Abstract
During amino acid limitation, the protein kinase Gcn2 phosphorylates the α subunit of eIF2, thereby regulating mRNA translation. In yeast Saccharomyces cerevisiae and mammals, eIF2α phosphorylation regulates translation of related transcription factors Gcn4 and Atf4 through upstream open reading frames (uORFs) to activate transcription genome wide. However, mammals encode three more eIF2α kinases activated by distinct stimuli. Did the translational control system involving eIF2α phosphorylation evolve from so simple (as found in yeast S. cerevisiae) to complex (as found in humans)? Recent genome-wide translational profiling studies of amino acid starvation response in the fission yeast Schizosaccharomyces pombe provide an unexpected answer to this question.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In response to diverse cellular stresses, ribosomes reprogram global protein synthesis to optimize the utilization of nutrients and energy and reconfigure the proteome to mitigate stress damage (Asano 2013; Dever 2002). For example, during amino acid limitation, the protein kinase Gcn2 (Eif2ak4 in mammals) is activated by uncharged tRNAs that accumulate due to the amino acid undersupply. The activated Gcn2 then phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2), thereby reducing delivery of initiator tRNAs to ribosomes which results in the inhibition of global protein synthesis. Concurrently, phosphorylation of eIF2α (eIF2α-P) enhances translation of select mRNAs, such as Saccharomyces cerevisiae GCN4 and mammalian atf4, which direct gene expression for stress adaptation (Hinnebusch et al. 2007; Vattem and Wek 2004) in both normal and cancer cells (Wek and Staschke 2010; Ye et al. 2010). In mammals, however, other diverse stress stimuli activate three more eIF2α kinases, Hri (Eif2ak1), Pkr (Eif2ak2), and Perk (Eif2ak3), thereby deploying a similar transcriptional response (Young and Wek 2016). How is translation regulated differentially by eIF2α phosphorylation? Of the four eIF2α kinases, the role of Hri in stress response remains a mystery in particular. How are these distinct kinases utilized to integrate the various stress stimuli? How did this translational control system evolve?
A critical cis-acting regulator for control by eIF2α-P is upstream ORFs (uORFs) present in the 5′-leaders of mRNAs. uORFs are suggested to be present in over 50% of mammalian and 10% of yeast mRNAs (Asano 2013). The prototypical example of uORF-dependent translational control is found for yeast GCN4 encoding a basic leucine-zipper transcription factor (bZIP) (Hinnebusch 1997). This system takes advantage of the scanning mechanism, which normally allows a mRNA to produce only a single protein (Asano 2014; Hinnebusch et al. 2007). In eukaryotes, ribosomes associated with eIF2 and its bound initiator tRNA are loaded to the 5′-cap of mRNAs and then processively scanned 5′ to 3′ in search of an initiation codon. Often uORFs are bypassed due to the poor context of its AUG start codon or, when translated, inhibit downstream initiation of the main coding sequence (CDS).
In the case of GCN4, however, the first of its four uORFs (uORF1) is fully translated, but allows ribosomes to reinitiate translation of another ORF located downstream; the ribosome remains linked to the mRNA after translation termination and resumes scanning for subsequent translation initiation (Grant and Hinnebusch 1994) (Fig. 1a). In the absence of the stress, the ribosome re-initiates at one of the three other uORFs, typically uORF4, and hence no translation of the main CDS (Fig. 1a, panel 1). Upon stress and eIF2α-P induced by Gcn2, however, delayed translation re-initiation allows for ribosomes to scan through the inhibitory uORFs, and instead initiates translation at the CDS. In this way, the GCN4 is well translated only during cellular stress and induced eIF2 α-P (Fig. 1a, panel 2). Gcn4 is the master regulator of yeast general amino acid control (GAAC) response, driving transcription of the majority (50–60%) of the genome (Natarajan et al. 2001). A mere deletion of uORFs of this gene completely abolishes the yeast’s ability to induce this response, emphasizing the crucial role played by this mechanism at the translational level (Mueller and Hinnebusch 1986).
