Abstract
Cohesin is essential for sister chromatid cohesion, which ensures equal segregation of the chromatids to daughter cells. However, the molecular mechanism by which cohesin mediates this function is elusive. Scc3, one of the four core subunits of cohesin, is vital to cohesin activity. However, the mechanism by which Scc3 contributes to the activity and identity of its functional domains is not fully understood. Here, we describe an in-frame five-amino acid insertion mutation after glutamic acid 704 (scc3-E704ins) in yeast Scc3, located in the middle of the second armadillo repeat. Mutated cohesin-scc3-E704ins complexes are unable to establish cohesion. Detailed molecular and genetic analyses revealed that the mutated cohesin has reduced affinity to the Scc2 loader. This inhibits its enrichment at centromeres and chromosomal arms. Mutant complexes show a slow diffusion rate in live cells suggesting that they induce a major conformational change in the complex. The analysis of systematic mutations in the insertion region of Scc3 revealed two conserved aspartic acid residues that are essential for the activity. The study offers a better understanding of the contribution of Scc3 to cohesin activity and the mechanism by which cohesin tethers the sister chromatids during the cell cycle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Author summary
Scc3/SA/STAG is an essential cohesin subunit, but its specific contribution to the activity of the complex is elusive. We identified a function of the second armadillo repeat of Scc3, which mediates interaction with the Scc2 loader. As a result, cohesin complexes that carry the mutated Scc3 fail to reach known cohesin sites on chromosomes and to establish cohesion. Interestingly, these mutants have a slower diffusion in live cells suggesting that the mutation in Scc3 induces a global conformational change. Finally, we identified two aspartic acid residues in Scc3 that are essential for cohesin activity. Our detailed characterization of Scc3 mutants revealed important information on the molecular basis of cohesin activity and dynamics. The findings will elucidate the mechanism by which cohesin mediates higher-order organization of chromatin, and may contribute to the understanding of Scc3-related disorders in humans.
Introduction
The cohesin complex belongs to the Structural Maintenance of Chromosome (SMC) family of protein complexes that is conserved from bacteria to humans. Cohesin mediates long-range chromatin interactions and plays a central role in maintaining genome stability (Nasmyth and Haering 2005; Onn et al. 2008; Uhlmann 2016). During mitosis, cohesin tethers the newly replicated DNA molecules, called sister chromatids, and thus ensures their proper segregation during cell division. In addition, cohesin regulates chromosome condensation, DNA repair and gene expression (Morales and Losada 2018; Onn et al. 2008; Uhlmann 2016). The structural and functional integrity of cohesin is important for human health (Cucco and Musio 2016; Mannini et al. 2010; Remeseiro et al. 2013; Singh and Gerton 2015).
The cohesin core is composed of a heterodimer of Smc1 and Smc3 proteins that form a ring. The SMCs are connected by a kleisin subunit called Mcd1/Scc1/Rad21 (hereon Mcd1), which also mediates interaction with a regulatory subunit called Scc3/SA/STAG (hereon Scc3) (Gruber et al. 2003; Haering et al. 2002). While yeast contains a single SCC3 gene, humans contain two mitotic paralogs called STAG1 and STAG2. These genes have been associated with distinct functions and human diseases (Cuadrado et al. 2019; Edungbola et al. 1987; Kojic et al. 2018; Mullegama et al. 2019; Romero-Perez et al. 2019; Viny et al. 2019; Wutz et al. 2020). Scc3 forms a sub-complex with two cohesin regulators called Wpl1 and Pds5 (Gandhi et al. 2006; Hartman et al. 2000; Kueng et al. 2006; Panizza et al. 2000; Rolef Ben-Shahar et al. 2008; Sumara et al. 2000).
Scc2 and Scc4 form a protein dimer, known as the loading complex, which mediates cohesin loading onto the chromatin (Ciosk et al. 2000). Scc4 interacts with the chromatin while Scc2 recruits cohesin (Bernard et al. 2006; Hinshaw et al. 2015; Ladurner et al. 2014; Shwartz et al. 2016; Watrin et al. 2006). In addition Scc2 activates the ATPase activity of cohesin which is essential for its loading and translocation along the chromatin (Lengronne et al. 2004; Petela et al. 2018). It has been shown that Scc2 interacts with several subunits of cohesin, including the SMC proteins, Mcd1 and Scc3 (Murayama and Uhlmann 2014; Shi et al. 2020). However, the functional importance of these multiple contacts has not been determined.
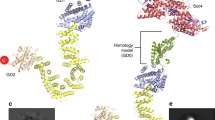
Scc3 contains two armadillo (ARM) repeats, which are associated with protein–protein interactions (Fig. 1a, b). The crystal structure of Scc3 from yeasts S. cerevisiae and Z. rouxii, and also the structure of the human Scc3 homolog STAG2, have been solved (Hara et al. 2014; Roig et al. 2014). The structure of all Scc3 homologs is similar and reveals an all-alpha elongated and crescent-shaped protein. The interface between Scc3 and Mcd1 was recently reported to mediate DNA binding (Li et al. 2018). However, the molecular details of the contribution of Scc3 to cohesin activity are elusive.
