Abstract
Environmental stress influences the cellular physiology in multiple ways. Transcription by all the three RNA polymerases (Pols I, II, or III) in eukaryotes is a highly regulated process. With latest advances in technology, which have made many extensive genome-wide studies possible, it is increasingly recognized that all the cellular processes may be interconnected. A comprehensive view of the current research observations brings forward an interesting possibility that Pol II-associated factors may be directly involved in the regulation of expression from the Pol III-transcribed genes and vice versa, thus enabling a cross-talk between the two polymerases. An equally important cross-talk between the Pol I and Pol II/III has also been documented. Collectively, these observations lead to a change in the current perception that looks at the transcription of a set of genes transcribed by the three Pols in isolation. Emergence of an inclusive perspective underscores that all stress signals may converge on common mechanisms of transcription regulation, requiring an extensive cross-talk between the regulatory partners. Of the three RNA polymerases, Pol III turns out as the hub of these cross-talks, an essential component of the cellular stress-response under which the majority of the cellular transcriptional activity is shut down or re-aligned.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eukaryotic gene expression is a highly complicated but precisely regulated process. Transcription, the first requirement for the gene expression, is carried out by the three RNA polymerases (Pols), each dedicated to a specific set of genes and assisted by its own set of transcription factors. These polymerase-specific factors are required for the basal transcription of all the target genes, whereas regulatory factors work in a gene and context-specific manner. With the recent advances in the field of genome-wide experimentations, it has become evident that RNA Pols work in harmony with one another, cross-talking with each other to co-ordinate the whole-genome transcription. This article presents a comprehensive view of mostly the recent literature with this perspective, focusing particularly on the Pol II and Pol III connectivities, which have seen a recent surge of global studies demonstrating the same.
Of the three RNA polymerases (Pols), Pol I transcribes the rDNA and synthesizes the ribosomal RNAs (25S, 18S, and 5.8S rRNAs in the budding yeast). Pol II transcribes the protein-coding genes to synthesize the mRNAs and many non-coding RNAs with regulatory functions. Pol III synthesizes the small/stable RNA species from a set of house-keeping genes coding for the 5S rRNA, all tRNAs, U6 snRNA, and several other types of small RNA species in different organisms (Dieci et al. 2007). Short Pol III transcripts are generally non-coding and devoid of polyA tails. Their genes characteristically consist of intra-genic promoter elements and generally no upstream, gene-specific regulatory sequences. With recent research, there has been accumulation of observations supporting a cross-talk including the genetic and physical interactions between not only the three polymerases but also the components of the three transcription machineries.
Common control on levels of functionally complete Pol complexes
All protein-coding genes are transcribed by the Pol II, yet the genes coding for the individual subunits of the RNA polymerases may have their own regulatory context. The three RNA polymerases of eukaryotes are multi-subunit complexes with at least five subunits common between them. It is possible that a common regulator of the levels of any one of these five subunits effectively controls the functional levels of the three enzymes in a coordinated manner. An mRNA-binding protein in the budding yeast, Rbs1, is shown to interact with several Pol III subunits and affect the levels of Rpb10, a small, shared subunit of the three Pols (Ciesla et al. 2020). The enzyme assembly, which is assisted by several factors, transporters, and chaperones, is proposed to proceed with multiple steps involving several discrete sub-complexes of distinct subunits (Wild and Cramer 2012). It is possible that the intermediate assembly sub-complexes compete with each other for the shared common subunits, in return controlling and coordinating the regulations of levels of a functionally active enzyme complex for all the three polymerases.
Bud27, an unconventional prefoldin, required for the bud site selection in yeast cells, was found necessary for the assembly of the three Pols and shows physical interaction with Rpb5, one of the five common subunits of the three Pols (Mirón-García et al. 2013). Bud27 is also required for Pol III biogenesis and activity. It modulates the relative level and nuclear localization of the two largest core subunits (Rpc160 and Rpc128) of Pol III and the cytoplasmic assembly of the other Pol III-specific subunits with the core subunits (Vernekar and Bhargava 2015). The Rpc128 subunit of the Pol III is at the core of the correct assembly of the 17-subunit Pol III enzyme (Ciesla et al. 2015). A reduced assembly of the functional enzyme complex eventually leads to reduced transcription activity. The transcriptional level control of the Rpc128 protein levels allows Pol II to in effect, control the Pol III assembly (Vernekar and Bhargava 2015).