Translational control by paired uORFs. a Model of GCN4 translational control. The schematics on the top describe the structure of GCN4 mRNA with boxes indicating uORFs. Table below describes the uORF-dependent delayed re-initiation model for GCN4 translation in unstressed cells (panel 1) and in stressed cells (panel 2). Gray ovals in the schematics represent ribosomes with 40S (smaller oval) and 60S (larger oval) subunits or the subunit alone. Black straight arrows indicate 40S ribosome scanning. Brown rounded arrows indicate ribosome dissociation. See text for details. b–d uORFs found in the leader regions of GCN4/atf4/cpc1 homologs (b), S. pombe fil1 and gcn5 (c), and fungal hri (d). See Fig. 2 for the classification of species shown. Boxes indicate uORFs (yellow, positive or blue, negative element, respectively) or the main CDS (gray). For dipeptide-coding uORFs, the amino acid sequences (MC, MM, and MI) of dipeptides originating from the uORFs are shown below. Overlapping uORF has been considered characteristic of the negative element, as found with Homo sapiens atf1 uORF2. However, this does not appear to be the case in diverse fungi including S. cerevisiae bearing GCN4 uORF4
Humans have a similar system targeting a homologous bZIP transcription factor Atf4 (Vattem and Wek 2004). Thus, the leader region of atf4 mRNA carries two uORFs: uORF1 serving as the positive element allowing downstream re-initiation and uORF2 serving as the negative element inhibiting translation of the main CDS in the absence of the stress (Fig. 1b). During amino acid starvation, Gcn2 (EIF2AK4) is fully responsible for expressing a set of genes including those encoding amino acid synthesis enzymes. However, various other stress stimuli—for example, heme depletion during erythrocyte development, oxidative stress (Hri), RNA virus infection (Pkr), and endoplasmic reticulum (ER) stress (Perk)—can similarly lead to eIF2α phosphorylation, thereby expressing a common set of genes termed the integrated stress response (ISR) through translational activation of atf4 mRNA (Ron and Harding 2007).
Did the translational control system involving eIF2α phosphorylation evolve from so simple (as found in yeast S. cerevisiae) to complex (as found in humans)? Recent genome-wide translational profiling studies of starvation response in the fission yeast Schizosaccharomyces pombe provide an unexpected answer to this question (Chikashige et al. 2020; Duncan et al. 2018).
Origin and diversity of translational control by eIF2α-P: an overview
As shown in Fig. 2, the budding yeast S. cerevisiae belongs to one of the two major fungal phyla termed Ascomycota (most yeasts and filamentous fungi). Similar to S. cerevisiae and other members of the subphylum Saccharomycotina, Gcn2 is the sole eIF2α kinase and a Gcn4 homolog is its effector in the other major phylum Basidiomycota (mushrooms). However, some filamentous fungi encode both Hri and Gcn2, along with Gcn4 homologs (Fig. 2, row 8), so do primitive groups of fungi termed Mucoromycota and Taphrinomycotina, although Gcn4 homolog is found in only a subset of its members (Fig. 2, rows 2–5). Importantly, the mRNAs coding for the Gcn4 homologs in this kingdom have uORFs similar to those found in GCN4 or atf4 mRNAs (Fig. 1b). Thus, the common ancestor of this kingdom (hence that of Metazoa and Fungi) had an intermediate level of complexity with two eIF2α kinases, Gcn2 and Hri, as well as a Gcn4 homolog with uORFs as their effector in a primitive “integrated” response.
Conservation and diversity of translational control in fungi. Left diagram depicts a simplified fungal tree of life with a branch on top representing Metazoa. Divergence time is indicated at branch points (Hoffman et al. 2015; Taylor and Berbee 2006; Wang et al. 1999); My, million years ago; By, billion years ago. Columns 1–4 indicate the presence ( +) or absence ( −) of eIF2α kinases, Gcn2 and Hri, 5MP, or Gcn4/Atf4/Cpc1 homolog. ± , Gcn4 homolog is found in L. transversale, but only a handful of members belonging to the phylum Mucoromycota have this homolog. Partially adapted from Fig. 7A of Chikashige et al. (2020). Identity of the proteins present in the indicated species is described in the legends of this figure, except for L. transversale proteins (Gcn2, XP_021880655; Hri, XP_021881008; 5MP, XP_021884142; and Gcn4, XP_021881329)
The glimpse of fungal diversity as shown in Fig. 2 locates the fission yeast S. pombe near the root of fungal evolution as the member of the subphylum Taphrinomycotina. The antiquity of the members of this group was recently underscored by the discovery that its member, Saitoella complicata, encodes 5MP (Riley et al. 2016) (highlighted in red in Fig. 3). This is the sole 5MP identified in the phylum Ascomycota so far (Fig. 3) (Hiraishi et al. 2014). 5MP (also known as BZW or eIF5C) is an ancient translational regulator capable of interacting with eIF2 and other eIFs, thereby inhibiting translation (Singh et al. 2011). By physically inhibiting interactions involving eIF2, 5MP can delay re-initiation and thereby induce Atf4 translation in mammalian cells (Kozel et al. 2016). By binding the ribosome through its eIF partners, 5MP can also increase the accuracy of translation initiation (Tang et al. 2017). Thus, in the last eukaryotic common ancestor, translation was controlled by eIF2α kinases, Gcn2 and Hri, and the eIF2-binding regulator, 5MP.