Bioinformatics analysis of Scc3. a Schematic of Scc3. The armadillo (ARM) repeats and Stromal Conserved Domain (SCD) are indicated. The location of the insertion after E704 is indicated. b Structure of S. cerevisiae Scc3 with a fragment of Mcd1 and bound to DNA (PDB 6H8Q) (Li et al. 2018). The nose and SCD are indicated. E704, D712 and D713 are colored in red and marked by arrows. c Multiple sequence analysis of Scc3 proteins generated by ClustalX. The arrow indicates S. cerevisiae E704. H human, Mm Mus Musculus, Xl Xenopus laevis, Dm Drosophila melanogaster, Sc Saccharomyces cerevisiae, Zr Zygosaccharomyces rouxii, Sp Schizosaccharomyces pombe
We previously described a transposon-based (RID) genetic screen that we performed in S. cerevisiae to identify functional domains in cohesin subunits (Matityahu et al. 2019; Orgil et al. 2015; Shwartz et al. 2016). In the current work, we describe scc3-E704ins, a new mutant of Scc3, which has a five-amino acid insertion after glutamic acid 704 in the second ARM repeat. This mutant was used to delineate the functions of Scc3 and its impact on the cohesin mechanism of action. The results advance our understanding of the mechanistic basis of cohesin activity.
Materials and methods
Yeast strains and media
The yeast strains used in this study are listed in Supplementary Table 1. Yeast strains were grown, as described, in YPD, SD-URA, or SD-LEU media, supplemented with 2% glucose (Guthrie 1991).
Cell synchronization
Cells were arrested in the G1 phase by the addition of alpha-factor (1.5 × 10−8 M final). Thereafter, to release them, the cells were washed twice with media containing pronase E (0.1 mg/ml; Sigma) and twice with media not containing pronase E. Exponentially grown cultures were arrested in G2/M using nocodazole (15 μg/ml final) in the indicated media. For early S arrest, hydroxyurea (200 mM; Sigma) was added.
Immunoprecipitation and Western blotting analysis
Cells were grown to mid-log phase, pelleted and washed with dH2O, and frozen in liquid nitrogen. Pellets were resuspended in 350 μl IP50/150 buffer (50 mM Tris, pH 8.0, 50/150 mM NaCl,1 mM EDTA, 5 mM MgCl2, 10% glycerol, 0.4% NP-40, protease inhibitor cocktail (Sigma)). For Smc3 acetylation experiments, IP50 was supplemented with 10 mM sodium butyrate (Sigma). Cells were lysed by adding glass beads (Sigma) to the resuspended pellets, followed by 4 working cycles of 1 min in a bullet blender (Next Advance). The lysates were cleared by two centrifugations of 5 and 15 min at 15,000 g at 4 °C. The lysate was supplemented with 2 U per 100 ml DNase I (New England Biolabs) for 20 min at 4 °C, and clarified by centrifugation. Immunoprecipitations were performed at 4 °C, and the appropriate antibodies were added for 1 h. The antibodies were collected on protein A magnetic beads (Bio-Rad) 1 h later and washed 3 times with IPH50/IPH150, and resuspended in 32 μl Laemmli buffer. Standard procedures for sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting were followed to transfer proteins from gels to a polyscreen PVDF membrane (Millipore). Membranes were blotted with the primary antibodies. Antibodies were detected using SuperSignal West Pico (Thermo) and LAS 4000 (GE). Antibodies used in this study were: mouse anti-HA (Roche), mouse anti-V5 (Invitrogen/Millipore), rabbit anti-Mcd1 (Rb555), which was a gift from Vincent Guacci, and rat anti-tubulin (Abcam).
Cohesion assay and chromatin immunoprecipitation (ChIP)
Cohesion at LYS4 was assayed using the LacI-GFP/LacO array. Cells were treated as described in the text and processed as described previously [33] to visualize GFP foci by microscopy. Each experiment was repeated three times and at least 300 cells were counted for every time-point in each experimental condition. ChIP was performed as previously described (Orgil et al. 2015). The primers used for qPCR are listed in Supplementary Table 2.
Chromosome spreads and immunofluorescence
Strains containing SCC3-GFP or scc3-E704-ins-GFP were grown at permissive temperature (23 °C) and arrested at G1 phase. The cells were shifted to restrictive temperature (35 °C) and rearrested in the G2/M phase. Chromosome spreads were prepared from 10 ml cell suspensions on superfrost slides (Thermo Scientific). Chromatin was probed with mouse anti-dsDNA (Abcam) followed by goat anti-mouse IgG conjugated to Alexa 647 (Invitrogen); Scc3-GFP was stained with rabbit anti-GFP (Abcam) followed by goat anti-rabbit IgG conjugated to Alexa 488 (Invitrogen). Slides were mounted with prolonged diamond anti-fade solution and visualized in Leica SPi8 Super-Resolution gSTED inverted confocal microscope through Plan apochromatic oil immersion objective (63X, NA = 1.40). HyVolution setup was used to capture images for fast real-time deconvolution and high-resolution images, better intensity and spatial distribution. Fluorescence intensity was calculated in Image J software.
Fluorescence correlation spectroscopy in living yeast
Live yeast cell FCS and imaging were performed using the Alba FLIM-Confocal Lifetime Imaging (ISS) system. Briefly, a 488 nm picosecond pulsed laser was used to excite the GFP-tagged protein with ~ 2 μW power measured at the back aperture. The excitation beam was delivered to the sample stage through an apochromatic water-immersion objective (60x, NA = 1.2) and the photons were collected by the same objective, and filtered through a 560 dichroic filter (Chroma). A 50 μm pinhole was employed to block off-focus photons, and the final signal was filtered through a band-pass filter (520/43, Chroma) before detection by a photon avalanche photodiode detector (SPCM-AQRH-15, Excelitas). Photons were recorded using the FastFLIM (ISS) module in the time-tagged time-resolved (TTTR) mode. Data were analyzed using VistaVison (ISS) and OriginPro software. The experimental procedure and analysis are detailed in the Supplementary Materials and Methods section.