Cross-transcription of genes by different RNA polymerases
The idea that the RNA polymerases may engage in a cross-transcription activity of different genes has been around for some time, as demonstrated by some earlier studies. One of the earliest examples could be the transcription of exogenously added protein-coding genes by Pol I in Trypanosoma. The 5′ cap feature of a pre-mRNA was found added to the nascent transcripts by trans-splicing a short (39 nucleotides), capped exon to the 5′ end (Lee and Ploeg 1997). The observation appeared to be an exception, but more examples of different polymerases cross-transcribing genes under different conditions and in different species followed. Just as all RNA polymerases show non-specific, promoter-independent transcription activity in vitro, transcription specificity under some conditions does not appear to be stringent in vivo. Comparatively recent studies have shown that RNA Pol III can initiate transcription from RNA Pol II core promoters under specific conditions in vitro, when assayed with crude human nuclear extracts (Duttke 2014). The selenocysteine tRNA (tRNA-Sec) gene (not found in yeast) is transcribed by Pol III in human cells but transcribed by Pol II in Trypanosoma brucei (Aeby et al. 2010). In Leishmania major, unlike other tRNAs, it is encoded by a single gene located in a Pol II polycistronic unit. Nuclear run-on assays with RNA pol inhibitors and with the cells that were previously exposed to UV showed that the tRNA-Sec gene is transcribed by both Pol III and Pol II (Padilla-Mejía et al. 2015), showing that RNA polymerase specificity is not absolute in vivo in Leishmania. However, a very recent study found that transcription of the same gene by Pol II interferes with its transcription by Pol III in the human cell lines (Gerber et al. 2020). Apart from these RNAs, the U3 snoRNA and the telomerase RNA genes have also been found transcribed by different RNA polymerases in different organisms. Most of the snoRNA genes are generally transcribed by Pol II in most of the organisms but not in plants, where many dicistronic tRNA–snoRNA units are transcribed by Pol III and the transcript is cleaved by a specific RNase to separate the two RNA types (Kruszka et al. 2003). The telomerase RNA gene is transcribed by Pol II in yeast and vertebrates (Chapon et al. 1997; Hinkley et al. 1998) but Pol III in the ciliates (Aigner et al. 2003). Similarly, the U3 snRNA gene is transcribed generally by Pol II in most of the organisms. Its known transcription by Pol III in plants suggests a co-evolution of the Pol II and Pol III transcription systems (Kiss et al. 1991).
Co-regulation of transcription by different RNA polymerases
Recent research shows a prevalence of the co-regulation of specific transcription of genes transcribed by different RNA polymerases. A tight coupling of the rRNA and tRNA production in cells has been known for many years now. Mutation in the largest subunit of Pol III or Pol I resulted in concerted alterations of rRNA and tRNA synthesis (Briand et al. 2001). In this context, ribosomal proteins were shown to promote the transcription of tRNA genes by Pol III in vitro (Dieci et al. 2009). A recent paper has found involvement of transcription activity of all the three RNA polymerases in the process of ribogenesis (Martínez-Fernández et al. 2020). A close link was found in the processing of pre-tRNA and pre-mRNAs, as well (Paushkin et al. 2004). Pol II and some of its transcription factors (c-myc, c-jun, and c-Fos) were found to bind near Type II and Type III Pol III-transcribed genes in human cell lines. Inhibition of Pol II activity by α-amanitin resulted in reduced expression of a number of Pol III genes as well (Raha et al. 2010). In another study, loss of Pol II found 300 bp upstream of Pol III-transcribed U6 snRNA gene and simultaneous inhibition of the U6 transcription with α-amanitin treatment underscore the requirement of Pol II activity for Pol III transcription (Listerman et al. 2007). Nevertheless, despite earlier demonstrations that the Pol specificity for target genes may not be absolute in vivo, mechanisms selecting Pol II/Pol III recruitment to a gene are more specific (Faresse et al. 2012; Dergai et al. 2018). TFIIS, an elongation factor of Pol II, was unexpectedly found on almost all Pol III-transcribed genes (Ghavi-Helm et al. 2008). While the study provided in vitro evidence that TFIIS may affect yeast Pol III start site selection, TFIIS has been proposed to act as a Pol III transcription factor in even mammalian cells (Carriere et al. 2012). A large number of new, potentially regulatory protein factors of Pol II-transcribed genes have been found near the Pol III-transcribed genes (Venters et al. 2011) in an earlier genome-wide study. A detailed compilation of the proteins found on/near the Pol III-transcribed genes, which could be the potential regulators of the yeast Pol III transcription, may be found in earlier reports (Acker et al. 2013; Nguyen et al. 2014).
