discussion
Following the description of the first identified NAC gene, NAM of petunia (Souer et al. 1996), multiple studies have expanded our knowledge of NAC TFs. Many NACs have been identified, including 138 NACs in Arabidopsis (http://planttfdb.cbi.pku.edu.cn/), 151 in rice (O. sativa; Nuruzzaman et al. 2010), 152 in soybean (Glycine max; Le et al. 2011), 79 in grape (Vitis vinifera; Wang et al. 2013), and 118 in Brachypodium distachyon (Zhu et al. 2015). The NAC family, one of the largest TF families in plants, is divided into 14 subfamilies. Although the exact functions of CUC (development-related NACs), VND (secondary wall-synthesis NACs), TIP (membrane-associated NACs), TERN (tobacco elicitor-responsive NAC) and SNAC subfamilies are unknown, they may have similar regulatory roles (Shen et al. 2009; Zhu et al. 2015). Because NAC TFs have multiple functions related to the regulation of plant growth and development, much attention has focused on their functional identification. The availability of the draft genome of moso bamboo (Peng et al. 2013) and BambooGDB database (http://www.bamboogdb.org/) have facilitated further analysis of bamboo TFs and miRNAs. All 125 NAC TFs and 93 conserved miRNAs have been functionally annotated in BambooGDB. Bioinformatics analysis of NACs in moso bamboo allowed us to verify the function of the ped-miR164b target gene PeSNAC1 in this study.
miRNAs play vital regulatory roles in both animals and plants, mainly by promoting cleavage or translation inhibition of targeted mRNAs (Bartel 2004). The biological functions of miRNAs are intimately relevant to the functions of their target genes. Identification of potential target genes is therefore an effective and essential approach for in-depth investigation of complex miRNA-mediated regulatory mechanisms. In plants, miR164 was first predicted in Arabidopsis, with its mature sequence found to be fairly conserved (Rhoades et al. 2002); this finding has been confirmed by the identification of miR164 key recognition sites in OMTN genes of 158 rice varieties (Fang et al. 2014). Truncation of PeSNAC1 mRNA by ped-miR164b revealed that the cleavage site was between nucleotides 11 and 12 of the target site, consistent with findings in rice (Fang et al. 2014) and maize (Li et al. 2012). This result is consistent with the principle that miRNAs recognize their targets based on near-perfect sequence complementarity (Rhoades et al. 2002). The evolutionary conservation of miR164 suggests that the target sites may have important functions.
Different miR164 family members are present in plants, such as three members (ath-miR164a/b/c) in Arabidopsis (Moldovan et al. 2010), six (osa-miR164a/b/c/d/e/f) in rice (Sunkar et al. 2008), and two (ped-miR164a/b) in moso bamboo. Previous studies have demonstrated that miR164 can target five NAC domain-encoding mRNAs, namely, NAC1, CUC1, CUC2, At5g07680, and At5g61430 mRNAs. In Arabidopsis, boundary establishment and maintenance is controlled by three partially redundant genes, CUC1, CUC2, and CUC3 (Rhoades et al. 2002). miR164 is reported to target CUC1 and CUC2 mRNAs, but not that of CUC3 for degradation in plants (Laufs et al. 2004). In addition, a single mutation of either CUC gene has been found to have no major effect on boundary formation, whereas double mutants have fused cotyledons that reflect abnormal boundary specification during embryo development (Raman et al. 2008). In our study, the overexpression of ped-miR164b in Arabidopsis resulted in phenotypes similar to those of cuc2 mutants. We thus hypothesized that ped-miR164b constrains the boundary formation by randomly reducing the expression of some related targets including CUC1 and CUC2. This prediction was confirmed by the down-regulation of CUC1 and CUC2 in OX-ped-miR164b plants.