Scc2 Overexpression
RNA was isolated using the RNeasy Mini Kit (QIAGEN), and cDNA synthesis was carried out according to the manufacturer's protocol using the High-Capacity RNA-to-cDNA kit (Applied Biosystems). qPCR primers for SCC2 and ACT1 were designed by Primer-BLAST software (Supplementary Table 2). 5 ng cDNA was used as a template for a qPCR reaction in a Step-One Plus RT-PCR (Applied Biosystems). The relative expression was calculated and shown as the fold change of SCC2 expression in cells growing in galactose/raffinose containing media.
Site-directed mutagenesis was performed on pIO97 (SCC3-3HA, URA3) using the Q5 Site-Directed Mutagenesis Kit (NEB) according to the manufacturer's instructions. Primers used for the reactions are listed in Supplementary Table 3.
Results
Scc3-E704ins does not support sister chromatid cohesion
Previously, we described a genetic screen aimed to identify functional domains in Scc3 by random insertion of transposon, followed by isolation of a dominant negative mutant (RID) screen (Orgil et al. 2015). We isolated a mutant containing 15-bp in-frame insertions after nucleotide 2112 in the SCC3 coding gene. The mutated gene encodes a protein that contains a five-amino acid insertion, CGRIE, located after glutamic acid (E) 704, which is at the end of a short alpha-helix (Fig. 1b). Hereon, this mutant will be referred to as scc3-E704ins. The insertion is located in the middle of the second armadillo domain of the protein (Fig. 1a, b). The α-helix domain is followed by a five-amino acid disordered loop and a second, short α-helix domain. Multiple sequence alignment of the insertion site revealed that the immediate insertion region is partially conserved (Fig. 1c) (Ashkenazy et al. 2010, 2016). The outcome of the short insertion in the first helix is likely to be its distortion and repositioning of the following loop.
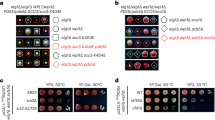
To examine the involvement of scc3-E704ins in cohesion activity, wild-type SCC3 and scc3-E704ins were cloned under the control of the SCC3 native promoter, and integrated into the URA3 locus. These cells carry the scc3-6 thermosensitive allele, which supports growth at 23 °C but neither at 30 °C nor 35 °C (Fig. 2a). Strains yIO112 (scc3-6), yIO113 (SCC3 scc3-6) and yIO114 (scc3-E704ins scc3-6) were grown to saturation, serially diluted, spotted on YPD plates and incubated at either permissive or restrictive temperatures for scc3-6 (Fig. 2a). Strains grew equally well at 23 °C, at which scc3-6 is active. In contrast, a sole copy of either scc3-6 or scc3-E704ins was unable to support cell growth at the restrictive temperatures of 30 °C and 35 °C.
scc3-E407ins does not support cell viability and cohesion. a Strains yIO112 (scc3-6), yIO113 (SCC3-6 scc3-6) and yIO114 (scc3-E704ins-6HA scc3-6) were grown to saturation in YPD media. Tenfold serial dilutions of each strain were plated on YPD plates and grown at either the permissive (23 °C) or restrictive (35 °C) temperature for scc3-6. b Strains yEB-009 (SCC3-AID), yAP-1029 (SCC3 SCC3-AID), and yAP-1030 (scc3-E704ins SCC3-AID) were grown to saturation in YPD media. Tenfold serial dilutions of each strain were plated on YPD plates without or with IAA at 23 °C. c Flowchart of the experimental design. d Strains as indicated in A were grown at 23 °C to mid-log phase and arrested in G1 using α-factor. Cells were then shifted to 32.5 °C for 1 h, released into the cell cycle and rearrested in G2/M with nocodazole and processed for cohesion assay
Next, we explored the possibility that scc3-E704ins is a thermosensitive allele using a strain in which the native SCC3 was fused to an auxin-induced degron (AID) that induces degradation of the fused protein in the presence of 3-Indoleacetic acid (IAA) in the growth medium. Strains yEB-009 (SCC3-AID), yAP-1029 (SCC3 SCC3-AID), and yAP-1030 (scc3-E704ins SCC3-AID) were grown to saturation, serially diluted, and spotted on plates without or with IAA and grown at 23 °C (Fig. 2b). All strains grew equally without IAA. In the presence of IAA, cells carrying the wild-type SCC3 grew well. However, scc3-E704ins did not support cell viability. These results indicate that at normal expression levels, scc3-E704ins does not support cell viability.
We examined if scc3-E704ins supports cohesion activity by monitoring premature chromosome separation. We used strains yIO112 (scc3-6), yIO113 (SCC3 scc3-6) and yIO114 (scc3-E704ins scc3-6), expressing LacI-GFP, and containing a LacO array integrated at the LYS4 locus on chromosome 4, about 40 kbp from the centromere. These were grown at 23 °C to mid-log phase and arrested in the G1 stage of the cell cycle. The temperature was raised to 35 °C to inactivate scc3-6, and cells were released into the cell cycle and rearrested at the G2/M phase (Fig. 2c). Sister chromatid cohesion was maintained in cells that expressed SCC3, while premature cohesion loss was detected in scc3-6, both alone and in the presence of scc3-E704ins (Fig. 2d). Cell cytometry did not reveal any cell cycle delays between strains (Supplementary Fig. S1). We, therefore, concluded that cohesin is inactive in the presence of a sole copy of scc3-E704ins.
