Abstract
Several studies have suggested the association between ADAM 12 polymorphisms and the risk of osteoarthritis (OA), but the results remained controversial. Therefore, we designed a meta-analysis to systematically evaluate the association on this issue. A literature search for eligible studies was conducted in PubMed, Web of Science and Google Scholar databases. The association between ADAM 12 polymorphisms and knee OA risk was calculated by odds ratios (ORs) and 95% confidence intervals (CIs). Study heterogeneity, sensitivity and publication bias analyses were also conducted. Ten articles covering 5048 cases and 6848 controls met our criteria for the final analysis. We found that the rs1871054 was significantly associated with the risk of knee OA (allele model OR 1.72, 95% CI 1.43–2.07, P < 0.001; additive model: OR 2.06, 95% CI 1.19–3.56, P = 0.010; dominant model: OR 2.45, 95% CI 1.85–3.25, P < 0.001; recessive model: OR 1.54, 95% CI 1.13–2.10, P = 0.007). rs1044122 was significantly associated with knee OA susceptibility in recessive model (OR 1.45, 95% CI 1.03–2.04, P = 0.031). For rs3740199 and rs1278279, no significant associations with knee OA were found. In the stratified analysis by gender, significant association was identified with the risk of knee OA for rs3740199 in men in allele model (OR 2.41, 95% CI 1.51–3.84, P < 0.001), dominant model (OR 2.68, 95% CI 1.17–6.14, P = 0.02) and recessive model (OR 3.51, 95% CI 1.68–7.36, P = 0.001), but not for additive model (OR 1.30, 95% CI 0.81–2.08, P = 0.28). This meta-analysis suggests that the ADAM 12 genetic polymorphisms rs1871054 and rs1044122 might be associated with risk of knee OA; rs3740199 might be associated with risk of knee OA in men. Further well-designed and large scale studies are warranted to validate these associations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is one of the most common rheumatic diseases and remains a leading cause of musculoskeletal disability [1]. In clinical practice, the most significant affected site of OA is in the knee [2]. Although some well-established facts, such as aging, obesity, female, gout, diabetes, some genetic profiles, and poor subchondral bone quality are associated with the increased risk of OA [3], the etiology underlying OA is not completely understood. Recently, several genome-wide linkage analyses and numerous studies have been performed to disclose potentially polymorphisms in various genes of OA pathogenesis [4, 5]. Of the identified polymorphisms, ADAM 12 (a disintegrin and metalloproteinase domain 12) polymorphisms seem to be promising but contradictory OA contributors [6].
ADAM 12 is an active proteinase, which belongs to the greater ADAM family of enzymes that regulate extracellular matrix turnover in cartilage [7]. Numerous researchers have suggested ADAM 12 is an important regulator in the pathogenesis of OA. Four of the important polymorphisms (rs3740199, rs1871054, rs1278279 and rs1044122) have been reported within the ADAM 12 gene and have been identified as contributors to the risk of OA [6]. However, the results remained controversial, and there is no systematic evidence on this issue. Therefore, we conducted a meta-analysis to systematically summarize the available evidence and evaluate the relationship between the ADAM 12 polymorphisms and knee OA risk.
Materials and methods
Search strategy
According to the observational studies in epidemiology guidelines [8], we performed a systematic search of studies in PubMed, Web of Science and Google Scholar, up to February 20, 2017. The following literature search terms were used: “ADAM12” OR “rs3740199” OR “rs1871054” OR “rs1278279” OR “rs1044122” AND “polymorphism” OR “mutation” OR “variant” OR “variation” OR “genotype” AND “osteoarthritis” OR “OA” OR “degenerative joint disease”. Additional relevant eligible studies were identified by a search of the references articles and review articles.
Inclusion and exclusion criteria
The inclusion criteria of the identified articles in our meta-analysis were as follows: (1) studies evaluating the association between the ADAM12 polymorphism and OA risk that were published; (2) a case–control or cohort studies addressing the numbers of affected and unaffected human control subjects or the total cohort sample size; and (3) studies with sufficient data in any of the four polymorphisms (rs3740199, rs1871054, rs1278279, rs1044122) to calculate the odds ratio (OR) with its 95% confidence interval (CI). The exclusion criteria were the studies published as abstract, editorial, summary, review, comment letter, or case report.
Data extraction
Two investigators independently extracted the data, and reached a consensus on all the studies. We extracted the first author’s name, year of publication, countries and ethnicities of participants, genotyping method, number of cases and controls, genotype or per-allele risk OR and 95% CI from each study. The data were extracted separately by disease or gender, if these were explicitly given. In addition, Hardy–Weinberg equilibrium (HWE) of controls was also collected.
