Abstract
In fungi, O-mannosylation is one type of conserved protein modifications that add the carbohydrate residues to specific residues of target proteins by protein O-mannosyltransferases. Previously, three members of O-mannosyltransferases were identified in Magnaporthe oryzae, with MoPmt2 playing important roles in fungal growth and pathogenicity. However, the biological roles of the rest Pmt proteins remain unclear. In this study, to understand if O-mannosyltransferases are crucial for fungal pathogenicity of M. oryzae, the Pmt-coding genes MoPmt1 and MoPmt4 were separately disrupted and their roles in pathogenesis were analyzed. Of the two genes, only MoPmt4 is specifically required for full virulence of M. oryzae. Deletion of MoPmt4 resulted in defects on radial growth, with more branching hyphae and septa as compared to Guy11. The MoPmt4 mutant was severely impaired not only in conidiation, but also in both penetration and biotrophic invasion in susceptible rice plants. This mutant also had defects in suppression of host-derived ROS-mediated plant defense responses that might be ascribed from the reduced activities of extracellular enzymes. Furthermore, like their fungi counterparts, MoPmt4 localized in the ER and had O-mannosyltransferase activity. Domain disruption analysis indicated that mannosyltransferase activity regulated by PMT domain of MoPmt4 is crucial for fungal development and pathogenicity of M. oryzae. Taken together, these data suggest that MoPmt4 is a protein O-mannosyltransferase essential for fungal development and full virulence of M. oryzae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ascomycete fungus Magnaporthe oryzae is one of the most significant fungal pathogens for the cultivated rice plants and is responsible for annual losses of up to 30% of the worldwide rice harvest, which otherwise would feed more than 60 million people (Talbot 2003; Dean et al. 2012). The rice blast pathosystem has been emerged as an ideal model to study fungus–plant interactions due to its economic importance and genetic tractability of both the fungus and host (Ebbole 2007). Like many other phytopathogens, the penetration of host plant cuticle, an important physical barrier against plant pathogens, is the initially critical step required for disease development of M. oryzae (Matar et al. 2017; Wang et al. 2016). To this end, the blast fungus has developed a specialized cell called an appressorium (Howard and Valent 1996), which could use mechanisms including osmotic pressure, hydrolytic enzymes, and the degradation of reactive oxygen species (ROS) to break into host plant tissue (Howard et al. 1991; Egan et al. 2007; Guo et al. 2011, 2017). During plant infection, sufficient turgor pressure (> 8 MPa) accumulated in mature appressorium is vital for the penetration of host cell barriers by M. oryzae (Talbot 2003; Ebbole 2007), and an intact fungal cell wall guarantees the proper function of appressorium during plant infection (Jeon et al. 2008; Guo et al. 2015, 2016a). In M. oryzae, disruption of genes like Mck1, MoGrr1, Mosec22, and MoSOM1 could result in defects of cell wall integrity (CWI), which, as a result, lead to failure of accumulation of sufficient turgor pressure and the loss of pathogenicity (Jeon et al. 2008; Song et al. 2010; Yan et al. 2011; Guo et al. 2015). In addition, extracellular enzymes like peroxides help scavenge of ROS at primary infection sites by depressing the plant immunity, and thereafter promote the infection of M. oryzae (Chi et al. 2009; Guo et al. 2010). Hence, identification of novel genes involved in cell wall development, enzymes secretion, and appressorium-mediated penetration would accelerate the understanding of molecular mechanisms of fungal pathogenicity, and provide clues to explore the potential of those proteins as a novel target for antifungal strategies in M. oryzae.
Glycosylation is one type of highly conserved protein modifications that involved the addition of carbohydrate residues to specific residues of target proteins, and is essential for determining the proper function of a newly synthesized protein (Willer et al. 2003). In eukaryotic cells, the N- and O-glycosylation are two most frequent glycosylation occurring within the endoplasmic reticulum (ER) and Golgi apparatus to introduce carbohydrate moieties to different amino acid residues (Strahl-Bolsinger et al. 1999). O-mannosylation is a type of O-glycosylation catalyzed by protein mannose transferases (PMTs), which perform the initial O-mannosyltransferase reaction and add the first mannose to Ser or Thr residues in the nascent protein chain at ER (Loibl and Strahl 2013). Thereafter, the O-mannosylated proteins have been moved into the Golgi apparatus for additional glycosylation, which continue to add more mannoses to target proteins. Over the last decades, three subfamilies of PMTs (Pmt1, Pmt2, and Pmt4) were identified with diverse roles in fungi, including in CWI, cell morphology, and the stability, sorting, and secretion of proteins (Strahl-Bolsinger et al. 1999; Lommel and Strahl 2009; Mouyna et al. 2010; Harries et al. 2015; Guo et al. 2016a). In filamentous fungi, only one member for each PMT subfamily has been identified, and the requirement for pmt genes in pathogenesis is variable and depends on the specific pathosystem (Goto et al. 2009; Fernandez-Alvarez et al. 2009; Mouyna et al. 2010; Gonzalez et al. 2013; Harries et al. 2015; Guo et al. 2016a). In Botrytis cinerea, the three pmt genes are essential for growth and pathogenesis of different plants (Gonzalez et al. 2013). In contrast, the pmt genes of Aspergillus fumigatus differentially affect growth, morphogenesis, and viability, but none of them are involved in pathogenicity (Mouyna et al. 2010). In Ustilago maydis, pmt4 is the unique PMT protein involved in fungal virulence and its disruption affected appressorium-mediated penetration and virulence in maize (Fernandez-Alvarez et al. 2009).
In M. oryzae, three Pmt genes were identified in our previous study, with MoPmt2, which was grouped into Pmt2 subfamily, playing important roles in fungal development and pathogenicity to rice plant (Guo et al. 2016a). However, the biological function of the other Pmt proteins remains unclear; to this end, we individually deleted the two Pmt genes (MoPmt1, MGG_02954; MoPmt4, MGG_04427) in M. oryzae, and found that the Pmt4 subfamily member MoPmt4 is essential for growth, morphogenesis, and pathogenicity of rice blast fungus.