The interaction of cohesin-scc3-E704ins complexes with the Scc2 loader is compromised
The E704 region in Scc3 has not been documented with cohesin complex assembly or interaction with cohesin regulators (Li et al. 2018; Orgil et al. 2015; Roig et al. 2014; Zhang et al. 2013). We verified the integrity of the complex by a series of co-immunoprecipitation assays (co-IP). These experiments were done in the presence of DNase I to ensure direct protein–protein interaction. Strains yIO92 (SCC3-6HA) and yIO538 (scc3-E704ins-6HA) were grown to mid-log phase and processed to co-IP with anti-HA antibodies. The steady-state levels of Scc3-6HA and scc3-E704ins-6HA were similar, indicating that the mutant protein is stable (Fig. 3a and Supplementary Fig. S2). Co-precipitation of Mcd1 was tested. No change in the level of Mcd1 protein was found, thus indicating that the mutation in Scc3 does not affect the integrity of the core complex.
scc3-E704ins mediates interaction with Scc2. a Cells of strains JH5257 (SCC3), yIO92 (SCC3-6HA) or yIO538 (scc3-E704ins-6HA) were grown to mid-log phase in YPD media, lysed and subjected to immunoprecipitation against the HA tag (Scc3). Precipitated proteins were analyzed by Western blot using antibodies against HA (IP) and Mcd1 (co-IP) (n = 3). b Strains as in A were grown to mid-log phase in YPD media, lysed and subjected to immunoprecipitation against the HA tag (Scc3). Precipitated proteins were analyzed by Western blot using antibodies against HA (IP) and V5 (co-IP). A representative blot is shown. c. The intensity ratio of 3 independent Scc3-HA/Scc2-V5 experiments was quantified by Image J and normalized. Statistical significance of p = 0.000249 was determined by two-tailed student's t test. d. Strains yAP1008 (scc3-6 pGAL-SCC2), yAP1009 (scc3-6 SCC3 pGAL-SCC2) and yAP1010 (scc3-6 scc3-E704ins pGAL-SCC2) were grown to saturation in SD-LEU media. Tenfold serial dilutions of each strain were plated on SD-LEU/galactose plates and grown at either the permissive (23 °C) or restrictive (30 °C) temperature for scc3-6
Next, we explored the effect of the mutation on the interaction of cohesin with the Scc2 loader by co-IP experiment of either Scc3-6HA (yIO92) or scc3-E704ins-6HA (yIO538) with Scc2-V5 (Fig. 3b). Samples were treated with DNase I and the salt concentration of the IP buffer was 150 mM NaCl. We noted that higher salt concentrations compromised Scc3–Scc2 interactions. Under these conditions, the interaction of scc3-E704ins with Scc2 was significantly and repeatedly weakened, as demonstrated by a 40% reduction in the co-precipitated Scc2 with the mutant protein, compared to the wild type (Fig. 3c). Therefore, we concluded that the E704 region of Scc3 is important but not essential for the interaction of cohesin with Scc2. Most recently, this finding was supported by a cryo-EM study of human STAG1 in complex with NIPBL (homologs of yeast Scc3 and Scc2, respectively). The E704 region in yeast Scc3 corresponds to a region that is located in proximity to the interaction domain between the proteins (Shi et al. 2020).
Assuming that the weaken Scc2–scc3-E704ins interaction is the molecular event that compromises cohesin activity, we predicted that overexpression of Scc2 might suppress the phenotype. For this purpose, we transformed cells with a centromeric plasmid in which SCC2 was inserted under the control of the GAL promoter. Overexpression of about twofold of Scc2 was validated by RT-qPCR in strains yAP1008 (scc3-6/pGAL-SCC2), yAP1009 (scc3-6 SCC3/pGAL-SCC2) and yAP1010 (scc3-6 scc3-E704ins/pGAL-SCC2) (Supplementary Fig. S3). These strains were grown to saturation, serially diluted and spotted on SD-LEU/galactose plates (Fig. 3d). The plates were incubated at 23 °C and 30 °C. All the strains grew equally well at 23 °C, indicating that SCC2 overexpression does not affect cell growth. At 30 °C, cells containing only scc3-6 did not grow, while cells containing the wild-type allele of SCC3 grew well. Overexpression of SCC2 partially suppressed the growth defect of the scc3-E704ins allele, indicating that this mutant indeed affects the integrity of the cohesin–loader interaction.