Proteins with similar domains may have identical or similar biological functions (Lin et al. 2007). miR164-tartgeted NAC genes have been reported to participate in the regulation of growth and development in Arabidopsis (Mallory et al. 2004; Guo et al. 2005; Li et al. 2012). Our alignment indicated that PeSNAC1 share a highly conserved domain with NACs in other plants, which implies that PeSNAC1 may also function in growth and developmental processes. Small-leaf, dissymmetric-leaf and afoliate phenotypes were observed in antisense OX-PeSNAC1 plants, very similar to OX-ped-miR164b plant phenotypes. Antisense OX-PeSNAC1 plants had fewer lateral roots than Col-0 plants, while sense OX-PeSNAC1 plants had more. On the basis of these observations, we speculate that PeSNAC1 together with ped-miR164b participates in the regulation of organ boundary maintenance and normal development in bamboo—an idea that requires further confirmation.
To fully understand whether ectopic expression of ped-miR164b or PeSNAC1 alters the seedling tolerance, we analyzed survival rates under salinity and drought stresses. Sense OX-PeSNAC1 plants exhibited a higher survival rate than Col-0 on NaCl and PEG6000 media, while rates of OX-ped-miR164b and antisense OX-PeSNAC1 plants were relatively lower. Chlorophyll fluorescence measurement is an effective internal probe for the study of plant photosynthesis under stress. Measurement of F v/F m under drought stress indicated that the intrinsic efficiency of photosystem II in OX-ped-miR164b and antisense OX-PeSNAC1 plants fell more rapidly, while that of sense OX-PeSNAC1 plants fell more slowly compared with Col-0, indicating that sense OX-PeSNAC1 plants are more stress tolerant than Col-0. However, further researches are required to identify the detailed function of TFs and miRNAs in bamboo.
Author contribution statement
Conceived and designed the experiments: ZMG. Performed the experiments: LLW, HSZ, DLC. Analyzed the data: HYS, LCL, LLW. Contributed reagents/materials/analysis tools: YFL, ZMG. Contributed to the writing of the manuscript: LLW, ZMG.
Abbreviations
- ATAF:
-
Arabidopsis transcription activation factor
- BLAST:
-
Basic local alignment search tool
- CaMV35S :
-
Cauliflower mosaic virus 35S
- cDNA:
-
Complementary DNA
- F v/F m :
-
Maximal quantum yield of PSII
- ORF:
-
Open reading frame
- CUC:
-
Cup-shaped cotyledon (CUC), a NAC protein
- miRNA:
-
MicroRNA, small regulatory RNAs
- NAC:
-
Abbreviation of NAM, ATAF1/2 and CUC2, a plant-specific TF family
- NAM:
-
No apical meristem (NAM), a NAC protein
- PCR:
-
Polymerase chain reaction
- qPCR:
-
Real time quantitative PCR
- RACE:
-
Rapid-amplification of cDNA ends
- RLM-5′ RACE:
-
RNA ligase-mediated 5′ RACE
- TF:
-
Transcription factor
References
Ahuja I, de Vos RC, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15(12):664–674
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler K (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Cui K, He CY, Zhang JG, Duan AG, Zeng YF (2012) Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J Proteome Res 11(4):2492–2507
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39(Web Server issue):W155–W159
Demmig-Adams B, Adams WWI (1996) Chlorophyll and carotenoid composition in leaves of Euonymus kiautschovicus acclimated to different degrees of light stress in the field. Aust J Plant Physiol 23(5):649–659
Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62(10):3563–3573
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8(2):e56573
Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65(8):2119–2135
Gao ZM, Fan SH, Gao J, Li XP, Cai CJ, Peng ZH (2006) Extract genomic DNA from Phyllostachys edulis by CTAB-based method. For Res 19(6):725–728 (in Chinese)
Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36(Web Server issue):W70–W74
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to down regulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
He CY, Cui K, Zhang JG, Duan AG, Zeng YF (2013) Next-generation sequencing-based mRNA and microRNA expression profiling analysis revealed pathways involved in the rapid growth of developing culms in Moso bamboo. BMC Plant Biol 13:119
Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18(11):2946–2957
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Huang Q, Wang Y, Li B, Chang J, Chen M, Li K, Yang G, He G (2015) TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol 15:268