Adapted from Fig. S10B of Chikashige et al. (2020)
5MP phylogenetic tree from diverse eukaryotes. The tree was generated by MAFFT version 7, with 100 bootstrap replicates on (https://mafft.cbrc.jp/) using sequences obtained from Genbank. Boxes to the right indicate the kingdoms to which the organisms of interest belong to. Bars indicate their subphyla and, for fungi, their classes. Red box, the sole Ascomycota homolog. Bootstrap values are indicated at the nodes.
How do some primitive fungal species accommodate the lack of Gcn4, the major control unit in the starvation response? Is translational control of specific mRNAs still important in these organisms? Despite the lack of Gcn4, S. pombe deploys a starvation response similar to the GAAC response in S. cerevisiae (Nemoto et al. 2010; Udagawa et al. 2008). Thus, the study on this yeast not only provides insights into cross-talk between the distinct eIF2α kinases, Gcn2 and Hri (Zhan et al. 2002, 2004), but also into the plasticity of regulatory networks that may operate at the level of transcription as well as translation.
Polysome profiling versus ribosome profiling
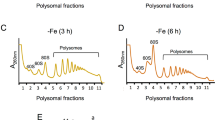
There are two fundamentally distinct methods of genome-wide translational profiling, polysome profiling and ribosome profiling (Ingolia et al. 2009; Piccirillo et al. 2014) (Fig. 4). In polysome profiling, cell lysates containing poly-ribosomes (polysomes) are fractionated by density gradient-velocity sedimentation, typically using a sucrose gradient and ultracentrifugation. Polysome fractions are collected, and the abundance of mRNA in each fraction is measured by DNA microarray hybridization or RNA sequencing (RNAseq) (Fig. 4a). Translational control is determined by the change in the number of ribosomes loaded per mRNA that is defined as translation efficiency (TE) of the mRNA (Chikashige et al. 2020).
Translational profiling methods. a Polysome profiling. Top indicates a typical A254 profile of polysomes. Bars indicate boundaries of fractions taken with their numbers indicated in-between. Numbers below the graph indicate the number of translating ribosomes bound to mRNAs in each fraction. Schematics below depict mRNA (brown line) loaded with different numbers of ribosomes (depicted as ovals as in Fig. 1a) found in each fraction. RNA from each fraction is quantified with DNA microarray (Microarray) or sequenced (RNAseq). The graph below shows the simulated ribosome mass in each fraction using the ribosome numbers as assigned above and mRNA abundance values obtained by microarray hybridization. b Ribosome profiling. The flowchart depicts the method of generation and sequencing of ribosome-protected mRNA fragments (RPF, short brown lines) from polysomes (the schematics on top, depicted as in panel A). Bottom graph presents an example of RPF read mapping and shows the protection patters in the 5′ half of fil1 mRNA adapted from Fig. S6A of Chikashige et al. (2020). Plots are color-coded by three reading frames presented on bottom. Schematics in the middle represent uORF structures, and the main CDS also color-coded similarly. Asterisk, the green peak located before uORF1 represents a possible additional uORF initiated by a CUG codon (Duncan et al. 2018)
In ribosome profiling, cell lysates are treated with RNases, and the ribosome-protected fragments (RPF) are collected for RNAseq after ribosome purification followed by the release and gel purification of the fragments (Fig. 4b). The read sequences are computationally mapped onto the transcriptome to determine the occupancy of ribosome in every transcript at a single nucleotide resolution. To determine TE, RNAseq of total RNA samples is additionally required. TE is then defined by the ratio of RPF sequencing reads to total RNAseq reads that were mapped to the gene of interest. The strong merit of this method is its ability to determine the precise location of coding regions through RPF distribution patterns (Fig. 4b, bottom).
Ribosome profiling of fission yeast reveals a new transcription factor regulated through uORFs
Duncan et al. employed ribosome profiling to study translational control of fission yeast during amino acid starvation (Duncan et al. 2018). By analyzing genes whose RPF distribution is changed upon the stress, they identified a noncanonical transcription factor of GATA zinc-finger type regulated depending on gcn2+. Subsequent knock-out studies indicate that the gene they named fil1 (for gcn Four-Induction Like) was shown to be responsible for at least a part of the GAAC response in S. pombe; fil1 deletion reduced expression of 15% of the starvation-induced genes including those encoding amino acid biosynthesis enzymes. Consistently, chromatin immunoprecipitation sequencing (Chip seq) analyses showed that Fil1 binds 10% of the starvation-induced genes. Importantly, significant amounts of RPF reads were mapped onto its 5′ leader region, in support of translation of four of its five uORFs (Fig. 1c, 4b, bottom). Starvation did not decrease the ribosome protection (hence translation) of uORF1, but decreased that of uORF4, and instead increased that of the fil1 CDS, as expected for the model that uORF1 and uORF4 serve as the positive and negative elements analogous to uORF1 and uORF4, respectively, of S. cerevisiae GCN4 (Duncan et al. 2018).