Materials and methods
Fungal strains and culture conditions
Magnaporthe oryzae strain Guy11 was used as wild-type strain and recipient for genetic modifications throughout this research. This strain and its transformants were cultured on RDC (Guo et al. 2011) at 28 °C under continuous fluorescent light for promoting conidiation and were cultured in liquid complete medium (Talbot 2003) at 25 °C for 48 h with agitation (150 rpm) for genomic DNA and total RNA extraction. To observe vegetative mycelial growth under stress, Congo Red (CR, 200 µg/mL) or Calcofluor White (CFW, 150 µg/mL) was individually added to CM agar medium (Guo et al. 2015). A mycelial disc (6 × 6 mm) of each strain was inoculated upside down on these stress media, and the growth rate was assessed by measuring culture diameters every 5th day. The inhibition rate was calculated following previously established methods (Guo et al. 2015). Conidia germination and appressoria development was performed by placing drops of conidial suspension on hydrophobic coverslip for 2–24 h. Conidiophore development and conidiation observation were performed as previously described (Guo et al. 2017).
Nucleic acid manipulation, qRT-PCR, and southern blotting
The standard molecular biology techniques, including preparation of plasmid DNA, restriction enzyme digestion, cloning, and DNA gel blot hybridization analysis, were performed as previously described (Sambrook and Russell 2001). Genomic DNA and total RNA were extracted following previously established methods (Guo et al. 2017). For the PCR screening of the transformants, genomic DNA was extracted by a quick and easy DNA extraction method (Jeon et al. 2008). Quantitative RT-PCR (qRT-PCR) was performed as described by Guo et al. (Guo et al. 2016a). To confirm the deletion of MoPmt4, DNA probes used for hybridization are amplified by primers Pmt4-2R/Pmt4-RF-1 and HPH-F/HPH-R (Table S1), respectively, and then labeled with digoxigenin-11-dUTP according to manufacturer instructions (11745832910, Roche, China). Southern hybridization procedures were performed following previously established methods (Guo et al. 2016a). The same strategy described above was also used to confirm the deletion of MoPmt1 in M. oryzae. Primers used in this section are listed in Table S1.
Gene replacement and complementation
According to the sequences at the locus MGG_04427 in the M. oryzae genome (http://fungidb.org/fungidb/), the 0.8-kb DNA fragments flanking the 5′ and 3′ regions of the gene were amplified using primers Pmt4-1F/Pmt4-1R and Pmt4-2F/Pmt4-2R, respectively, from genomic DNA of Guy11. They were then jointed together by overlap PCR with primers Pmt4-1F/Pmt4-2R to construct plasmid pMDT-MoPmt4. The 1.4-kb HPH marker cassette (Carroll et al. 1994) that amplified with primers HPH-F/HPH-R was inserted into the EcoRV site in plasmid pMDT-MoPmt4 to generate deletion construct pMDT-MoPmt4-HPH. A ~ 2.9-kb DNA fragment amplified with primers Pmt4-1F/Pmt4-2R from deletion construct was transformed into protoplasts of wild-type Guy11 as previously described (Guo et al. 2017). Transformants were selected on medium supplemented with 200-ppm hygromycin and thereafter screened by PCR using primer pairs Pmt4-P1/P2 and P3/Pmt4-P4, respectively. Transformants carrying the expected null mutation were further confirmed by Southern blot and RT-PCR. For complementation, 3.9-kb DNA fragments containing full-length of MoPmt4 gene (without stop codon) and 0.72 kb DNA fragments containing eGFP-coding sequence were amplified from Guy11 genomic DNA and pMoC-eGFP vector, using the primers Pmt4-pro/Pmt4C-R and eGFP-F/eGFP-R, respectively, and then cloned into BamHI digested pMoC-eGFP vector (Guo et al. 2016b) using yeast gap repair approach. The resulting plasmid pMoC::MoPmt4-eGFP was introduced to Agrobacterium tumefaciens strain EHA105, and ATMT of MoPmt4-4 mutants was performed as previously described (Guo et al. 2016b). Resulting transformants were selected on half CM agar medium amended with 50-µg mL−1 carboxin and the confirmed by PCR and RT-PCR. The same strategy was used to generate the pMDT-MoPmt1-HPH vector and the expected null mutation of MoPmt1 was further confirmed following the above methods. Primers used in this section are listed in Table S1.
Cellular localization of MoPmt4 in M. oryzae
To observe the subcellular localization of MoPmt4, we fused ER retention signal HEEL with a red fluorescent protein (RFP) based on the established vector described by Yi et al. (2009) using yeast gap repair approach, and then transformed it (pYF11-ProLHS1-LHS1SP-RFP-HEEL) into complemental strain, at which the MoPmt4 protein was labeled with a GFP tag. Green and red fluorescence of conidia, appressorium, vegetative, and infectious hypha were examined under confocal fluorescence microscope (Zeiss LSM710, 63× oil). Primers used in this section are listed in Table S1.
Generation of the MoPmt4 ΔPMT-GFP and MoPmt4 ΔMIR-GFP constructs
To generate the MoPmt4ΔPMT-GFP construct, PCR products containing the native promoter of MoPmt4 were amplified with primers Pmt4-pro/PMT-1 and PMT-2/Pmt4C-R (Table S1), and introduced by co-transformation with fragments amplified with primers eGFP-F/eGFP-R and Pmt4-T1/Pmt4-T2 (Table S1) into the yeast strain FY834 with BamHI digested vector pMoC-eGFP (Guo et al. 2016b). Plasmid pMoC-MoPmt4ΔPMT-eGFP was rescued from the resulting Ura + yeast transformants. The same strategy was used to generate the pMoC-MoPmt4ΔMIR-eGFP vector, PCR products were amplified with primers Pmt4-pro/MIR-1, MIR-2/Pmt4C-R, eGFP-F/eGFP-R, and Pmt4-T1/Pmt4-T2 (Table S1), respectively. The corresponding transformants were validated by primers Pmt4-qRT1/Pmt4-qRT2 and Pmt4-RT1/Pmt4-RT2 (Table S1), respectively.