Jefferson RA (1987) Assaying for chimeric genes in plants: the GUS fusion system. Plant Mol Biol Rep 5(4):387–405
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23(12):4185–4207
Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131(17):4311–4322
Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18(4):263–276
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li J, Guo G, Guo W, Guo G, Tong D, Ni Z, Sun Q, Yao Y (2012) MiRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol 12:220
Li B, Hu S, Cao Y, Xu G (2015) Bioinformatics analysis of NAC gene family in moso bamboo. Genomics Appl Biol 34(8):1769–1777 (in Chinese)
Lin R, Zhao W, Meng X, Wang M, Peng Y (2007) Rice gene OsNAC19 encodes anovel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci 172(1):120–130
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14(12):1035–1046
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence–a practical guide. J Exp Bot 51(345):659–668
Meshi T, Iwabuchi M (1995) Plant transcription factors. Plant Cell Physiol 36(8):1405–1420
Moldovan D, Spriggs A, Yang J, Pogson BJ, Dennis ES, Wilson IW (2010) Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J Exp Bot 61:165–177
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465(1–2):30–44
Pastori GM, Foyer CH (2002) Common components, networks, and pathways of cross-tolerance to stress. The central role of “Redox” and abscisic acid-mediated controls. Plant Physiol 129(2):460–468
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, Li Y, Fan D, Guo Y, Li W, Lu Y, Weng Q, Zhou C, Zhang L, Huang T, Zhao Y, Zhu C, Liu X, Yang X, Wang T, Miao K, Zhuang C, Cao X, Tang W, Liu G, Liu Y, Chen J, Liu Z, Yuan L, Liu Z, Huang X, Lu T, Fei B, Ning Z, Han B, Jiang Z (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45(4):456–461
Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55(1):65–76
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110(4):513–520
Shen H, Yin YB, Chen F, Xu Y, Dixon RA (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res 2:217–232
Song JB, Gao S, Sun D, Li H, Shu XX, Yang ZM (2013) miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol 13:210
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85(2):159–170
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25
Koyama T, Mistuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22(11):3574–3588
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta 230(4):659–669
Wang N, Zheng Y, Xin H, Fang L, Li S (2013) Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep 32(1):61–75
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14(23):024–3036
Xu P, Mohorianu I, Yang L, Zhao H, Gao Z, Dalmay T (2014) Small RNA profile in moso bamboo root and leaf obtained by high definition adapters. PLoS ONE 9(7):e103590
Yang L, Lou Y, Peng Z, Zhao H, Sun H, Gao Z (2015) Molecular characterization and primary functional analysis of PeMPEC, a magnesium-protoporphyrin IX monomethyl ester cyclase gene of bamboo (Phyllostachys edulis). Plant Cell Rep 34(11):2001–2011
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8(6):R96
Zhao H, Chen D, Peng Z, Wang L, Gao Z (2013) Identification and characterization of microRNAs in the leaf of ma bamboo (Dendrocalamus latiflorus) by deep sequencing. PLoS One 8(10):e78755
Zhao H, Peng Z, Fei B, Li L, Hu T, Gao Z, Jiang Z (2014a) BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database (Oxford) 2014:bau006
Zhao H, Wang L, Dong L, Sun H, Gao Z (2014b) Discovery and comparative profiling of microRNAs in representative monopodial bamboo (Phyllostachys edulis) and sympodial bamboo (Dendrocalamus latiflorus). PLoS One 9(7):e102375
Zhu G, Chen G, Zhu J, Zhu Y, Lu X, Li X, Hu Y, Yan Y (2015) Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS One 10(10):e0139794
Acknowledgments
This work received financial support from the Special Fund for Forest Scientific Research in the Public Welfare from State Forestry Administration of China (No. 201504106) and the Sub-Project of National Science and Technology Support Plan of the Twelfth Five-Year in China (No. 2015BAD04B01 and No. 2015BAD04B03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by B. Li.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Zhao, H., Chen, D. et al. Characterization and primary functional analysis of a bamboo NAC gene targeted by miR164b. Plant Cell Rep 35, 1371–1383 (2016). https://doi.org/10.1007/s00299-016-1970-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-1970-6