The reduced affinity of scc3-E704ins-cohesin to Scc2 suggests that chromatin binding might be affected by the Scc3 mutation. This possibility was examined using the chromatin immunoprecipitation (ChIP) assay. Strains yIO113 (SCC3-HA) and yIO114 (scc3-E704ins-HA) were grown to mid-log phase, arrested at G2/M phase and processed for ChIP with anti-HA antibodies. We examined cohesin binding at CARC1 on chromosomes III and V arms, centromeres of chromosomes III, IV, V, and the rDNA locus on chromosome XII (Fig. 4a–f). Wild-type Scc3 was enriched at all loci, while DNA binding was residual with the scc3-E704ins.
scc3-E704ins is not enriched on chromosomal loci a-e. Strains yIO113(SCC3-6HA) and yIO114 (scc3-E704ins-6HA) were processed for chromatin immunoprecipitation analysis. HA-tagged proteins were immunoprecipitated. Precipitated DNA was analyzed by quantitative PCR for a CARC1 on chromosome III, b Chromosome V arm, c centromere III, d centromere IV, e centromere V, and f the rDNA locus on chromosome XII (n = 3). g Strains yAP01 (SCC3-GFP scc3-6) and yAP03 (scc3-E704ins-GFP scc3-6) were grown at the permissive temperature (23 °C) and arrested at G1 using α-factor. The cells were shifted to restrictive temperature (32.5 °C) and rearrested in the G2/M phase using nocodazole. Nuclei were spread and Scc3 was detected by immunofluorescence. The bulk DNA was stained with anti-dsDNA antibody. h The intensity ratio of the Scc3/dsDNA fluorescence described in G. At least 100 nuclei were measured from each strain. ***p < 0.001
The reduced enrichment of cohesin-Scc3-E704ins at centromeres and chromosome arms may be the result of its failure to load onto the chromatin or, alternatively, its increased dissociation and failure to accumulate on the chromatin. To discriminate between these possibilities, we performed analyses of SCC3 and scc3-E704ins via immunofluorescence on mitotic chromosome spreads. These were prepared from strains yAP01 (SCC3-GFP) and yAP03 (scc3-E704ins-GFP), which were arrested at either the G1 or G2/M phases of the cell cycle, followed by Scc3 immunofluorescence (Fig. 4g). As anticipated, Scc3 was not detected on the chromatin spreads of cells at G1. Both wild-type and mutant Scc3 were found on the mitotic chromatin, although the intensity of scc3-E704 was significantly weaker (Fig. 4h). These results suggest that cohesin is loaded, but its chromatin tethering activity is compromised.
scc3-E704ins reduces cohesin diffusion in live cells
We recently showed by fluorescence correlation spectroscopy (FCS) that depletion of Scc3 in live cells leads to slow diffusion of the depleted complex with respect to the holocomplex (Liu et al. 2020). To explore the effect of the scc3-E704ins mutant on cohesin diffusion, we analyzed by FCS strains yAP01 (SCC3-GFP) and yAP03 (scc3-E704ins-GFP) in S phase cells, as described in (Liu et al. 2020) (Fig. 5, S4; Table 1). Using epifluorescent microscopy, we validated that the GFP-fused, wild-type and mutant Scc3 proteins are localized to the nucleus in S and G2/M phases of the cell cycle (Supplementary Fig. S5). The diffusion time of scc3-E704ins-GFP was found to be slower than that of the wild type, as demonstrated by fitting with a one-component anomalous diffusion model (Fig. 5). The degree of anomalous behavior (α term in Eq. 3, see supplementary information for details) was found to be significantly different. scc3-E704ins-GFP fitted with an α value that was higher on average than that of SCC3-GFP (~ 0.75 vs ~ 0.55). This indicates a greater restriction in the mobility of scc3-E704ins-GFP (Cui and Irudayaraj 2015). We further investigated the diffusion properties with a two-component 3D diffusion model. The slow diffusion component of both strains exhibited similar behavior, with a diffusion time of ~ 15 ms. This may indicate the conditions in which Scc3-GFP and scc3-E704ins-GFP behave similarly, and presumably with cohesin binding to larger complexes such as chromatin (Table 1). Interestingly, the fast diffusion component of scc3-E704ins-GFP is about 2.5-fold slower than that of Scc3-GFP. The difference in mobility between scc3-E704ins-GFP and SCC3-GFP is within this range and possibly associated with changes in cohesin-scc3-E704ins functions. Molecular diffusion is negatively proportional to the hydrodynamic radius, according to the Stoke–Einstein equation. Thus, our results suggest that scc3-E704ins-GFP hinders the cohesin function through association with and recruitment of other large biomolecules or that it undergoes conformational changes.
Scc3 and scc3-E704ins diffusion in live cells. a Representative normalized fluorescence correlation spectroscopy data of SCC3-GFP and scc3-E704ins-GFP at 27 °C. b Diffusion time is analyzed from both strains with a one-component anomalous diffusion model. Data are shown as mean ± s.d., n ~ 20 for each group. *p < 0.0005 (Welch's t test)
Targeted mutagenesis of residue near E704 reveals key functional residues
We aimed to identify the key residues in the E704 region that contributed to the loss-of function of the scc3-E704ins allele. We used ConSurf to identify tentative functional residue in this region (Ashkenazy et al. 2010, 2016) (Fig. 6a). Based on this analysis, we mutated E704, S705, D712, D713, W714 and E720 to alanine, and integrated the mutated genes into the URA3 locus of scc3-6 cells (Fig. 6a). The viability of these strains was examined by a semi-quantitative spot assay (Fig. 6b). No effect on cell viability was observed in cells carrying scc3-E704A, W714A or E720A, when the scc3-6 allele was inactivated at 35 °C. However, cells carrying scc3-S705A were sick, as evidenced by a 100-fold decrease in growth; and scc3-D712A and scc3-D713A were sick at 30 °C. S705 is buried and, therefore, expected to be structural. In contrast, D712 and D713 are exposed and located in a loop, implying their functional importance. We tested if D712A and D713A are thermosensitive alleles by their ability to complement SCC3-AID allele at 23 °C (Fig. 6c). scc3-D712A and scc3-D713 are expressed in cells (Supplementary Fig. S6). We repeated the growth assay in cells spotted on YPD plates without or with 2 mM IAA. scc3-D712A and SCC3 cells grow equally well on plates with IAA at 23 °C. However, cells carrying scc3-D713A were sick. This analysis suggests that D712A is a temperature-sensitive allele, while D713A is a mild functional allele.