Accordingly, it was concluded that the ancestor of the S. pombe lineage was able to acquire an unrelated transcription factor of zinc-finger type in place of a bZIP (Gcn4 homolog), yet organizing orthologous genes under its control. This shows the plasticity of regulatory networks related to the starvation response during the course of evolution.
Polysome profiling of fission yeast provides an overview of genome-wide translational control during amino acid starvation
Since Fil1 controls only a subset of GAAC genes, are there other genes bearing uORFs or even distinct nucleotide motifs responsible for GAAC induction? To address this, we performed polysome profiling of fission yeast treated similarly with amino acid starvation (Chikashige et al. 2020). By performing polysome profiling followed by microarray hybridization of all seven gradient fractions, ~ 2000 genes were found to be translationally up-regulated in response to starvation in a Gcn2-dependent manner. We found that these genes are regulated by functional groups. During the stress, mRNAs encoding chromatin components and RNA regulation are preferentially translated, and yet, those encoding ribosomal proteins are modestly depleted of ribosomes, in agreement with the cellular needs of transcriptional regulation (GAAC response) and the slower rate of protein biosynthesis by ribosomes during starvation. Importantly, these functional groups are not the well-known Gcn4 or Fil1 targets (Duncan et al. 2018; Natarajan et al. 2001).
The ~ 2000 translationally regulated genes include mRNAs with evidence for uORF translation, including gcn5 mRNA (Fig. 1c). gcn5 encodes a histone acetyl transferase involved in the starvation response in both S. cerevisiae and S. pombe (Udagawa et al. 2008). Our re-analysis of ribosome profiling data by Duncan et al. indicated that the starvation represses translation of uORF3 relative to that of the gcn5 CDS (Chikashige et al., 2020), suggesting that uORF3 serves as the negative element for gcn5 induction. Further analysis using luciferase reporter constructs showed that uORF1 serves as the positive element (Chikashige et al., 2020). Moreover, the re-analysis listed many other mRNAs with the uORF–CDS translation ratio decreased upon the stress, suggesting that the CDS translation is induced by alleviating the inhibitory effect of the uORF, similar to Gcn4 or Atf1. These mRNAs include those encoding putative transcription factors, Prt1, Prz1, and SPCC777.02, which are also included in the list of translationally controlled genes in this work. Some of the listed uORFs, including that of prt1, are the sole uORF of the mRNA, suggestive of a mechanism distinct from the paired uORF system as found in gcn5 or fil1 (Hood et al. 2009). Interestingly, the uORF regulation of Gcn5 (Chikashige et al. 2020) or direct Fil1 homolog itself is not found outside of Taphrinomycotina (Todd et al. 2014). Thus, the lack of Gcn4 homolog in the ancestor of the S. pombe lineage appears to have brought an opportunity to invent uORF-mediated regulation of several transcription factors including Gcn5 and Fil1.
An unexpected finding of this work was that mRNA with introns are better translated during the starvation dependent on Gcn2 (Chikashige et al. 2020). Introns are known to prevent genotoxic DNA:RNA hybrids (a.k.a. R-loops) generated during DNA transcription (Bonnet et al. 2017). In agreement, the work confirmed that mRNAs with three or more introns are expressed better during the stress, which would likely contribute to preventing R-loop formation in the time of insult. However, the effect of the stress on translation of intron-containing mRNA was even more dramatic, as it was seen with mRNAs with one or more introns. Given the recent discovery that introns protect budding yeast from starvation (Parenteau et al. 2019), the work forms the basis for further studies on relationship among splicing, translation, and stress response.