Phenotype assays
For growth assay, mycelia plugs of Guy11 and its derivative mutants were placed onto fresh CM, minimal agar medium (MM), V8 agar medium (V8), and RDC agar medium (Guo et al. 2017), respectively, and then incubated in dark at 28 °C for 5 days. To evaluate sporulation, conidia, which were harvested from RDC agar plates, were counted using a hemacytometer, and size was measured using a microscope following the described methods (Guo et al. 2016a). Conidial germination and appressorium formation, appressorium turgor, were determined by previously established methods (Guo et al. 2017). The evaluation of porosity of appressorial wall was performed using a solute exclusion technique and screened for cytorrhysis and plasmolysis (Jeon et al. 2008). The assay for protoplast release was carried out following the methods described by Guo et al. (2015).
Pathogenicity assay
For plant pathogenicity assays, equal volume of conidial suspensions (1 × 105 conidia mL−1) of Guy11 and its derivative mutants were incubated to 14-day-old rice seedlings (Oryza sativa cv CO-39) and 7-day-old detached barleys (Hordeum vulgare cv Golden Promise), respectively. The development disease lesions were examined at 5 days after inoculation. To test pathogenicity on abraded rice leaves, drops of conidia suspension (20 µL, 1.0 × 105 conidia mL−1) of Guy11 and its derivative mutants were placed on wounded sites of the rice leaves, and virulence was evaluated at 5 dpi. For plant penetration assays, drops of conidia suspension (20 µL, 1.0 × 104 conidia mL−1) of Guy11 and its derivative mutants were incubated to 7-day-old detached barleys in darkness. At 32 or 48 hpi, plant penetration and IH in host cells were observed using a light microscopy.
Host-derived ROS examination
ROS was detected by staining with DAB (Sigma-Aldrich) following the conidia incubating rice sheaths 48 h. DPI (0.4 µM) was mixed with the conidial suspensions to suppress host-derived ROS on detached barleys. The detailed procedures were described (Guo et al. 2017).
Transmission electron microscopy
Mycelia used for transmission electron microscopy (TEM) were grown in liquid CM for 48 h and fixed in 2.5% (v/v) glutaraldehyde and 1% (v/v) osmium tetroxide. Sections were prepared and visualized using an H-7650 transmission electron microscope (Hitachi, Tokyo, Japan) as previously described (Xu et al. 2010).
Complementation of the S. pombe Ogm4 mutant
The open-reading frames of MoPmt4 gene from M. oryzae were amplified with primers Pmt4-YC1/Pmt4-YC2 (Table S1) by PCR using Guy11 cDNA as template. The fragments were then cloned into the expression vector pREP41X, obtaining the plasmids pREP41X-MoPmt4. The resulting plasmid was transformed into S. pombe Ogm4 cells and grown on EMM lacking Leu. As a control, the wild-type strain and Ogm4 mutant were transformed with the empty pREP41X vector, respectively. Cell growth was compared after all strains were individually grown at 28 °C in EMM liquid media supplemented with adenine, histidine, and uracil to exponential phase.
Results
Deletion of MoPmt1 and MoPmt4 genes in M. oryzae
In M. oryzae, three conserved PMT proteins classified into three subfamilies were identified in the previous study, and the PMT2 subfamily member MoPmt2 played important roles in fungal morphogenesis, CWI, and full virulence to rice plants (Guo et al. 2016a). However, the functions of the other two PMT proteins remain unclear. To investigate the roles of MoPmt1 and MoPmt4 in fungal growth and pathogenicity, we individually deleted the MoPmt1 and MoPmt4 genes in the genome of M. oryzae utilizing the deletion vector which contains the hygromycin-resistance gene (Fig. S1A, S1B). Two hygromycin B-resistant transformants from each PMT gene were first verified by diagnostic PCR (Fig. S1C, S1D), and then were further confirmed by Southern blot and semiquantitative RT-PCR (RT-PCR) (Fig. S1E, S1F, S1G, S1H). To ensure the biological roles of each Pmt genes, the complemented strains MoPmt1c and MoPmt4c, which the MoPmt1 and MoPmt4 genes under their native promoters, were introduced into MoPmt1-10 and MoPmt4-4 mutants, respectively, were generated and verified by RT-PCR (Fig. S1G, S1H).
MoPmt genes are necessary for vegetative growth in M. oryzae
To assess the role of MoPmt genes in fungal growth, wild-type Guy11, MoPmt1 mutants, MoPmt2 mutants, MoPmt4 mutants, and complemented strain MoPmt4c were compared on complete medium (CM), oatmeal agar medium (OM), minimal medium (MM), and RDC medium (Guo et al. 2011). All the MoPmt mutants showed significantly retarded radial growth on tested media (Fig. 1a; Fig. S2) at 5 dpi. In liquid CM, both MoPmt1 mutants and MoPmt2 mutants showed significant reduction in fungal biomass in comparison with Guy11 (Fig. 1b, c). By contrast, more compact mycelial growth was observed following incubation in liquid CM for 48 h (Fig. 1b), with significantly more fungal biomass in MoPmt4 mutants than in Guy11 and MoPmt4c after incubation time increased to 72 and 120 h, respectively (Fig. 1c). To determine the reason, we compared hyphae branches of Guy11, MoPmt4 mutants and MoPmt4c under microscopy, and found the MoPmt4 mutants presenting more branches on hyphae than that of by Guy11 and MoPmt4c (Fig. 1d, e).