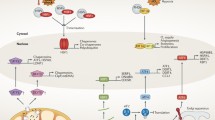
The superimposition of different RNA polymerase activities observed in the above-mentioned findings indicates that Pol II and Pol I play an important role in regulating Pol III transcription. Such a co-ordination of transcriptional output may not be possible without an extensive interaction network of the protein factors associated with each of the polymerases. The transcription complex (TC) of Pol III is constituted by the Pol III and its basal factors TFIIIC and TFIIIB. A recently published interactome of the budding yeast pol III TC, captured using proteomic approaches, consists of Pol I/Pol II subunits and their transcription factors, several chromatin modifier complexes, as well as many other regulatory complexes associated with components of the Pol III TC (Bhalla et al. 2019a). The Pol III TC was found to interact with a large number of proteins involved in various cellular functions, suggesting that they may be involved in regulating transcription by Pol III (Fig. 1). Genetic and even physical interactions of Pol III machinery with many of them have been documented by now. These interactions enabling the cross-talk between the partners may be manifested in several ways in a cell. In this context, the recent study with mammalian cell lines (Gerber et al. 2020) has shown that Pol II can regulate the transcription factor MYC-dependent tDNA transcription indirectly by controlling the expression of the MYC gene at the transcription level.
STRING network analysis of the Pol III TC interactome reveals a network of the several macromolecular complexes with specialized functions. Pol III TC interactome data (Bhalla et al. 2019a) were used to find the functional protein association networks using the STRING v. 10.0 database (http://version10.string-db.org). Network retrieval was set to display not more than 20 interactions with the highest confidence score (0.90). The parameters selected as pieces of evidence are shown at the bottom right corner of the network. All disconnected nodes are removed for the sake of clarity. Several nodes representing a basic cellular function are seen interconnected with each other
Co-localization of Pol II and Pol III on mutual targets
Just as many of the regulatory factors (Pol II-transcribed gene products) regulate transcription by Pol III, there is a possibility of regulation of Pol II-transcribed genes by Pol III and its transcription. The presence of active Pol III transcription complexes near Pol II promoters in the budding yeast inhibits pol II transcription, a well-documented phenomenon known as tRNA gene-mediated (tgm) silencing (Hull et al. 1994). Consistent with this, depletion of TFIIIC (an initiation factor of Pol III) resulted in deprepression of intergenic regions and the Pol II genes near the TFIIIC binding sites (Wang et al. 2014). On the other hand, TFIIIC auto-regulates the transcription of TFC6 gene (coding for a subunit of TFIIIC) by RNA Pol II in the budding yeast (Kleinschmidt et al. 2011).
Several genome-wide studies have found human Pol III presence near the active Pol II gene promoters while Pol II was also found near the majority of Pol III-occupied loci (Oler et al. 2010; Barski et al. 2010; Moqtaderi et al. 2010; Canella et al. 2012). Association of the two Pols with common loci housing the histone modifications which mark transcriptionally active chromatin suggests that by being together at a locus with open, active chromatin, both Pols may engage in a cross-talk. A recent study exploring the relevance of this Pol II presence near highly transcribed Pol III genes found that depletion of the RPB1 (largest) subunit of Pol II leads to increase in Pol III occupancy at the tRNA genes, repressed under normal conditions. A transcriptional interference by Pol II could be confirmed as direct cause of the Pol III repression on these loci (Gerber et al. 2020).
In comparison to human cells, although the Pol II-transcribed genes are found near many Pol III-transcribed genes, the Pol III occupancy and activity of genes do not show any dependence on the neighboring genes in yeast (Bhalla et al. 2019b; Shukla et al. 2021). A re-look at the earlier genome-wide ChIP-seq data on yeast Pol III occupancy (Kumar and Bhargava 2013) found low levels of yeast Pol III not associated with TFIIIB or TFIIIC close to + 150 bp position of several Pol II-transcribed genes (unpublished). With validations by the ChIP-Real Time PCR, the associations may not be non-specific, though they were several-fold lower than the Pol III levels at the canonical target genes. No other Pol III peaks were found within 1 kb of the occupied gene ends or on other than these 92 Pol II-transcribed genes. While no special category could be assigned to group all these genes together, many of them are related to stress-management or required for normal transcription by Pol II. Under nutrient starvation, Pol III peaks could be found on different genes. These observations raised the possibility of Pol III involvement in the regulation of Pol II transcription under certain conditions.