Statistical analysis
The strength of association between the ADAM12 polymorphisms (rs3740199, rs1871054, rs1278279, rs1044122) and the OA risk was calculated by OR and 95% CI. The pooled OR was estimated under four genetic models: allele model (C vs. G or T vs. C or A vs. G or C vs. T), additive model (CC vs. GG or TT vs. CC or AA vs. GG or CC vs. TT), dominant model (CC + GC vs. GG or TT + CT vs. CC or AA + GA vs. GG or CC + TC vs. TT), and recessive model (CC vs. GC + GG or TT vs. CT + CC or AA vs. GA + GG or CC vs. TC + TT). The statistical significance of the combined OR was determined using the Z test. Stratified analyses were performed based on gender.
The heterogeneity among the included studies was calculated by Cochran’s Q test, in which P < 0.10 was considered significant [9]. If the heterogeneity was not significant, the fixed-effects model was applied [10], otherwise the random-effects model was adopted [11]. The I 2 statistic was also adopted to quantitatively measure the heterogeneity (I 2 > 50% indicated evidence of heterogeneity) [12]. The HWE among the control participants was assessed with a Chi square test.
Furthermore, in order to assess the stability of the results, sensitivity analysis was performed by sequentially omitting each included study. Potential publication bias was examined with Begg’s funnel plot and Egger’s regression test [13]. Statistical analyses were performed using STATA version 12.0 software (Stata Corporation, College Station, Texas). For all the analyses, P < 0.05 was considered to be significant and all the tests were two-sided, unless otherwise indicated.
Results
Characteristics of eligible studies
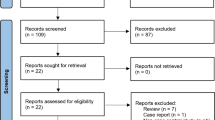
After a comprehensive systematic search, 82 relevant articles were retrieved. Screening of the abstracts excluded 66 articles. Following a full text review and detailed evaluations, 10 eligible articles covering 5048 cases and 6848 controls met our criteria for the final analysis (Fig. 1) [14,15,16,17,18,19,20,21,22,23]. The baseline characteristics of the eligible studies included in the meta-analysis are summarized in Table 1. The genotype and allele distributions or per-allele risk OR and 95% CI of the polymorphisms are shown in Supplementary Tables 1 and 2. All the distributions of genotype in controls were consistent with the HWE.
Quantitative synthesis
The meta-analysis results of association between the ADAM12 polymorphisms (rs3740199, rs1871054, rs1278279, rs1044122) and knee OA susceptibility are shown in Table 2. Subgroup analysis was conducted in six studies stratified analyses by gender, and the stratified analysis results of the association between rs3740199 polymorphism and knee OA risk are shown in Table 3.
Analysis of rs3740199 and OA susceptibility
Nine studies covering 4823 cases and 6635 controls assessed the potential influence of the rs3740199 polymorphism on knee OA susceptibility. Pooling data did not show any associations in all genetic models. However, in the stratified analysis by gender, significant associations were identified in men in allele model (OR 2.41, 95% CI 1.51–3.84, P < 0.001), dominant model (OR 2.68, 95% CI 1.17–6.14, P = 0.02) and recessive model (OR 3.51, 95% CI 1.68–7.36, P = 0.001), but not found an association in additive model (OR 1.30, 95% CI 0.81–2.08, P = 0.28). The results are shown in Tables 2 and 3 and Fig. 2a.
Analysis of rs1871054 and OA susceptibility
Five studies were included in the analysis, including 1036 knee OA patients and 1118 controls. We found that the rs1871054 was significantly associated with the risk of knee OA in all genetic models (allele model: OR 1.72, 95% CI 1.43–2.07, P < 0.001; additive model: OR 2.06, 95% CI 1.19–3.56, P = 0.010; dominant model: OR 2.45, 95% CI 1.85–3.25, P < 0.001; recessive model: OR 1.54, 95% CI 1.13–2.10, P = 0.007). The results are shown in Table 2 and Fig. 2b.
Analysis of rs1278279 and OA susceptibility
As for rs1278279, there were three studies involving 919 cases and 975 controls for data syntheses in the meta-analysis. The results suggested that rs1278279 polymorphism was not significantly associated with the risk of knee OA in all genetic models. The results are shown in Table 2 and Fig. 2c.
Analysis of rs1044122 and OA susceptibility
As for rs1044122, four studies consisted of 1104 cases and 1184 controls were included in the meta-analysis. Rs1044122 was found significantly associated with the risk of knee OA susceptibility in recessive model (OR 1.45, 95% CI 1.03–2.04, P = 0.031). No significant association was found in the other three models. The results are shown in Table 2 and Fig. 2d.