MoPmt4 is essential for normal mycelial growth. a Colony diameters of the tested strains on different media were measured at 5 dpi and statistically analyzed. b Tested strains were inoculated in liquid CM for 48 h at 28 °C in darkness and then photographed. c Fungal mycelia of tested strains were collected at 3 dpi and 5 dpi, freezing dried, and statistically analyzed. d Hyphae branches of Guy11, MoPmt4 mutants and MoPmt4c were examined under light microscope. e Hyphae length between the branches of the tested strains were measured and statistically analyzed. Asterisks in Fig. 2a, c, e indicate significant differences. Error bar represents standard deviation
MoPmt genes are involved in asexual development in M. oryzae
To assess the role of MoPmt genes in asexual development, conidiation was compared among Guy11, MoPmt1 mutants, MoPmt2 mutants, MoPmt4 mutants and MoPmt4c. The MoPmt mutants produced less conidia on the conidiophores than that of Guy11 (Fig. 2a). Quantitative measurement of conidia ensured that conidiation was reduced by approximately 1.5-fold in MoPmt1 mutants, 10.5-fold in MoPmt2 mutants, and 2.4-fold in MoPmt4 mutants (MoPmt4-4, 2.5-fold; MoPmt4-10, 2.3-fold), compared with Guy11 (Fig. 2b). Of those spores that formed by MoPmt4 mutants, approximately 36.2% of conidia exhibited short in length, compared to 14.1 and 14.3% of that by Guy11 and MoPmt4c, respectively (Fig. 2c, d). For those conidia in small size, the average length was 1.56 micrometer shorter than that of Guy11 and MoPmt4c (Fig. 2e), indicating that MoPmt4 is essential for normal conidial development in M. oryzae.
Deletion of MoPmt4 results in abnormal conidial morphology and reduced conidiation. a Tested strains were grown on RDC medium for 8 days and the conidia on conidiophores were examined by light microscopy. Scale bar = 50 µm. b Wild-type Guy11, mutants, and complemented strains were grown on RDC medium for 8 days and conidia produced by those strains were collected, counted, and statistically analyzed (P < 0.01). c Conidia shape of Guy11, mutants and complemented strains was examined under light microscopy and photographed. d Percentage of abnormal conidia was recorded and statistically analyzed. e Conidia sizes were determined as width by length from 150 conidia of each strain. Asterisks in Fig. 3b, e indicate significant differences. Error bar represents standard deviation
MoPmt4 is essential for the pathogenic development of M. oryzae. a Conidial suspensions (1 × 105 conidia mL−1) of Guy11, MoPmt1 mutants, MoPmt4 mutants, and complemented strain MoPmt4c were inoculated on 14-day-old rice seedlings (Co-39). The infected leaves were evaluated at 5 dpi. b Pathogenicity was tested on barley leaves with conidia (1 × 105 conidia mL−1) and photographed at 5 dpi. c Microscope observation of the conidia development on 14-day-old lesions after illuminated for 24 h
MoPmt4 is required for virulence on rice plants
Since Pmt genes differentially affect growth, morphogenesis and pathogenesis in phytopathogens (Mouyna et al. 2010; Gonzalez et al. 2013; Le et al. 2018), to verify if both MoPmt1 and MoPmt4 are required for virulence as the MoPmt2 gene, leave from both 14-day-old rice seedlings and 7-day-old barley seedlings inoculated with conidial suspension (1 × 105 conidia mL−1) of wild-type strain, MoPmt1 mutants, MoPmt2 mutant, and MoPmt4 mutants, respectively, were evaluated 5-day post-inoculation (dpi). Disease symptoms caused by MoPmt1 mutants were comparable to wild-type Guy11 (Fig. 3a, b). By contrast, both the MoPmt2 mutant and MoPmt4 mutants severely reduced virulence on rice leaves, with most of the leaves developing small and restricted lesions, compared with that of by Guy11 (Fig. 3a, b). When extended the incubation time to 14 days to compare conidia production on disease lesions, we found that a large mount of conidia were produced on infectious lesions by Guy11, whereas seldom conidium was observed on lesions by MoPmt4 mutants (Fig. 3c). The loss of pathogenicity was fully recovered, while the MoPmt4 gene was reintroduced into MoPmt4 mutant (Fig. 3a–c), suggesting a potential role of MoPmt4 in fungal virulence. Since it aims to identify genes involved in pathogenicity in M. oryzae, MoPmt4 was thus selected to investigate the roles on fungal development and pathogenicity.
MoPmt4 encodes protein O-Mannosyltransferase
In Schizosaccharomyces pombe, OGM4 is a PMT4 subfamily member and encodes protein with O-mannosyltransferase activity (Willer et al. 2005). To determine whether MoPmt4 encodes a functional homolog of S. pombe OGM4, we tested the capacity of MoPmt4 to complement a yeast strain lacking ogm4. The cDNA of MoPmt4 was cloned into pREP41X to generate the plasmid pREP41X-MoPmt4 (Maundrell 1993). Both pREP41X-MoPmt4 and pREP41X were individually transformed into ogm4 mutant. As a control, the empty vector pREP41X was also transformed into wild-type strain FY527. Under inducible condition, the S. pombe ogm4 mutant forms aggregates, while the wild-type strain shows individual cell growth. When we expressed MoPmt4 in ogm4 mutant, the aggregates growth disappeared and the transformants restore the single-cell growth shown by FY527 (Fig. S3), indicating that MoPmt4 is homologous to OGM4 in S. pombe and has O-mannosyltransferase activity in M. oryzae.
MoPmt4 is essential for appressorium development, lipid, and glycogen metabolism
Development of melanin appressorium is essential for effective penetration of rice blast fungus. Therefore, the process of appressorium formation was carefully examined among Guy11, MoPmt4 mutants, and MoPmt4c. Conidia germination and melanin appressorium formation of the MoPmt4 mutants were significantly delayed at 2 and 8 h, but restored to the wild-type levels at 8 and 24 h, respectively (Fig. 4a, b). Rapid degradation of lipid droplets and glycogen is essential for generation of turgor pressure during appressoria development. However, the MoPmt4 mutants showed a retardation in translocation of lipid droplets and glycogen to appressoria during appressorium formation, in comparison with Guy11 and MoPmt4c (Fig. 4c–f), indicating a critical role of MoPmt4 in regulating glycogen and lipids metabolization in M. oryzae.