Analysis of alanine point mutations in the Scc3 E704 region. a The conservation and exposure of the E704 region were determined by ConSurf server. The indicated residues were replaced with alanine. b Cells of strain yIO168 (scc3-6), carrying an ectopic copy of SCC3-6HA (yIO113), scc3-E704A-6HA (yAP015), scc3-S705A-6HA (yAP016), scc3-D712A-6HA (yAP017), scc3-D713A-6HA (yAP018), scc3-W714A-6HA (yAP1011) or scc3-E720A-6HA (yAP019) were grown to saturation in YPD media. Tenfold serial dilution of the strains was plated on YPD plates and grown at either permissive (23 °C) or restrictive (30 °C, 35 °C) temperature. c Strains yEB-009 (SCC3-AID), yAP-1029 (SCC3 SCC3-AID), and yAP-1032 (scc3-D712A SCC3-AID) and yAP-1033 (scc3-D713A SCC3-AID) were grown to saturation in YPD media. Tenfold serial dilutions of each strain were plated on YPD plates without or with IAA at 23 °C. d Strains yIO112 (scc3-6), yIO113 (SCC3-6HA scc3-6), yAP017 (scc3-D712A scc3-6) and yAP018 (scc3-D713A-6HA scc3-6) were analyzed for sister chromatid cohesion using the GFP dot assay as described in Fig. 2c, d (n = 3)
We analyzed sister chromatid cohesion in strains yIO113 (SCC3 scc3-6), yIO112 (scc3-6), yAP017 (scc3-D712A scc3-6) and yAP018 (scc3-D713A scc3-6) (Fig. 6d). Cells were grown at 23 °C to mid-log phase and arrested in the G1 stage of the cell cycle. The temperature was raised to 35 °C to inactivate scc3-6, and cells were released into the cell cycle and rearrested at the G2/M phase. About 20% of the cells carrying scc3-D712A and 40% of the cells carrying scc3-D713A showed premature cohesion loss, implying that these residues are important for the activity of Scc3.
Next, we analyzed associations of the mutants with the chromatin. Strains yIO113 (SCC3 scc3-6), yAP017 (scc3-D712A-HA scc3-6) and yAP018 (scc3-D713A-HA scc3-6) were grown to mid-log phase at 23 °C, and arrested at the G1 stage of the cell cycle. The temperature was raised to 35 °C to inactivate scc3-6. Cells were released into the cell cycle and rearrested in G2/M by nocodazole, and processed for ChIP by anti-HA antibodies. We analyzed cohesin enrichment at chromosome arm, centromeres, and the rDNA locus (Fig. 7). The binding of Scc3-D712A was about fourfold less than the wild-type Scc3, while Scc3-D713A revealed a milder chromatin binding defect. These results indicate that D712 and D713 play a functional role in Scc3.
Reduced levels of scc3-D712A and scc3-D713A on chromosomes. Strains yIO113 (SCC3 scc3-6), yAP017 (scc3-D712A-HA scc3-6) and yAP018 (scc3-D713A-HA scc3-6) were grown in YPD at 23 °C, and arrested at the G1 stage of the cell cycle. The temperature was raised to 35 °C to inactivate scc3-6. Cells were released into the cell cycle and rearrested in G2/M by nocodazole, and processed for ChIP analysis with anti-HA antibodies. Precipitated DNA was analyzed by quantitative PCR for the a CARC1 on chromosome III, b centromere III, c centromere V, (d) rDNA locus on chromosome XII. (n = 3)
Discussion
We identified, by transposon mutagenesis, the E704ins region in Scc3 as essential for cohesin activity. The mutated allele causes loss of function when expressed at normal levels. The residue is located in the second ARM repeat. Insertion mutation at this domain compromised the interaction of cohesin with Scc2 to affect proper loading onto the chromatin. We also found that aspartic acid residues D712 and D713, which are located in this region, are essential for cell viability and cohesion. These results highlight the significance of the second ARM repeat of Scc3 to cohesin activity and assign a new function to it.
The affinity of Scc2 to the scc3-E704ins-cohesin complexes is reduced but not abolished, suggesting that the interaction is mediated by several sites. This property is conserved, as evident by a previous study in S. pombe, which suggested that the E704 region interacts with the Scc2 loader (Murayama and Uhlmann 2014). Most recently, the structure of Smc1-Smc3-Rad21-Nipbl-Stag1 was solved by cryo-EM (Shi et al. 2020). The structure reveals Stag1 E647-Q660 region as an important interaction interface with NIPBL/Scc2. However, this interface is not exclusive as Stag1 has multiple interaction interfaces with Nipbl and the SMC proteins (Supplementary Fig. S7). In humans, a 50% reduction in the level of Scc2 is associated with Cornelia de Lange syndrome, a developmental disorder. The cellular phenotype of such decline in Scc2 includes alternations in gene expression, protein translation, and chromatin organization. The minimal Scc2 expression level that is required for cohesin in yeast is yet to be determined. However, we suggest that even if Scc2 remains bound to some of the cohesin-scc3-E704ins complexes, the corrupt interaction between Scc3 and Scc2 is sufficient to destabilize cohesin association with chromatin even if Scc2 is still bound to cohesin through other sites.