Control of Pol III transcript levels by the RNA surveillance machinery
Primary transcript from tDNA undergoes several defined steps of processing before mature tRNA is produced. Primary processing of Pol II transcripts is known to be co-transcriptional. One of the subunits of RNaseP, the protein complex responsible for 5′ end processing of tRNA in yeast, has been shown to interact with a subunit of TFIIIB, the transcription initiation factor for Pol III (Ishiguro et al. 2002). The interactions of various components of Pol III transcription machinery and the tRNA processing complex with each other during the transcription of Pol III genes suggest that the Pol III transcript processing may be co-transcriptional. Thus, a co-ordination at the transcription level may have a direct impact on the production of the mature RNA transcript levels. In addition to its role in pre-tRNA processing, RNase P was found to enable nuclear unstable RNA turnover probably due to its property of somewhat non-specific cleavage of RNA (Marvin et al. 2011). A defective RNaseP led to the accumulation of even unspliced mRNAs. This suggests a 5′-end processing defect of pre-tRNA and mRNA turnover under a stress condition may be linked via RNaseP, which may be seen in the cellular transcriptome alterations under that condition. This may be an added level of cross-talk between the two transcription machineries.
The tRNAs are known to be some of the most stable RNA species in the cell. Recent research has found the involvement of RNA surveillance machinery in controlling mature tRNA levels in cells (Wlotzka et al. 2011). Nab2, a poly(A)-RNA-binding protein, has additional roles in mRNA transcription, tRNA metabolism, and ribosomal subunit export as it is associated with the entire open-reading frame of actively transcribed RNA polymerase II and III genes (González-Aguilera et al. 2011). Many of the RNA surveillance machinery components and RNA processing complex subunits known for targeting the Pol II transcripts have been found to interact with the Pol III TC (Bhalla et al. 2019a). Nab2 has also been shown to influence the Pol III transcription by stabilizing the TFIIIB and Pol III binding at the gene promoters (Reuter et al. 2015). The presence of these RNA metabolism proteins known for targeting the Pol II transcripts (like Nrd1-Nab3-Sen1 complex, Nab2, exosome, and TRAMP complex components) in the list of Pol III interactors suggests a common control mechanism of maintaining the transcript levels via cross-talk between Pol II and Pol III and establishing a direct relation between the transcription status of the two RNA polymerases.
Extensive communication between Pol II and Pol III transcription complexes
In human embryonic stem cells, a peak of H3K4me3 (epigenetic mark for active state) and/or RNA Pol II were found between the H3K27me3 (epigenetic mark for repressed state) and Pol III-binding peaks, indicating that H3K4me3 and Pol II activity may insulate Pol III targets from neighboring repressive effects of H3K27me3 on the chromatin (Alla and Cairns 2014). In comparison, most of the histone modifications are not found near yeast tRNA genes. Some of the multi-subunit complexes are known to have both global and local chromatin-modifying activities. One of such complexes in yeast, the abundant chromatin remodeler, RSC is known to associate with many Pol III-transcribed genes (Ng et al. 2002). RSC maintains the nucleosome density at pol III- and nucleosome positioning at pol II-transcribed genes (Parnell et al. 2008). The Rsc4 subunit makes a physical contact with the common subunit of three Pols, Rpb5; a mutation in Rsc4 (rsc4-∆4) was found to compromise transcription by Pol II and Pol III (Soutourina et al. 2006). RSC was found to have gene-specific effects at Pol III-transcribed loci (Kumar and Bhargava 2013), distinguishable from its global influence in the gene-flanking regions (Fig. 2). In an average plot of nucleosome occupancies of the 120 genes showing nucleosome dynamics in the gene region, a clear shift of the downstream (DS) nucleosome toward the gene body and NFR (nucleosome-free region) shrink without affecting the upstream (US) nucleosome position or gene-flanking nucleosomal arrays was seen (Kumar and Bhargava 2013). As opposed to this, both the profiles in Fig. 2 show lower average occupancy on both sides of the NFR due to movement of flanking nucleosomes toward the gene, but without any significant shrinkage of NFR or loss of nucleosomal periodicity. This suggests that the loss of RSC-Pol III interaction in rsc4-∆4 affects the nucleosome occupancy on the genes in two ways. Shrinkage of NFR may be the result of its gene-targeted action and lower nucleosome occupancy in the flanking regions may be part of the global effect of its activity. Pol III and RSC are not lost from the genes in this mutant, indicating that the dynamics of the DS nucleosome seen in this mutant is guided by a cross-talk between RSC and Pol III while bound to these genes (Arimbasseri and Bhargava 2008; Mahapatra et al. 2011; Kumar and Bhargava 2013).