MoPmt4 is essential for lipid and glycogen metabolism. a Droplets of conidial suspension (5 × 104 spores mL−1) were inoculated on the hydrophobic coverslips for 2 and 8 h, and then, the germinated conidia were counted and statistically analyzed. Bars = 10 µm. b Conidial suspension (5 × 104 spores mL−1) was inoculated on the hydrophobic coverslips for 8 and 24 h, and then, the melanin appressoria were counted and statistically analyzed. c Germinated conidia stained with iodine solution were visualized under a light microscope at 12 hpi. Bars = 20 µm. d Germinated conidia contained the glycogen deposits were recorded in the indicated strains and statistically analyzed. e Germinated conidia stained with Nile red solution were visualized under a Nikon inverted Ti-S epifluorescence microscope at 12 hpi. Bars = 20 µm. f Germinated conidia contained the lipid droplets were counted in the indicated strains and statistically analyzed. Asterisks in a, b, and f indicate significant differences. Error bar represents standard deviation
MoPmt4 is required for penetration and colonization of host tissues
As the MoPmt4 mutants displayed attenuated virulence on host plants (Fig. 1a), the appressoria-mediated penetration by Guy11 and MoPmt4 mutants was examined. The MoPmt4 mutants could less effectively penetrate into the epidermal cells of barley leaves at 32 and 48 hpi, respectively, compared to Guy11 and MoPmt4c (Fig. 5a, b). The primary invasive hyphae (IH) developed by MoPmt4 mutants showed swollen morphology, compared with that of Guy11 and MoPmt4c (Fig. 5c). Given that penetration defects ascribed to reduced virulence, the invasive growth of MoPmt4 mutants was examined by inoculating the conidia of MoPmt4 mutants on abraded rice leaves, which allowed direct entry of the fungus into host tissues through a wound site without appressorium. However, smaller necrotic lesions were observed on the inoculation sites by the MoPmt4 mutants, in contrast to extendible and necrotic lesions by wild-type Guy11 and MoPmt4c (Fig. 5d). Most of the IH developed by the mutants displayed extremely retarded growth in host cells and failed to colonize the leaf epidermis beyond the first cell (Fig. 5e). On the contrary, the wild-type Guy11 and MoPmt4c could successfully colonized the host cells and freely expand to new cells adjacent to initially invaded cells (Fig. 5f). These findings suggest a potential role for MoPmt4 in plant penetration and colonization.
MoPmt4 is critical for penetration and colonization in M. oryzae. a Conidial suspension (5 × 104 spores mL−1) was inoculated on barley leaves for 32 h and appressoria-mediated penetration by Guy11, MoPmt4 mutants, and MoPmt4c was examined under light microscope. White asterisks indicated swollen primary IH, Bars = 40 µm. b The appressoria penetrated into barley epidermal cells were measured at 32 and 48 hpi, and the data were statistically analyzed. c Swollen primary IH in barley epidermal cells was counted at 32 hpi in indicated strains and statistically analyzed. d Droplets of conidial suspension of indicated strains were inoculated on abraded rice leaves and diseased leaves were harvested at 5 dpi. e Infectious hyphae growth in barley epidermal cells was observed at 48 hpi. White asterisks indicated swollen primary IH, Bars = 40 µm. f Percentage of infected cells occupied by IH of indicated strains at 48 hpi was measured and statistically analyzed. Asterisks in b and c indicate significant differences. Error bar represents standard deviation
MoPmt4 is necessary for turgor generation and cell wall integrity
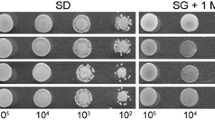
Sufficient turgor pressure within appressorium guarantees the penetration of M. oryzae (Jeon et al. 2008). In view of penetration defects by MoPmt4 mutant, appressorium turgor pressure was measured using the incipient cytorrhysis assay. The collapsed appressoria were less frequently observed in MoPmt4 mutants than in Guy11 and MoPmt4c over the different concentrations of glycerol (Fig. 6a, b), suggesting higher turgor pressure within mutant appressoria. As retarded degrading of lipid droplets and glycogen during appressorium development (Fig. 4c–f), turgor pressure within the mutant appressoria should not be higher than that within wild-type appressoria. Therefore, to delve into the lower cytorrhysis rate of the mutant, we next examine the CWI by testing both the sensitivity to cell wall-perturbing agents and the porosity of cell wall by previously described methods (Money 1989; Guo et al. 2017). The MoPmt4 mutants exhibit a significantly increased sensitivity to CR and CFW, compared to those of Guy11 and MoPmt4c (Fig. 6c, d). The ratios of plasmolysis to cytorrhysis in wild type were 0.56, 1.44, and 0.29 in glycerol, PEG 400 and 1000, respectively (Fig. 6e), in contrast to average ratios of 5.6, 4.9, and 2.5 in glycerol, PEG 400 and 1000 by MoPmt4 mutants. Furthermore, protoplast release by MoPmt4 mutants was much faster than that of Guy11 and MoPmt4c after enzyme treatment for 30, 60, and 90 min (Fig. 6f), suggesting that CWI was impaired in MoPmt4 mutants. TEM assay further confirmed the development of potholed cell wall ultrastructures in the MoPmt4 mutant, compared to intact cell wall by wild-type Guy11 (Fig. S4).
MoPmt4 is required for CWI and turgor pressure accumulation. a Appressoria at 24-h post-germination were treated with different concentrations of glycerol (2M, 3M, and 4M), and examined by microscopy after 10 min. Appressorial collapse is illustrated here for four molar glycerols. Bars = 20 µm. b Appressorium of the indicated strains was treated with given concentration of glycerol solution for 10 min, and the numbers of collapsed appressoria (> 100) were counted and statistically analyzed. c All tested strains were inoculated on CM containing cell wall-perturbing agents with the final concentrations of 150 (CFW) and 200 (CR) µg mL−1, and then photographed at 5dpi. d Statistical analysis of mycelial growth of the indicated strains under CR and CFW. e Appressorium induced on hydrophobic coverglass was treated with different Stokes’ radii of PEG. Cell wall porosity was measured by comparing the ratio of plasmolysis to cytorrhysis. f Equal amount of fungal mycelia of the tested strains was enzyme treatment and the released protoplasts were collected, counted, and statistically analyzed at each given time. Asterisks in b, d, and e indicate significant differences. Error bar represents standard deviation
MoPmt4 deletion activates plant defense responses
Since the MoPmt4 mutant developed resistant-type lesions and restrained infectious hyphae growth on rice seedlings, we thus compared the defense responses against the wild type and the mutant. As the conidia of MoPmt4 mutant are sensitive to hydrogen peroxide (Fig. S5), the ability to degrade host-derived ROS at infection sites was examined by staining with 3, 3′-diaminobenzidine (DAB) 48 h after inoculation. Rice cells containing wild-type IH were seldom stained with DAB, whereas a majority of primary infected rice cells with MoPmt4 mutant were strongly stained with DAB, indicating the accumulation of H2O2 at infection sites (Fig. 7a, b).