Based on our sequence alignment (Fig. 1a), our analysis identified the negatively charged aspartic acid residue located at the flanking of the interface is essential for the activity. We identified D712A as a thermosensitive allele while D713A as a weak functional allele. We suggest that these residues play a role in either intra- or inter-molecular interaction mediated by this loop. However, our analysis shows that their overall effect on cohesion is limited.
The loading mechanism of cohesin is unclear. All current models suggest that it involves the opening of the DNA entry gate. Binding of Scc2 to two cohesin subunits followed by a conformational change may provide the mechanical force for such opening (Orgil et al. 2015; Shi et al. 2020). Both our previous (Liu et al. 2020) and current FCS studies showed that Scc3 affects the diffusion of cohesin in live cells. We suggest that this change in cohesin dynamics is due to a conformational change. It has been suggested that DNA binding is associated with a fold-back of the SMC coiled-coil domain (Orgil et al. 2015; Shi et al. 2020). This is congruent with the location of one of the binding sites in the E704 region of Scc3, as evident from our results.
The five-amino acid insertion is located at the end of an α-helix and probably distorts the helix. Searching for the functional residues in this region revealed that D712 and D713 are essential for cell viability, cohesin loading, and cohesion. These residues are evolutionarily conserved from yeast to humans. Notably, yeast glutamic acids are replaced with aspartic acids in higher eukaryotes that maintain a strong negative charge. The human homologs of Scc3, STAG1 and STAG2 have been shown to be associated with cancer (Hill et al. 2016). A search of genome databases revealed a small number of missense mutations in the region corresponding to the yeast E704. It would be interesting to examine if these mutations are drivers of cancer development. Altogether, this work elucidated the contribution of Scc3 to cohesin activity and advances our understanding of the mechanistic details of cohesin loading and cohesion establishment.
References
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529-533. https://doi.org/10.1093/nar/gkq399
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344-350. https://doi.org/10.1093/nar/gkw408
Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP (2006) A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol 16:875–881. https://doi.org/10.1016/j.cub.2006.03.037
Guthrie CaF (1991) Guide to yeast genetics and molecular biology. Methods Enzymol 194: 1–863
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 doi:
Cuadrado A, Gimenez-Llorente D, Kojic A, Rodriguez-Corsino M, Cuartero Y, Martin-Serrano G, Gomez-Lopez G, Marti-Renom MA, Losada A (2019) Specific contributions of cohesin-SA1 and cohesin-SA2 to TADs and polycomb domains in embryonic stem cells. Cell Rep 27(3500–3510):e3504. https://doi.org/10.1016/j.celrep.2019.05.078
Cucco F, Musio A (2016) Genome stability: what we have learned from cohesinopathies. Am J Med Genet C Semin Med Genet 172:171–178. https://doi.org/10.1002/ajmg.c.31492
Cui Y, Irudayaraj J (2015) Dissecting the behavior and function of MBD3 in DNA methylation homeostasis by single-molecule spectroscopy and microscopy. Nucleic Acids Res 43:3046–3055. https://doi.org/10.1093/nar/gkv098
Edungbola LD, Watts SJ, Kayode OO (1987) Endemicity and striking manifestations of onchocerciasis in Shao, Kwara State, Nigeria. Afr J Med Med Sci 16: 147–156 doi:
Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16:2406–2417. https://doi.org/10.1016/j.cub.2006.10.061
Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112: 765–777
Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788
Hara K, Zheng G, Qu Q, Liu H, Ouyang Z, Chen Z, Tomchick DR, Yu H (2014) Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol 21:864–870. https://doi.org/10.1038/nsmb.2880
Hartman T, Stead K, Koshland D, Guacci V (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 151: 613–626
Hill VK, Kim JS, Waldman T (2016) Cohesin mutations in human cancer. Biochim Biophys Acta 1866:1–11. https://doi.org/10.1016/j.bbcan.2016.05.002
Hinshaw SM, Makrantoni V, Kerr A, Marston AL, Harrison SC (2015) Structural evidence for Scc4-dependent localization of cohesin loading. Elife 4:e06057. https://doi.org/10.7554/eLife.06057
Kojic A, Cuadrado A, De Koninck M, Gimenez-Llorente D, Rodriguez-Corsino M, Gomez-Lopez G, Le Dily F, Marti-Renom MA, Losada A (2018) Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol 25:496–504. https://doi.org/10.1038/s41594-018-0070-4
Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127:955–967. https://doi.org/10.1016/j.cell.2006.09.040
Ladurner R, Bhaskara V, Huis in 't Veld PJ, Davidson IF, Kreidl E, Petzold G, Peters JM (2014) Cohesin's ATPase activity couples cohesin loading onto DNA with Smc3 acetylation. Curr Biol 24: 2228–2237 https://doi.org/10.1016/j.cub.2014.08.011
Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578. https://doi.org/10.1038/nature02742
Li Y, Muir KW, Bowler MW, Metz J, Haering CH, Panne D (2018) Structural basis for Scc3-dependent cohesin recruitment to chromatin. Elife. https://doi.org/10.7554/eLife.38356