Global activity of RSC influences nucleosome density in the flanks of Pol III-transcribed genes. Normalized log2 (nucleosomal DNA/genomic DNA) intensity ratios for nucleosomes were binned around TSS and average nucleosome occupancy on the tRNA genes in the wild type and rsc4-∆4 mutant are compared as reported earlier (Kumar and Bhargava 2013). The gene body (blue box), upstream (US), and downstream (DS) nucleosomes are marked. a Average occupancy for all the 273 genes in the wild type and rsc4-∆4 mutant strain. b Average nucleosome occupancy profile on the 153 genes not showing a change within 100 bp on their both sides in the mutant. The highly similar profiles in both the panels show that the nucleosomal arrays in both gene-flanks have moved toward NFR in the rsc4-∆4 cells, which is reflected in loss of intensity but increase in nucleosome density near the genes. In the NFR housing the gene body, an increase of nucleosome occupancy is seen in the mutant. The denser packing does not disturb the US nucleosome position and nucleosomal periodicity in the flanking regions is maintained
Such activities of a chromatin modifier may have far-reaching effects on chromatin structure influencing both Pol II and Pol III-transcribed loci simultaneously. Recruitment of the Pol III transcription factor TFIIIC under serum starvation on a subset of the Alu elements (repetitive DNA) located near the cell cycle genes leads to hyperacetylation of H3K18 in nucleosomes on these elements in mammalian cell lines (Ferrari et al. 2020). With this acetylation, accessibility of the chromatin increases, enabling a TFIIIC-CTCF contact and looping-out of the acetylated chromatin region. This, in turn, ensures a basal transcription level of the nearby cell cycle genes, critical for their re-activation upon serum re-exposure (Ferrari et al. 2020). However, using Hi-C analyses of 3D chromatin organization, depletion of the Pols I/III or even Pol II had only small scale, local alterations in chromatin loop interactions of the mouse embryonic stem (mES) cells. Transcription inhibition due to degradation of the polymerases did not influence the large-scale 3D organization of chromatin (Jiang et al. 2020). This could be due to the known gene-specific effects of several chromatin modifiers on at least Pol III transcription (Shukla and Bhargava, 2018), as observed in case of RSC activity on Pol III-transcribed loci (Fig. 2). However, it is clear that gene-specific effects dominate over the global effects of chromatin modifiers.
The TATA-binding protein TBP is known to serve as a basal initiation factor for all the three RNA Pols in most of the species. Association of several Pol II transcription factors like TFIID subunits (Taf5/10/12/14), TFIIS and other elongation complexes like FACT, Paf1, etc. with Pol III TC (Bhalla et al. 2019a), suggest a signaling between the transcription complexes of two Pols under different physiological conditions. In this context, an earlier study probing the Pol II transcriptome in four mutants of Pol III TC components made two interesting observations. (i) The affected genes were not found near the Pol III-transcribed genes and (ii) the mutations resulted in up-regulation of a specific set of genes known to be under the regulation of a transcriptional activator of the amino-acid biosynthesis pathway, Gcn4. This generated a Gcn4-dependent reprogramming of the genome expression (Conesa et al. 2005). A simple interpretation of the results could be that the Pol III TC defects hamper the tRNA production causing a shortfall in the amino-acid carriers to ribosomes for use in general mRNA translation. Influencing the amino-acid biosynthesis in a gene-specific manner rather than a transcription of the whole-genome compensates the shortfall. The results reveal a close link between the gene-specific effects of Pol III activity on the Pol II transcription. Conversely, transcriptional interference by mammalian Pol II in serum-rich condition was found necessary for Maf1-dependent repression of the Pol III-transcribed genes (though a small number) under serum starvation (Gerber et al. 2020). In agreement with the observation that Pol II may be a gene-specific regulator of tDNA transcription by Pol III (Gerber et al. 2020), at least two chromatin-associated complexes Paf1 and FACT required for Pol II transcription in a gene-specific manner (Penheiter et al. 2005; Jimeno-González et al. 2006) have recently been reported to show the little or only gene-specific effect on Pol III transcription (Bhalla et al. 2019b; Shukla et al. 2021), but alter the local chromatin architecture in a subtle manner, which may have regulatory influences on other chromatin-affected non-transcriptional processes, if not transcription. One of the tested tRNA genes tT(CGU)K of yeast, which shows high transcription in the FACT mutant (Shukla et al. 2021), is also one of the genes which show increased transcription upon Pol II depletion in mammalian cells (Gerber et al. 2020). This may not be a co-incidence; rather it demonstrates that Pol II influences the Pol III transcription via a subtle mechanism across the species.