MoPmt4 is involved in scavenging of ROS. a 14-day-old rice sheaths were injected with related conidial suspensions. DAB was used to stain the sheaths for 8 h after 48 hpi. Bar = 20 µm. b DAB-stained rice sheath cells of the indicated strains were counted and statistically analyzed. Asterisks represent significant differences (p < 0.01). c Expression of rice pathogenesis-related (PR) genes over time after inoculation. The transcriptional expression of PR1a, PBZ1, and AOS2 in the infected rice was analyzed using quantitative RT-PCR
To determine whether plant defense was activated by infection with MoPmt4 mutant, the expression patterns of three PR genes, PR1a, PBZ1, and AOS2 were analyzed by quantitative RT-PCR. The expression of PR1a, PBZ1, and AOS2 in rice plants inoculated with wild-type Guy11 followed a typical pattern of compatible interaction, and the induction of these genes was delayed to 72 hpi. However, the activation of PR1a, PBZ1, and AOS2 was observed in higher level even at 24 and 48 hpi by inoculating with MoPmt4 mutant (Fig. 7c). This difference was more pronounced at 72 hpi than at 24 hpi, and the transcription levels of PR1a, PBZ1, and AOS2 were increased by approximate 6.0, 3.2, and 18.3 folds, respectively, in MoPmt4 mutant-infected rice leaves at 72 hpi, compared to those in wild-type challenged rice leaves (Fig. 7c).
MoPmt4 involves in suppressing ROS generation in plant cells
To determine whether impairment of plant colonization by MoPmt4 mutant is ascribed from plant defense response that caused by ROS accumulation at infection sites, we evaluated the effect of pretreatment with diphenyleneiodonium (DPI), an inhibitor of the plant NADPH oxidase, on plant infection. Under conditions without DPI treatment, average 75% of appressoria of wild type successfully developed IH that colonized more than three cells, compared with average 10% of that by MoPmt4 appressoria (Fig. 8a, b). Meanwhile, only 4% of appressoria of Guy11 developed swollen primary IH, in contrast to more than 72% of that by MoPmt4 mutant (Fig. 8c, d). Treatment of DPI significantly increased the frequencies of IH development by MoPmt4 mutant (Fig. 8a, b), and the IH morphology of MoPmt4 mutant was recovered as the same as wild-type Guy11 (Fig. 8c, d). On wounded rice leaves, DPI treatment could also obviously increase the virulence of MoPmt4 mutant (Fig. 8e), suggesting that MoPmt4 might help to evade of plant defense by suppression of ROS production in host cells during fungal penetration.
Suppression of host-derived ROS restores infectious growth of MoPmt4 mutant. a DPI treatment restored IH growth. Barley leaves were treated with or without DPI (0.4 µM) solution. Invasive growth was observed at 48 hpi. Bar = 40 µm. White asterisks indicate the swollen hyphae cells. b Percentage of infected barley cells occupied by infectious hyphae of indicated strains. Each of the strains was inoculated on barley leaves pretreated with or without DPI for 48 h, and then, the percentage of barley cells occupied by IH was statistically compared. The total number of appressorium is indicated above each column. c Tested strains were inoculated on barley leaves pretreated with or without DPI for 30 h, and then, the morphology of IH tips was observed and compared. Black asterisks indicate swollen IH tips. d Barley leaves pretreated with or without DPI were inoculated with indicated strains on for 30 h, and the numbers of swollen primary IH were counted and statistically analyzed. e Droplets of conidial suspension of indicated strains were inoculated on abraded rice leaves pretreated with or without DPI and diseased leaves were photographed at 5 dpi
Subcellular localization of MoPmt4 protein in M. oryzae
According to ProtComp 9.0 (http://www.softberry.com/), MoPmt4 is predicted to localize at ER apparatus in M. oryzae (Table S2). ER retention signal, such as HEEL from ER chaperone protein LHS1 in M. oryzae, has been validated to localize at ER apparatus and widely used as an ER marker in protein localization assay (Derkx and Madrid 2001; Yi et al. 2009). To investigate the localization of MoPmt4, an RFP-HEEL fusion construct was generated and transformed into the complemental strain, at which the MoPmt4 protein was labeled with a GFP tag. The resulting transformants with detectable RFP signals showed wild-type growth, and then were used for localization assay. In vegetative hyphae, conidia and initial IH, co-localization of MoPmt4-GFP with RFP-HEEL was observed (Fig. 9a–c). However, in appressorium development, the fluorescent signal of MoPmt4-GFP was only partially colocalized with signal of RFP-HEEL in the central region of appressorium (Fig. 9a).
MoPmt4 localizes to ER apparatus. a Localization pattern of MoPmt4 in the conidia and mature appressorium in M. oryzae. The confocal fluorescent images (Zeiss LSM710) indicate the co-localization of MoPmt4 with MoLHS1, which was expressed as an ER marker. b, c Localization pattern of MoPmt4 in the vegetative hypha and invasive hypha. The confocal fluorescent images (Zeiss LSM710) indicate the co-localization of MoPmt4 with MoLHS1. DIC indicates differential interference contrast image
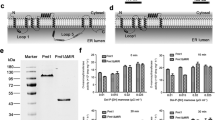
PMT motif is essential for the full virulence of M. oryzae
Bioinformatic analysis suggests that MoPmt4 contains PMT and MIR motifs at the N-terminal regions (Guo et al. 2016a). To further assess the role of the respective motif in MoPmt4 protein, motif deletion constructs (Fig. 10a), MoPmt4ΔPMT-GFP and MoPmt4ΔMIR-GFP, were introduced into the MoPmt4-4 mutant, respectively. The MoPmt4/MoPmt4ΔPMT-GFP transformant expressing MoPmt4 lacking PMT motif and the MoPmt4/ MoPmt4ΔMIR-GFP transformant lacking MIR motif were confirmed by RT-PCR (Fig S6) and fluorescent microscopy (Fig. 10b). Thereafter, phenotypes of each strain were compared with wild-type Guy11 and MoPmt4-4 mutant. Strain lacking of PMT motif results in defects on growth, conidiation, appressoria-mediated penetration, and full virulence of M. oryzae (Fig. 10c–g), which are similar to that observed in MoPmt4 mutant. On the contrary, strain deletion of MIR motif could only results in slight defects on growth and conidiation, but not the ability of penetration and pathogenicity on host plants (Fig. 10c–g). These results suggest that PMT motif in MoPmt4 protein is essential for the development and pathogenicity of M. oryzae.