Liu W, Biton E, Pathania A, Matityahu A, Irudayaraj J, Onn I (2020) Monomeric cohesin state revealed by live-cell single-molecule spectroscopy. EMBO Rep 21:e48211. https://doi.org/10.15252/embr.201948211
Mannini L, Menga S, Musio A (2010) The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum Mutat 31:623–630. https://doi.org/10.1002/humu.21252
Matityahu A, Shwartz M, Onn I (2019) Identifying functional domains in subunits of structural maintenance of chromosomes (SMC) complexes by transposon mutagenesis screen in yeast. Methods Mol Biol 2004:63–78. https://doi.org/10.1007/978-1-4939-9520-2_6
Morales C, Losada A (2018) Establishing and dissolving cohesion during the vertebrate cell cycle. Curr Opin Cell Biol 52:51–57. https://doi.org/10.1016/j.ceb.2018.01.010
Mullegama SV, Klein SD, Signer RH, Center UCG, Vilain E, Martinez-Agosto JA (2019) Mutations in STAG2 cause an X-linked cohesinopathy associated with undergrowth, developmental delay, and dysmorphia: expanding the phenotype in males. Mol Genet Genomic Med 7:e00501. https://doi.org/10.1002/mgg3.501
Murayama Y, Uhlmann F (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505:367–371. https://doi.org/10.1038/nature12867
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648. https://doi.org/10.1146/annurev.biochem.74.082803.133219
Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24:105–129. https://doi.org/10.1146/annurev.cellbio.24.110707.175350
Orgil O, Matityahu A, Eng T, Guacci V, Koshland D, Onn I (2015) A conserved domain in the scc3 subunit of cohesin mediates the interaction with both mcd1 and the cohesin loader complex. PLoS Genet 11:e1005036. https://doi.org/10.1371/journal.pgen.1005036
Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 10: 1557–1564
Petela NJ, Gligoris TG, Metson J, Lee BG, Voulgaris M, Hu B, Kikuchi S, Chapard C, Chen W, Rajendra E, Srinivisan M, Yu H, Lowe J, Nasmyth KA (2018) Scc2 is a potent activator of cohesin’s ATPase that promotes loading by binding Scc1 without Pds5. Mol Cell 70(1134–1148):e1137. https://doi.org/10.1016/j.molcel.2018.05.022
Remeseiro S, Cuadrado A, Losada A (2013) Cohesin in development and disease. Development 140:3715–3718. https://doi.org/10.1242/dev.090605
Roig MB, Lowe J, Chan KL, Beckouet F, Metson J, Nasmyth K (2014) Structure and function of cohesin’s Scc3/SA regulatory subunit. FEBS Lett 588:3692–3702. https://doi.org/10.1016/j.febslet.2014.08.015
Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321:563–566. https://doi.org/10.1126/science.1157774
Romero-Perez L, Surdez D, Brunet E, Delattre O, Grunewald TGP (2019) STAG Mutations in Cancer. Trends Cancer 5:506–520. https://doi.org/10.1016/j.trecan.2019.07.001
Shi Z, Gao H, Bai XC, Yu H (2020) Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science. https://doi.org/10.1126/science.abb0981
Shwartz M, Matityahu A, Onn I (2016) Identification of functional domains in the cohesin loader subunit Scc4 by a random insertion/dominant negative screen. G3 (Bethesda) 6: 2655–2663. https://doi.org/10.1534/g3.116.031674
Singh VP, Gerton JL (2015) Cohesin and human disease: lessons from mouse models. Curr Opin Cell Biol 37:9–17. https://doi.org/10.1016/j.ceb.2015.08.003
Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762
Uhlmann F (2016) SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol 17:399–412. https://doi.org/10.1038/nrm.2016.30
Viny AD, Bowman RL, Liu Y, Lavallee VP, Eisman SE, Xiao W, Durham BH, Navitski A, Park J, Braunstein S, Alija B, Karzai A, Csete IS, Witkin M, Azizi E, Baslan T, Ott CJ, Pe’er D, Dekker J, Koche R, Levine RL (2019) Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell 25(682–696):e688. https://doi.org/10.1016/j.stem.2019.08.003
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM (2006) Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16:863–874. https://doi.org/10.1016/j.cub.2006.03.049
Wutz G, Ladurner R, St Hilaire BG, Stocsits RR, Nagasaka K, Pignard B, Sanborn A, Tang W, Varnai C, Ivanov MP, Schoenfelder S, van der Lelij P, Huang X, Durnberger G, Roitinger E, Mechtler K, Davidson IF, Fraser P, Lieberman-Aiden E, Peters JM (2020) ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesin(STAG1) from WAPL. Elife. https://doi.org/10.7554/eLife.52091
Zhang N, Jiang Y, Mao Q, Demeler B, Tao YJ, Pati D (2013) Characterization of the interaction between the cohesin subunits Rad21 and SA1/2. PLoS ONE 8:e69458. https://doi.org/10.1371/journal.pone.0069458
Acknowledgments
We thank Ilia Mistesky for providing reagents and Vincent Guaaci and Doug Koshland for providing yeast strains and reagents. We also thank members of the Onn Lab for their support and Steve Spencer for his editorial review and corrections.
Funding
This work was supported by Israel Science Foundation Grants 1099/16 (IO) and 987/20. JI thanks the W.M. Keck Foundation for their support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pathania, A., Liu, W., Matityahu, A. et al. Chromosome loading of cohesin depends on conserved residues in Scc3. Curr Genet 67, 447–459 (2021). https://doi.org/10.1007/s00294-020-01150-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01150-3