PAF1 complex occupies the tRNA genes in gene-specific manner (Bhalla et al. 2019b). Though it associates with the Pol III TC in both active and repressed states, most of its known roles on Pol II-transcribed genes are not found on the Pol III-transcribed genes. Instead, it helps resolve the steric conflict of replication and transcription machineries on these short genes (Bhalla et al. 2019b). This may be one of the novel ways of maintaining genomic integrity under replication stress using the complex, which facilitates transcription by Pol II, for countering replication stress on the Pol III-transcribed genes. On the other hand, the FACT complex, a factor known to facilitate chromatin transcription by Pol II, has also been found to affect Pol III transcription in only gene-specific manner. It was found to participate in a novel, global stress-sensor mechanism which links transcriptional regulation and nucleosome dynamics to environmental stress conditions (Shukla et al. 2021). Binding of both, the PAF1 or FACT complexes to the acidic patch contributed by the histone H2A-H2B dimer on the nucleosome surface (Cucinotta et al. 2019), gives them a handle to alter the local nucleosome profile of the chromatin regions.
Perspective
The above perceptions support the idea that perturbations under a physiological state could include an involvement of the transcription activity of all the three Pols. Apart from the basal transcription factors and DNA-binding classical activators/repressors of transcription; the chromatin-associated transcription regulators, which generally work via protein–protein interactions, are classified as co-regulators. A recent review of the studies on transcription regulation by steroid hormone receptors presents the evidences that gene-specific actions of the co-regulators create a ‘physiological coregulator code’ for every pathway, enabling regulation of a specific set of genes under a condition (Stallcup and Poulard 2020). Such a strategy of regulation in higher species offers a pool of therapeutic targets for future research. Stress can modulate the transcription through its effect on transcription machinery as well as chromatin structure. The earlier data on the Pol III TC interactome changes under a stress condition like nutrient starvation (Bhalla et al. 2019a) show the possibility of a larger network of the cross-talk between the three polymerases, which may undergo pervasive but very subtle changes under a variety of challenges. The current literature also gives compelling pieces of evidence to conclude that Pol III TC acts as a hub for a highly extensive network of protein–protein interactions (see Fig. 1) underlying different physiological activities, along with mutually responsive transcription by the three Pols to facilitate a cross-talk between the three transcriptional machineries of a eukaryotic cell. More studies validating the physical and genetic interactions between the components of a variety of stress modules would be required to decipher their connections with transcription regulation under a given condition. With emergence of more evidences in future, this perspective with potential for a change in paradigm is likely to become stronger. The viewpoint will also help to explain why and how one stress signal often leads to multiple physiological abnormalities.
References
Acker J, Conesa C, Lefebvre O (2013) Yeast RNA polymerase III transcription factors and effectors. Biochim Biophys Acta 1829:283–295
Aeby E, Ullu E, Yepiskoposyan H, Schimanski B, Roditi I, Muhlemann O, Schneider A (2010) tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res 38:5833–5843
Aigner S, Postberg J, Lipps HJ, Cech TR (2003) The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42:5736–5747
Alla RK, Cairns BR (2014) RNA Polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS ONE 9:e85648
Arimbasseri AG, Bhargava P (2008) Chromatin structure and expression of a gene transcribed by RNA polymerase III are independent of H2A.Z deposition. Mol Cell Biol 28:2598–2607
Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K (2010) Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 17:629–634
Bhalla P, Vernekar DV, Gilquin B, Couté Y, Bhargava P (2019a) Interactome of the yeast RNA polymerase III transcription machinery constitutes several chromatin modifiers and regulators of the genes transcribed by RNA polymerase II. Gene 702:205–214
Bhalla P, Shukla A, Vernekar DV, Arimbasseri AG, Sandhu KS, Bhargava P (2019b) Yeast PAF1 complex counters the pol III accumulation and replication stress on the tRNA genes. Sci Rep 9:12892