PMT motif is essential for the full function of MoPmt4. a Diagrams of gene deletions used in complementation experiments. b Conidia of Guy11, MoPmt4 mutants, MoPmt4ΔPMT-GFP, MoPmt4ΔMIR-GFP, and MoPmt4c were collected and observed under an epifluorescence microscope. c Mycelia growth on the CM media. The tested strains were inoculated on CM media to grow for 5 days and then photographed. d Indicated strains were grown on RDC medium for 7 days and the conidia on conidiophores were examined by light microscopy. Scale bar = 50 µm. e Conidia suspensions (1 × 105 conidia mL−1) of the indicated strains were sprayed on rice seedlings and disease leaves were photographed at 5 dpi. f, g Plant penetration assays. Conidia suspensions (1 × 105 conidia mL−1) of the indicated strains were inoculated on barley leaves for 30 and 48 h, respectively, and then, fungal penetration (f) and IH growth (g) were examined under a light microscope and statistically analyzed
Discussion
Protein glycosylation is the most common post-translational modification conserved in eukaryotic organisms, and plays a critical role in determining the structure and function of numerous proteins, including receptors, cell wall components, and secreted proteins (Lehle et al. 2006; Lommel and Strahl 2009). In eukaryotes, O-mannosylation is one type of glycosylation (N- and O-glycosylation) and is initially catalyzed by protein O-mannosyltransferases (PMTs) to add the mannose residues to target proteins (Lommel and Strahl 2009). In pathogenic fungi, many glycoproteins, which modified by the widely existing PMTs, are likely to involve in fungal pathogenic development, since mutants with altered O-mannosylation patterns showed defective CWI and attenuated virulence to host (Prill et al. 2005; Olson et al. 2007; Fernandez-Alvarez et al. 2009; Gonzalez et al. 2013; Guo et al. 2016a; Le et al. 2018). For M. oryzae, fungal cell wall is the primary contact surface between the fungus and plant, and the intact cell wall guarantees the fungal growth and pathogenic development during plant invasion (Guo et al. 2015, 2016a; Jeon et al. 2008). Previously, protein O-mannosylation regulated by MoPmt2 was identified to involve in development of fungal cell wall and pathogenicity, and deletion of MoPmt2 resulted in defective CWI and attenuated virulence, further suggesting the critical role of protein O-mannosylation in pathogenic development of M. oryzae (Guo et al. 2016a). In this study, both MoPmt1 and MoPmt4 were separately deleted from the genome of M. oryzae, and their roles on fungal growth and pathogenicity have been analyzed. Our results revealed that MoPmt proteins are essential for mycelial growth and conidiation of M. oryzae. Of the three mannosyltransferases tested, both MoPmt2 and MoPmt4 are required for full virulence of M. oryzae. Disruption of either MoPmt2 or MoPmt4 equally resulted in reduction of pathogenicity, with MoPmt2 mutants showing a weaker virulence than MoPmt4 mutants to rice seedlings. In addition, in most aspect assayed, such as fungal growth, septa distribution, conidiation, and CWI, MoPmt4 mutant seems to be half-way between the wild type and MoPmt2 mutants (Guo et al. 2016a). Deletion of MoPmt4 led to severe defects in radial growth, with more branching hyphae and septa as compared to Guy11, which are similar to the phenotypes of BcPmt4 mutants in B. cinerea and MoPmt2 mutant in M. oryzae (Gonzalez et al. 2013; Guo et al. 2016a). In addition, like MoPmt2 mutant, MoPmt4 mutants also showed defective appressorial CWI, which, as a result, lead to a drastic reduction in appressorium-mediated penetration in terms of plant penetration, indicating that protein substrates of MoPmt2 and MoPmt4 might include cell wall proteins. In other fungal pathogens like U. maydis, B. cinerea, and A. fumigatus, deletion of the homologous gene results always in similar phenotypes such as defective CWI, morphogenesis, and the virulence of fungal pathogens (Fernandez-Alvarez et al. 2009; Gonzalez et al. 2013; Goto et al. 2009), suggesting a conserved role of Pmt4 proteins in fungal cell wall development and pathogenicity. Besides of those conserved roles, we found in this study additional roles of MoPmt4 that may be unique to M. oryzae. These include roles of glycogen and lipid droplets metabolism, secretion of extracellular enzymes, scavenging of host-derived ROS, and suppression of plant defense responses during plant infection. With these functions taken into account, MoPmt4 is critical for fungal development, and for full virulence of M. oryzae.
In pathogenicity assay, the MoPmt4 mutants showed reduced virulence toward rice leaves, with most of them developing small and restricted lesions, which, as a result, greatly prohibit the potential for conidiation (Fig. 1a, c). The pathogenicity defect seems not ascribe from the malformed mutant conidia, which germinate in a slightly delayed manner. In plant infection, though melanized appressoria formed by the mutant showed slight delay at early stage, they thereafter restored to the similar level as the wild-type strain, excluding the defective appressoria development as a potential for attenuated virulence of MoPmt4 mutants. However, compared to wild-type strain Guy11, only ~ 19% of mutant appressoria penetrated the barley epidermal cells and developed IH in first-invaded cells. This penetration defect might at least partially attribute to less turgor pressure generated in mutant appressoria than in parental strain Guy11. After penetration, IH of the mutants that grow inside in first-invaded barley epidermal cells are often misshapen. Specifically, most of them appear almost round and shortened, and are always difficult to differentiate the secondary IH during fugal colonization compared to Guy11. Based on above, these phenotypes, including both penetration defects and abnormal IH growth in host cells, could together account for the pathogenic defects in the MoPmt4 mutants.