Conservation of uORF-dependent control of Hri translation across diverse fungi
Our work also revealed an unexpected cross-talk between Gcn2 and Hri through translational control (Chikashige et al. 2020). As shown in Fig. 1d, the 5′ leader region of S. pombe Hri2, one of the two Hri homologs in S. pombe (Zhan et al. 2002), carries four uORFs. Hri2 was a part of ~ 2000 genes regulated during starvation at the translational level. We recapitulated Gcn2-dependent Hri2 translational regulation using a luciferase reporter bearing the same uORFs as found in its leader region and identified through a mutational approach uORF3 as the positive element allowing downstream re-initiation and uORF4 as the negative element inhibiting the CDS translation (yellow and blue boxes in Fig. 1d) (Chikashige et al. 2020). Of note is the uORF3 (AUG-AUC) encoding a dipeptide MI. Our re-analysis of ribosome profiling data showed that, among various determinants of sequence motifs to allow re-initiation, pyrimidine-rich codons are enriched at the last and second to last codons of the uORFs, which display a low uORF/CDS translation ratio suggestive of high rate of re-initiation (Chikashige et al. 2020). This trend agrees with a previous mutational analysis of GCN4 uORF1 (Grant and Hinnebusch 1994) and helped to narrow down the candidates of positively acting uORFs in other systems (Chikashige et al. 2020) (also see yellow boxes in Fig. 1 representing the positive uORF element candidates).
More importantly, the MI motif, along with other uORF arrangements similar to the one found in S. pombe hri2 mRNA, is present in the leader region of A. nidulans hriA mRNA (Fig. 1d). This conservation is impressive, considering the large divergence between S. pombe and A. nidulans during fungal evolution (Fig. 2). Other Aspergillus species also carry uORF-bearing hri mRNA, as far as its leader regions are annotated (Chikashige et al. 2020). These facts strongly suggest that the coupling of the dual eIF2α kinases, Gcn2 and Hri, through uORF-dependent regulation of the latter is a conserved strategy found across diverse fungi. Since eIF2α-P was observed in fission yeast deleted for gcn2 during the starvation, but not in the mutant deleted additionally for hri1 and hri2, it is conceivable that Hri is involved in eIF2α-P during this response (Udagawa et al. 2008). Hri is proposed to be activated by oxidative stress associated with metabolic perturbation during the stress, and thereby enhance eIF2α phosphorylation (Nemoto et al. 2010) (Fig. 5a).

adapted from Molecular Biology of the Gene, 7th edition. Right, translational regulatory motifs mediated by uORF-dependent control. In (B), Coherent FFL with AND node makes a persistent detector that only responds to a long-lived signal (Alon, 2007). However, Coherent FFL with OR node works differently, as described in the text
Translational control by eIF2α phosphorylation. a Amino acid starvation pathway in S. pombe. Oxi, oxidative stress. Genes on the bottom are the targets of eIF2α-P. Toolkit (cis), the regulatory motifs used for translational control (arrow, positive; stopped bar, negative). Question mark on Gcn2 refers to evidence suggesting that oxidative stress activates Gcn2 (Anda et al. 2017). Question mark on M-Stop refers to its possible involvement in regulation of rps/rpl translation. b and c Translational regulatory circuits discussed in this review. Left, original definition by Alon (2007),
What is the implication of the control of Hri translation by Gcn2? In systems biology, the circuit made of Gcn2, Hri, and eIF2α-P makes the coherent feed forward loop (FFL) with OR logic (Fig. 5b) (Alon 2007). This switch allows continued production in the face of a transient loss of the input signal. In the context of amino acid starvation, it is interpreted that this unit allows continued eIF2α-P production, even if the starvation is resolved. As much as the oxidative stress input is present, eIF2α-P would continue to be produced. Martin et al. showed that fission yeast with both Hri1/2 and Gcn2, under nitrogen starvation, show an immediate on and a delayed off response, supporting the coherent FFL |OR| switch (Martín et al. 2013). In mammals, uORF-dependent translational activation (Lee et al. 2009; Novoa et al. 2001; Young et al. 2015) of GADD34, a regulatory subunit of eIF2α phosphatase (Choy et al. 2015) makes up the incoherent FFL (Alon 2007), that is supposed to generate a pulse of eIF2α phosphorylation soon after stress activates an eIF2α kinase (Fig. 5c). Thus, uORF-mediated regulation can be used to generate a unit of regulatory circuits.
Importantly, the aforementioned work of Martin et al. also showed that, when only gcn2 is present (in the case of hri1/2 mutant bearing no coherent FFL), there seems to be a quick pulse of eIF2α-P within 30 min which also disappears within the next 30 min (Martín et al. 2013). This result suggests an intriguing possibility of a mammalian-type incoherent FLL |AND| switch, which may also involve eIF2α dephosphorylation in the context of uORF-mediated translational regulation. Along with this work, the recent studies, therefore, highlight the conservation of certain modules between yeast, fungi, and mammals.