Briand JO, Navarro F, Gadal O, Thuriaux P (2001) Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 21:189–195
Canella D, Bernasconi D, Gilardi F, LeMartelot G, Migliavaca E, Praz V, Cousin P, Delorenzi M, Hernandez N (2012) The CyclixX Consortium, a multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res 22:666–680
Carriere L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, Jounier S, Aude J-C, Keime C, Murvai J, Foglio M, Gut M, Gut I, Lathrop M, Soutourina J, Ge´rard M, Werner M, (2012) Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res 40:270–283
Chapon C, Cech TR, Zaug AJ (1997) Polyadenylation of telomerase tRNA in budding yeast. RNA 3:1337–1351
Ciesla M, Turowski M, Nowotny M, Tollervey D, Boguta M (2020) The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase. Nucleic Acids Res 48:12252–12268
Ciesla M, Makała E, Płonka M, Bazan R, Gewartowski K, Dziembowski A, Boguta M (2015) Rbs1, a new protein implicated in RNA polymerase III biogenesis in yeast Saccharomyces cerevisiae. Mol Cell Biol 35:1169–1181
Conesa C, Ruotolo R, Soularue P, Simms TA, Donze D, Sentenac A, Dieci G (2005) Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription. Mol Cell Biol 25:8631–8642
Cucinotta CE, Hildreth AE, McShane BM, Shirra MK, Arndt KM (2019) The nucleosome acidic patch directly interacts with subunits of the Paf1 and FACT complexes and controls chromatin architecture in vivo. Nucleic Acids Res 47:8410–8423
Dergai O, Cousin P, Gouge J, Satia K, Praz V, Kuhlman V, Lhôte P, Vannini A, Hernandez N (2018) Mechanism of selective recruitment of RNA polymerases II and III to snRNA gene promoters. Genes Dev 32:711–722
Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23:614–622
Dieci G, Ruotolo R, Braglia P, Carles C, Carpentieri A, Amoresano A, Ottonello S (2009) Positive modulation of RNA polymerase III transcription by ribosomal proteins. Biochem Biophys Res Comm 379:489–493
Duttke SHC (2014) RNA Polymerase III accurately initiates transcription from RNA Polymerase II promoters in vitro. J Biol Chem 289:20396–20404
Faresse NJ, Canella D, Praz V, Michaud J, Romascano D, Hernandez N (2012) Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet 8:e1003028
Ferrari R, de Cucalon LIL, Di Vona C, Le Dilly F, Vidal E, Lioutas A, Oliete JQ, Jochem L, Cutts E, Dieci G, Vannini A, Teichmann M, de la Luna S, Beato M (2020) TFIIIC binding to Alu elements controls gene expression via chromatin looping and histone acetylation. Mol Cell 77:475–487
Gerber A, Ito K, Chu C-S, Roeder RG (2020) Gene-specific control of tRNA expression by RNA polymerase III. Mol Cell 78:765–778
Ghavi-Helm Y, Michaut M, Acker J, Aude J-A, Thuriaux P, Werner M, Soutourina J (2008) Genome wide location analysis reveals a role of TFIIS in RNA Polymerase III transcription. Genes Dev 22:1934–1947
González-Aguilera C, Tous C, Babiano R, de la Cruz J, Luna R, Aguilera A (2011) Nab2 functions in the metabolism of RNA driven by polymerases II and III. Mol Biol Cell 22:2729–2740
Hinkley S, Blasco MA, Funk WD, Feng J, Villeponteau B, Greider CW, Herr W (1998) The mouse telomerase RNA 5’-end lies just upstream of the telomerase template sequence. Nucl Acids Res 26:532–536
Hull MW, Erickson J, Johnston M, Engelke DR (1994) tRNA genes as transcriptional repressor elements. Mol Cell Biol 14:1266–1277
Ishiguro A, Kassavetis GA, Geiduschek EP (2002) Essential roles of Bdp1, a subunit of RNA polymerase III initiation factor TFIIIB, in transcription and tRNA processing. Mol Cell Biol 22:3264–3275
Jiang Y, Huang J, Lun K, Li B, Zheng H, Li Y, Zhou R, Duan W, Wang C, Feng Y, Yao H, Li C, Ji X (2020) Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II and Pol III. Genome Biol 21:158
Jimeno-González S, Gómez-Herreros F, Alepuz P, Chávez S (2006) A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol Cell Biol 26:8710–8721
Kiss T, Marshallsay C, Filipowicz W (1991) Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65:3517–3526
Kleinschmidt RA, LeBlanc KE, Donze D (2011) Autoregulation of an RNA Polymerase II promoter by the RNA Polymerase III transcription factor IIIC (TFIIIC) complex. Proc Nat Acad Sci USA 108:8385–8389
Kruszka K, Barneche F, Guyot R, Ailhas J, Menue I, Schiffer S, Marchfelder A, Echeverria M (2003) Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNaseZ. The EMBO J 22:621–632
Kumar Y, Bhargava P (2013) A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes. BMC Genomics 14:402
Lee MG-S, Van der Ploeg LHT (1997) Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu Rev Microbiol 51:463–489
Listerman I, Bledau AS, Grishina I, Neugebauer KM (2007) Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet 3:2268–2277