Heterogeneity analysis
By meta-regression analysis, significant heterogeneity was existed in the polymorphism rs3740199 under dominant model (P = 0.061), as well as for the polymorphism rs1871054 and rs1044122 under additive model (P = 0.001; P = 0.078; respectively). Moreover, the significant heterogeneity was also observed for the polymorphism rs3740199 in subgroup of gender under additive model (P = 0.004) of male and dominant model (P = 0.04) of female.
Sensitivity analysis and publication bias
Sensitivity analysis was performed to assess the influence of each study, by sequential excluding each eligible study. The results showed that no pooled ORs was substantially affected, indicating that the results were stability and liability. Publication bias of included studies was assessed with Begg’s funnel plots and Egger’s test. The shape of the funnel plots did not show any evidence of obvious asymmetry for all comparison models (Fig. 3), but the Egger’s test revealed the evidence for publication bias in rs1278279 (P = 0.025 for the additive model).
Discussion
The genetic background plays a crucial role in OA pathogenesis. Identification of polymorphism in susceptibility genes should, therefore, be considered as possibility to predict disease phenotypes as well to development prediction models for OA based on genotype information [24]. In recent years, several genome-wide association studies (GWAS) have already reported GDF5, BTNL2, SENP6, and FILIP1 were associated with the development of knee OA. However, the ADAM12 polymorphisms have not been confirmed in these GWAS analysis [25, 26]. The associations of the ADAM 12 polymorphisms with OA seem to be promising. Up to now, four of the important polymorphisms (rs3740199, rs1871054, rs1278279 and rs1044122) within the ADAM12 have been investigated in relation to OA [6].
The rs3740199 has been reported to be associated with the increased risk of the knee OA development in females [14]. However, another two studies found that the rs3740199 carried the risk for knee OA development only in men [16, 23]. In our study, which was based on a meta-analysis, we obtained the result that the polymorphism rs3740199 is only associated with increased risk of knee OA in men in allele model, dominant model and recessive model, but not for additive model. As for rs1871054, the impact of this polymorphism in knee OA pathogenesis has also remained controversial. Two studies found that rs1871054 has no significant association with knee OA [15, 16], but three studies found that rs1871054 has significant associated with knee OA [20,21,22]. In our meta-analysis, we found that the rs1871054 was significant associated with the risk of knee OA in all genetic models.
Moreover, the association of the polymorphisms rs1278279 and rs1044122 with knee OA risk has sparsely been investigated. We only found three studies for rs1278279 [15, 21, 22] and four studies for rs1044122 [15, 20,21,22] in our meta-analysis. The results indicated that rs1278279 polymorphism was not statistical significantly associated with the risk of knee OA in all genetic models, but rs1044122 was found significantly associated with knee OA susceptibility in recessive model. We recommend further researches to corroborate the relation of these two polymorphisms to knee OA risk.
In our meta-analysis, it should be pointed out that the heterogeneity between studies was also observed in several models, such as rs3740199 in dominant model, rs1871054 and rs1044122 in additive model. The common heterogeneity reasons of the study may be attributed to the number of studies, the diversity of ethnicity and genotyping quality [27]. Limited number of studies may be the main reason of the heterogeneity in our study, which restricted further exploration of the sources of heterogeneity. Moreover, the Egger’s test revealed the evidence for publication bias in rs1278279 in additive model. However, this bias has relatively small influence on the results of the present meta-analysis.
The current study also has some potential limitations that should be addressed. Firstly, not all of the included studies provided the exact genotype data. Therefore, in order to exactly evaluate the association between the genetic polymorphism and knee OA risk, we added the per-allele risk OR and 95% CI in several models, yet the analysis of the other models may lack these data in the analysis. Secondly, gene–gene and gene–environment potential interactions may have influenced our results, as genetic and environmental factors play a crucial role in OA pathogenesis. However, no information was available to test this. Thirdly, as many other factors such as age and body mass index may participate in the development of OA, we did not perform the stratified analysis based on these factors due to the limited data.
In conclusion, the results of this meta-analysis suggested that the ADAM 12 genetic polymorphisms rs1871054 and rs1044122 might be associated with risk of knee OA. For rs3740199 and rs1278279, no significant associations with knee OA were found. In the stratified analysis by gender, rs3740199 might be associated with the risk of knee OA in men. To the best of our knowledge, this is the first meta-analysis to assess the associations between ADAM12 polymorphisms and the risk of knee OA. As for the disadvantages in this meta-analysis, further well-designed and large-scale studies are warranted to validate these associations.