Other cis-regulatory motifs identified in polysome profiling studies of fission yeast amino acid starvation response
Through motif-enrichment analyses of 5′ UTR of genes regulated during amino acid starvation, we also identified the UGA(C/G)GG-like motif involved in Gcn2-mediated regulation independent of uORFs (Chikashige et al. 2020). The requirement for this motif in Gcn2-dependent translational control was verified by a reporter assay and subsequent deletion studies using the 5′ UTR of hrd1 carrying this motif. This motif is potentially found in 98 genes (~ 2%) of the fission yeast genome and is similar to but distinct from TGACGT motif defined as Atf1/Pcr1 transcription factor heterodimer binding site (Kato et al. 2013; Steiner and Smith 2005): Atf1 and Pcr1 enhance transcription of genes involved in core environmental stress response (CESR) mediated by the Sty1/Spc1 MAP kinase (Chen et al. 2003). Interestingly, the translational control of hrd1 depended on intact atf1 or pcr1 (Chikashige et al. 2020), suggesting that the starvation signal is integrated with the Sty1/Spc1-mediated response at the translational level. In agreement with this scenario, a strong amino acid starvation signal directly induces Sty1/Spc1 signaling in parallel with Gcn2 signaling that is activated through uncharged tRNAs (Udagawa et al. 2008).
Another motif identified in our recent work is the mono-peptide-coding uORF or M-Stop. This motif tends to inhibit downstream CDS translation more strongly after the stress (Chikashige et al. 2020). In contrast to the other modes of regulation discussed here, this regulation is independent of Gcn2. Since the translation of M-Stop does not involve the elongation phase, the regulation must either involve differential rates of initiation or termination (peptide release). The likely mechanism involving the former would postulate that the starvation differentially impacts translation initiation of M-Stop mRNA with poor or strong initiation contexts [analogous to the Kozak context of mammalian start codons (Asano 2014)]. However, our luciferase reporter assays indicate that the starvation does not alter the rate of translation from non-AUG codons (unpublished observations), whose alteration correlates globally with the change in the rate of translation from AUG codons under poor initiation contexts (Zhou et al. 2020). If so, this leaves us with the model that translation of M-Stop is controlled by modulating termination activity. In this scenario, starvation induces ribosome stalling at M-Stop coding region through the inhibition of release factor activities that might associate with reduced protein synthesis during the stress; the ribosome stalling would then inhibit translation of downstream CDS. Regardless of the mechanism, we propose that M-Stop provides the toolkit for negative regulation of mRNA translation in response to amino acid starvation, as observed, for example, for ribosomal protein mRNAs (Chikashige et al. 2020).
Conclusions and perspectives
The recent translational profiling studies of fission yeast starvation response conveyed two important messages related to the evolution of translational control by eIF2α phosphorylation (Chikashige et al. 2020; Duncan et al. 2018). First, the loss of Gcn4 was easily accommodated by acquiring additional, unrelated transcription factors (Fil1 and Gcn5) as response effectors, which somewhat work cooperatively. Second, they provided the first glimpse of how the two distinct eIF2α kinases, Gcn2 and Hri, cooperate in a relatively primitive metabolic regulatory system. In both the cases, uORF presented the flexible toolkits of cis-acting regulatory response elements. Essentially, the generation of this toolkit requires mere base substitutions to make a new start codon and a very broad consensus around the stop codon of positive uORF elements, taking advantage of a relatively long 5′-UTR characteristic of eukaryotic genomes.
This flexibility is in good contrast to uORFs found in prokaryotes, as it is often integrated with mRNA structures or peptides encoded (Asano et al. 1991; Yanofsky 2000). If it serves to induce translation, it must allow a second ribosome to be recruited to the site downstream of the uORF start codon through its specific structure (such as pseudoknot) (Asano and Mizobuchi 1998). Thus, for this system to serve as a toolkit, the whole region of the uORF must be transferred by recombination, and when it is observed to happen, the complexity of original regulation is sometimes lost, or substituted with another means such as transcriptional control (Asano and Mizobuchi 2000).
The work also uncovered a new use of the shortest uORF (M-Stop) or a nucleotide motif in translational regulation during the stress (Fig. 5a). The UGA(C/G)GG motif identified is similar to the transcription factor (Atf1/Pcr1) binding site (TGACGT) in the DNA sequence. Again, the acquisition of this motif apparently benefitted from the relatively long 5′-UTR often embedding transcriptional control signals transcribed as its part. Lastly, however, not discussed in the present work is the involvement of non-AUG start codons in eukaryotic translational control (Asano 2014; Kearse and Wilusz 2017). Base substitutions in 5′-UTR can readily generate near-cognate start codons that can start a new ORF. Non-AUG translation from these codons is usually weak and, therefore, allows downstream initiation of the same or distinct reading frames by leaky scanning. This adds an N-terminal peptide to an existing protein altering its cytoplasmic localization (Asano 2014) or allows polycistronic translation of viral mRNAs (Ogden et al. 2019) and of a human mRNA as recently reported (Loughran et al. 2020). In human cancer, 5MP appears to regulate the choice of start codon between an AUG codon and a CUG codon, altering the oncogenic property of the c-Myc transcription factor (Sato et al. 2019). With 5MP’s ability to cause delayed re-initiation and increase the initiation accuracy, it would be intriguing to investigate its basic eukaryotic role in, for example, basidiomycete fungal model Cryptococcus neoformans or even S. complicata (Fig. 3).