Mahapatra S, Dewari PS, Bhardwaj A, Bhargava P (2011) Yeast H2A.Z, FACT complex and RSC regulate transcription of tRNA gene through differential dynamics of flanking nucleosomes. Nucleic Acids Res 39:4023–4034
Martínez-Fernández V, Cuevas-Bermúdez A, Gutiérrez-Santiago F, Garrido-Godino AI, Rodríguez-Galán O, Jordán-Pla A, Lois S, Triviño JC, de la Cruz J, Navarro F (2020) Prefoldin-like Bud27 influences the transcription of ribosomal components and ribosome biogenesis in Saccharomyces cerevisiae. RNA 26:1360–1379
Marvin MC, Clauder-Munster S, Walker SC, Sarkeshik A, Yates JR, Steinmetz LS, Engelke DR (2011) Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 17:1441–1450
Mirón-García MC, Garrido-Godino AI, García-Molinero V, Hernández-Torres F, Rodríguez-Navarro S, Navarro F (2013) The Prefoldin Bud27 mediates the assembly of the eukaryotic RNA polymerases in an Rpb5-dependent manner. PLoS Genet 9:e1003297
Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K (2010) Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17:635–640
Ng HH, Robert F, Young RA, Struhl K (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16:806–819
Nguyen N-T-T, Saguez C, Conesa C, Lefebvre O, Acker J (2014) Identification of proteins associated with RNA polymerase III using a modified tandem chromatin affinity purification. Gene 556:1–60
Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR (2010) Human RNA polymerase III transcriptomes and relationships to pol II promoter chromatin and enhancer binding factors. Nat Struct Mol Biol 17:620–628
Padilla-Mejía NE, Florencio-Martínez LE, Moreno-Campos R, Vizuet-de-Rueda JC, Cevallos AM, Hernández-Rivas R, Manning-Cela R, Martínez-Calvillo S (2015) The selenocysteine tRNA gene in Leishmania major is transcribed by both RNA polymerase II and RNA polymerase III. Eukaryot Cell 14:216–227
Parnell TJ, Huff JT, Cairns BR (2008) RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27:100–110
Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR (2004) Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell 117:311–321
Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20:213–223
Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M (2010) Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Nat Acad Sci USA 107:3639–3644
Reuter LM, Meinel DM, Sträßer K (2015) The poly(A)-binding protein Nab2 functions in RNA polymerase III transcription. Genes Dev 29:1565–1575
Shukla A, Bhargava P (2018) Regulation of tRNA gene transcription by the chromatin structure and nucleosome dynamics. BBA-GRM 1861:295–309
Shukla A, Bhalla P, Potdar PK, Jampala P, Bhargava P (2021) Transcription-dependent enrichment of the yeast FACT complex influences nucleosome dynamics on the RNA polymerase III-transcribed genes. RNA (in Press). https://doi.org/10.1261/rna.077974.120
Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O (2006) Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26:4920–4933
Stallcup MR, Poulard C (2020) Gene-specific actions of transcriptional coregulators facilitate physiological plasticity: evidence for a physiological coregulator code. Trends Biochem Sci 45:497–510
Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41:480–492
Vernekar DV, Bhargava P (2015) Yeast Bud27 modulates the biogenesis of Rpc128 and Rpc160 subunits and the assembly of RNA polymerase III. Biochim Biophys Acta 1849:1340–1353
Wang Q, Nowak CM, Korde A, Oh D-H, Dassanayake M, Donze D (2014) Compromised RNA polymerase III complex assembly leads to local alterations of intergenic RNA polymerase II transcription in Saccharomyces cerevisiae. BMC Biol 12:89
Wild T, Cramer P (2012) Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci 37:99–105
Wlotzka W, Kudla G, Granneman S, Tollervey D (2011) The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. The EMBO J 30:1790–1803
Acknowledgements
I apologize to all researchers whose work is not cited inadvertently for lack of direct relevance. I owe thanks to all the past lab members for their work which enabled my writing this article.
Funding
Research in my lab was supported by the Department of Biotechnology (DBT) and ES scheme of Council of Scientific and Industrial Research (CSIR), Govt. of India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no competing interests.
Additional information
Communicated by Michael Polymenis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bhargava, P. Regulatory networking of the three RNA polymerases helps the eukaryotic cells cope with environmental stress. Curr Genet 67, 595–603 (2021). https://doi.org/10.1007/s00294-021-01179-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-021-01179-y