References
Tonge DP, Pearson MJ, Jones SW (2014) The hallmarks of osteoarthritis and the potential to develop personalised disease-modifying pharmacological therapeutics. Osteoarthr Cartil 22(5):609–621
Peat G, McCarney R, Croft P (2001) Knee pain and osteoarthritis in older adults: a review of the community burden and current use of primary health care. Ann Rheum Dis 60(2):91–97
Brandt KD, Dieppe P, Radin E (2009) Etiopathogenesis of osteoarthritis. Med Clin North Am 93(1):1–24
Zintzaras E, Kitsios GD, Ziogas DC, Rodopoulou P, Karachalios T (2010) Field synopsis and synthesis of genetic association studies in osteoarthritis: the CUMAGAS-OSTEO information system. Am J Epidemiol 171(8):851–858
Malemud C, Muoh O, Askari A (2014) Clinical significance and implications of genetic and genomic studies in patients with osteoarthritis. Adv Genom Genet 4:193–206
Yang CY, Chanalaris A, Troeberg L (2017) ADAMTS and ADAM metalloproteinases in osteoarthritis-looking beyond the ‘usual suspects’. Osteoarthr Cartil 25(7):1000–1009
Aigner T, Soeder S, Haag J (2006) IL-1beta and BMPs-interactive players of cartilage matrix degradation and regeneration. Eur Cell Mater 12:49–56 (discussion 56)
Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB (2000) Meta-analysis of observational studies in epidemiology. J Am Med Assoc 283(15):2008–2012
Cao JL, Yuan P, Abuduwufuer A, Lv W, Yang YH, Hu J (2015) Association between the TERT genetic polymorphism rs2853676 and cancer risk: meta-analysis of 76,108 cases and 134,215 controls. PLoS One 10(6):e0128829
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414):557–560
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Valdes AM, Hart DJ, Jones KA, Surdulescu G, Swarbrick P, Doyle DV, Schafer AJ, Spector TD (2004) Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum 50(8):2497–2507
Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, Loughlin J, Doherty M, Spector TD (2006) Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum 54(2):533–539
Kerna I, Kisand K, Tamm AE, Lintrop M, Veske K, Tamm AO (2009) Missense single nucleotide polymorphism of the ADAM12 gene is associated with radiographic knee osteoarthritis in middle-aged Estonian cohort. Osteoarthr Cartil 17(8):1093–1098
Limer KL, Tosh K, Bujac SR, McConnell R, Doherty S, Nyberg F, Zhang W, Doherty M, Muir KR, Maciewicz RA (2009) Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case–control population (the GOAL study). Osteoarthr Cartil 17(6):782–789
Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, Tsezou A, Blanco FJ, Meulenbelt I, Slagboom PE, Valdes AM, Spector TD, Gomez-Reino JJ, Gonzalez A (2009) Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthr Cartil 17(3):321–327
Shin MH, Lee SJ, Kee SJ, Song SK, Kweon SS, Park DJ, Park YW, Lee SS, Kim TJ (2012) Genetic association analysis of GDF5 and ADAM12 for knee osteoarthritis. Jt Bone Spine 79(5):488–491
Kerna I, Kisand K, Tamm AE, Kumm J, Tamm AO (2013) Two single-nucleotide polymorphisms in ADAM12 gene are associated with early and late radiographic knee osteoarthritis in Estonian population. Arthritis 2013:878126
Lou S, Zhao Z, Qian J, Zhao K, Wang R (2014) Association of single nucleotide polymorphisms in ADAM12 gene with susceptibility to knee osteoarthritis: a case-control study in a Chinese Han population. Int J Clin Exp Pathol 7(8):5154–5159
Wang L, Guo L, Tian F, Hao R, Yang T (2015) Analysis of single nucleotide polymorphisms within ADAM12 and risk of knee osteoarthritis in a Chinese Han population. Biomed Res Int 2015:518643
Poonpet T, Tammachote R, Tammachote N, Kanitnate S, Honsawek S (2016) Association between ADAM12 polymorphism and knee osteoarthritis in Thai population. Knee 23(3):357–361
Takahashi H, Nakajima M, Ozaki K, Tanaka T, Kamatani N, Ikegawa S (2010) Prediction model for knee osteoarthritis based on genetic and clinical information 2010. Arthritis Res Ther 12(5):R187
Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC et al (2012) Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380(9844):815–823
Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, Meulenbelt I et al (2011) Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis 70(2):349–355
Mao X, Ke Z, Liu S, Tang B, Wang J, Huang H, Chen S (2015) IL-1β +3953C/T, −511T/C and IL-6 −174C/G polymorphisms in association with tuberculosis susceptibility: a meta-analysis. Gene 573(1):75–83
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by the Natural Science Foundation of Zhejiang Province, China (Grant number LY13H270007).
Conflict of interest
Xuerong Hu, Guoli Sun and Weidong Wang declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, X., Sun, G. & Wang, W. Association of ADAM 12 polymorphisms with the risk of knee osteoarthritis: meta-analysis of 5048 cases and 6848 controls. Rheumatol Int 37, 1659–1666 (2017). https://doi.org/10.1007/s00296-017-3778-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-017-3778-2