In conclusion, the comprehensive understanding of fission yeast starvation response uncovered an evolutionary flexibility in the integration of translational regulatory networks utilizing common cis-regulatory toolkits apt for eukaryotic initiation mechanisms. I believe that the common ancestor of the animals (the phylum Metazoa) also took advantage of a similar flexibility to begin to evolve their appreciated complexity.
References
Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461
Anda S, Zach R, Grallert B (2017) Activation of Gcn2 in response to different stresses. PLoS ONE 12:e0182143
Asano K (2013) Translational Control. In: Dubitzky W, Wolkenhauser O, Cho K-H, Yokota H (eds) Encyclopedia of Systems Biology. Springer, New York, pp 2278–2282
Asano K (2014) Why is start codon selection so precise in eukaryotes? Translation 2:e28387
Asano K, Mizobuchi K (1998) An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIa plasmid ColIb-P9. J Biol Chem 273:11815–11825
Asano K, Mizobuchi K (2000) Structural analysis of late intermediate complex formed between plasmid ColIb-P9 Inc RNA and its target RNA: how does a single antisense RNA repress translation of two genes at different rates? J Biol Chem 275:1269–1274
Asano K, Moriwaki H, Mizobuchi K (1991) An induced mRNA secondary structure enhances repZ translation in plasmid ColIb-P9. J Biol Chem 266:24549–24556
Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Géli V, de Almeida SF, Palancade B (2017) Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol Cell 67:608-621.e606
Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J (2003) Global transcriptional response of fission yeast to environmental stress. Mol Biol Cell 14:214–229
Chikashige Y, Kato H, Thornton M, Pepper W, Hilgers M, Cecil A, Asano I, Yamada H, Mori C, Brunkow C et al (2020) Gcn2 eIF2α kinase mediates combinatorial translational regulation through nucleotide motifs and uORFs in target mRNAs. Nucleic Acids Res 48:8977–8992
Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, Peti W (2015) Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep 11:1885–1891
Dever TE (2002) Gene-specific regulation by general translation factors. Cell 108:545–556
Duncan CDS, Rodríguez-López M, Ruis P, Bähler J, Mata J (2018) General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc Natl Acad Sci USA 115:E1829–E1838
Grant CM, Hinnebusch AG (1994) Effect of sequence context at stop codons on efficiency of reinitiation in }U}GCN4}u} translational control. Mol Cell Biol 14:606–618
Hinnebusch AG (1997) Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem 272:21661–21664
Hinnebusch AG, Dever TE, Asano K (2007) Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In: Mathews MB, Sonenberg N, Hershey JWB (eds) Translational Control in Biology and Medicine. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY, pp 225–268
Hiraishi H, Oatman J, Haller S, Blunk L, McGivern B, Morris J, Papadopoulos E, Guttierrez W, Gordon M, Bokhari W et al (2014) Essential role of eIF5-mimic protein in animal development is linked to control of ATF4 expression. Nucl Acids Res 42:10321–10330
Hoffman CS, Wood V, Fantes PA (2015) An ancient yeast for young geneticists: a primer on the Schizosaccharomyces pombe model system. Genetics 201:403–423
Hood HM, Neafsey DE, Galagan J, Sachs MS (2009) Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol 63:385–409
Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223
Kato H, Kira S, Kawamukai M (2013) The transcription factors Atf1 and Pcr1 are essential for transcriptional induction of the extracellular maltase Agl1 in fission yeast. PLoS ONE 8:e80572
Kearse MG, Wilusz JE (2017) Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31:1717–1731
Kozel C, Thompson B, Hustak S, Moore C, Nakashima A, Singh CR, Reid M, Cox C, Papadopoulos E, Luna RE et al (2016) Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation. Nucl Acids Res 44:8704–8713
Lee YY, Cevallos RC, Jan E (2009) An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J Biol Chem 284:6661–6673
Loughran G, Zhdanov AV, Mikhaylova MS, Rozov FN, Datskevich PN, Kovalchuk SI, Serebryakova MV, Kiniry SJ, Michel AM, O’Connor PBF et al (2020) Unusually efficient CUG initiation of an overlapping reading frame in POLG mRNA yields novel protein POLGARF. Proc Natl Acad Sci USA 117:24936–24946
Martín R, Berlanga JJ, de Haro C (2013) New roles of the fission yeast eIF2α kinases Hri1 and Gcn2 in response to nutritional stress. J Cell Sci 126:3010–3020
Mueller PP, Hinnebusch AG (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45:201–207
Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368
Nemoto N, Udagawa T, Ohira T, Jiang L, Hirota K, Wilkinson CRM, Bähler J, Jones N, Ohta K, Wek RC et al (2010) The roles of stress-activated Sty1 and Gcn2 kinases and protooncoprotein homologue Int6/eIF3e in responses to endogenous oxidative stress during histidine starvation. J Mol Biol 404:183–201
Novoa I, Zeng H, Harding HP, Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153:1011–1022
Ogden PJ, Kelsic ED, Sinai S, Church GM (2019) Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 366:1139–1143
Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S (2019) Introns are mediators of cell response to starvation. Nature 565:612–617
Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O (2014) Translational control of immune responses: from transcripts to translatomes. Nat Immunol 15:503–511
Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH et al (2016) Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci USA 113:9882–9887
Ron D, Harding HP (2007) eIF2alpha phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB (eds) Translational Control in Biology and Medicine. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY, pp 345–386
Sato K, Masuda T, Hu Q, Tobo T, Gillaspie S, Niida A, Thornton M, Kuroda Y, Eguchi H, Nakagawa T et al (2019) Novel oncogene 5MP1 reprograms c-Myc translation initiation to drive malignant phenotypes in colorectal cancer. EBioMed 44:387–402
Singh CR, Watanabe R, Zhou D, Jennings MD, Fukao A, Lee B-J, Ikeda Y, Chiorini JA, Fujiwara T, Pavitt GD et al (2011) Mechanisms of translational regulation by a human eIF5-mimic protein. Nucl Acids Res 39:8314–8328
Steiner WW, Smith GR (2005) Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169:1973–1983
Tang L, Morris J, Wan J, Moore C, Gillaspie S, Aube E, Fujita Y, Nanda J, Anderson A, Cox C et al (2017) Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide. Nucl Acids Res 45:11941–11953
Taylor JW, Berbee ML (2006) Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98:838–849
Todd RB, Zhou M, Ohm RA, Leeggangers HA, Visser L, de Vries RP (2014) Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics 15:214
Udagawa T, Nemoto N, Wilkinson C, Narashimhan J, Watt S, Jiang L, Zook A, Jones N, Wek RC, Bähler J et al (2008) Int6/eIF3e promotes general translation and Atf1 abundance to modulate Sty1 MAP kinase-dependent stree response in fission yeast. J Biol Chem 283:22063–22075
Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274
Wang DY, Kumar S, Hedges SB (1999) Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc Biol Sci 266:163–171
Wek RC, Staschke KA (2010) How do tumors adapt to nutrient stress? EMBO J 29:1946–1947
Yanofsky C (2000) Transcription attenuation: once viewed as a novel regulatory strategy. J Bacteriol 182:1–8
Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis D, Bobrovnikova-Marjon E, Alan Diehl J, Ron D, Koumenis C (2010) The GCN2-ATF4 pathway is critical for tumor cell survival and proliferation in response to nutrient deprivation. EMBO J 29:2082–2094
Young SK, Wek RC (2016) Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291:16927–16935
Young SK, Willy JA, Wu C, Sachs MS, Wek RC (2015) Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J Biol Chem 290:28257–28271
Zhan K, Vattem KM, Bauer BN, Dever TE, Chen J-J, Wek RC (2002) Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stress. Mol Cell Biol 22:7134–7146
Zhan K, Narasimhan J, Wek RC (2004) Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics 168:1867–1875
Zhou F, Zhang H, Kulkarni SD, Lorsch JR, Hinnebusch AG (2020) eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide. RNA 26:419–438
Acknowledgements
I thank Hiroaki Kato (School of Medicine, Shimane University), Yuji Chikashige (National Institute of Information and Communications Technology, Kobe), Shintaro Iwasaki (RIKEN, Wako), and Richard Todd (Department of Plant Pathology, Kansas State University) for helpful discussion.
Funding
K-INBRE program Pilot Grant, National Institutes of Health [P20 GM103418]; National Institutes of Health R15 grant [GM125671]; National Science Foundation Research Grant [1412250]; JSPS KAKENHI [18K19963]; Kansas State University (KSU) Terry Johnson Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Asano, K. Origin of translational control by eIF2α phosphorylation: insights from genome-wide translational profiling studies in fission yeast. Curr Genet 67, 359–368 (2021). https://doi.org/10.1007/s00294-020-01149-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01149-w