Plant defense responses are often associated with the rapid generation of ROS and transcriptional activation of PR genes in the infected cells. Since the MoPmt4 mutant developed small and restricted resistant-type lesions on rice seedlings, and IH of the MoPmt4 mutant in the infectious cells was restrained, thus, the defense responses against the wild type and the mutant were compared. During plant infection, the earliest defense responses are the rapid accumulation of reactive oxygen species (ROS) at the infection site (Apostol et al. 1989), and the ability to detoxify those ROS permits the pathogenic development of M. oryzae (Chi et al. 2009; Guo et al. 2011). In this study, conidia of the MoPmt4 mutants were more sensitive to hydrogen peroxide than that of Guy11, and more DAB-stained leaf sheath cells were detected when challenged with MoPmt4 mutants compared with Guy11, which is similar to the phenotypes observed in DES1 and Moap1 mutants (Chi et al. 2009; Guo et al. 2011), making us assume that the H2O2-degrading ability was severely weakened in the MoPmt4 mutants. The DPI treatment, which prevents accumulation of host-derived ROS, significantly increased appressorium penetration and restores the normal infectious growth of MoPmt4 mutants in plant tissues, and further confirmed this assumption. Since host-derived ROS could activate the defense-gene expression during plant infection (Nurnberger et al. 2004), a stronger induction of PR1a, PBZ1, and AOS2 in MoPmt4-challenged rice tissues also suggests that plant defense responses, which might be activated by host-derived ROS, prevent the infection of MoPmt4 mutants. In M. oryzae, secreted peroxidases degrade ROS at infection site (Chi et al. 2009). The activity of extracellular peroxidases and laccases showed severe reduction in MoPmt4 mutant (Fig. S7). Since protein glycosylation plays a critical role in determining the function of cell wall components and secreted proteins, therefore, together with conserve roles of MoPmt4 protein, we deduce that deletion of MoPmt4 might block the modification of extracellular proteins such as peroxidases and laccases during fungal development in M. oryzae. This will ultimately impair the degrading of host-derived ROS, which, in turn, activate the plant defense response to prohibit the infectious of the mutant. However, we could not exclude the possibility that some other defects, such as abnormal cell wall, might also be partly responsible for the ROS resistance in MoPmt4 mutants.
In S. cerevisiae, it is known that the PMT1 and PMT2 subfamily members form heterodimerization, while the PMT4 protein forms homodimerization to have protein mannosylation activity (Girrbach and Strahl 2003). In M. oryzae, three putative PMT proteins, with one member in each Pmt subfamily, were identified (Guo et al. 2016a). However, deletion of MoPmt2 resulted in severe defects on virulence, while deletion of MoPmt1 did no alteration on pathogenicity in M. oryzae. If the dimerization described in S. cerevisiae was really necessary for the mannosylation activity of PMT proteins, then the MoPmt1 and MoPmt2 mutants would share similar phenotypes. Since this is clearly not the case, suggesting that the dimerization between MoPmt1 and MoPmt2 proteins is not true for M. oryzae. This result also agrees with the findings in other fungi, including B. cinerea, U. maydis, and A. fumigatus (Gonzalez et al. 2013; Fernandez-Alvarez et al. 2009; Goto et al. 2009). Like many other PMTs, MoPmt4 contained two large transmembrane domains locating at its N- and C-terminal regions and localized at ER (Fig. 9). In addition, two typical PMT and MIR domains, which shared strong sequence similarity with the corresponding domains in S. cerevisiae, are presented in MoPmt4 protein (Guo et al. 2016a). In this study, the PMT motif was identified to involve the fungal growth, conidiation, appressoria-mediated penetration, and full virulence of M. oryzae, while the MIR motif was approved to only play slight effects on growth and conidiation, but not penetration and pathogenicity. Since the PMT domain is located in the ER lumen and has been implicated in the mannosyltransferase activity. The essential DE motif (aa, 80–81) in PMT domain suggests to play a critical role in catalysis during protein mannosylation (Lommel et al. 2011). In view of similar phenotypes between MoPmt4ΔPMT-GFP and MoPmt4 mutant, we presumed that disruption of PMT motif in MoPmt4 might affect the enzymatic activity of MoPmt4, which blocked the addition of first mannose residue to pathogenicity-related proteins, and thus affected the fungal development and full pathogenicity of M. oryzae.
In conclusion, the protein O-mannosyltransferase MoPmt4 shares conserved roles, such as defective fungal growth, CWI, and appressorium penetration, with these homologues in other fungi, during the development of M. oryzae. Besides, additional roles of MoPmt4, including the glycogen and lipid droplets metabolism, secretion of extracellular enzymes, scavenging of host-derived ROS, and suppression of plant defense responses during plant infection, were also regarded as unique causes of this protein in determining the pathogenicity of M. oryzae.
References
Apostol I, Heinstein PF, Low PS (1989) Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiol 90:109–116
Carroll AM, Sweigard JA, Valent B (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet Newsl 41:22
Chi M-H, Park S-Y, Kim S, Lee Y-H (2009) A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog 5:e1000401
Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD et al (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Derkx PM, Madrid SM (2001) The foldase CYPB is a component of the secretory pathway of Aspergillus niger and contains the endoplasmic reticulum retention signal HEEL. Mol Genet Genomics 266:537–545
Ebbole DJ (2007) Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 45:437–456
Egan MJ, Wang Z-Y, Jones MA, Smirnoff N, Talbot NJ (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci U S A 104:11772–11777
Fernandez-Alvarez A, Elias-Villalobos A, Ibeas JI (2009) The O-mannosyltransferase PMT4 is essential for normal appressorium formation and penetration in Ustilago maydis. Plant Cell 21:3397–3412
Girrbach V, Strahl S (2003) Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J Biol Chem 278:12554–12562
Gonzalez M, Brito N, Frias M, Gonzalez C (2013) Botrytis cinerea protein O-mannosyltransferases play critical roles in morphogenesis, growth, and virulence. PLoS One 8:e65924
Goto M, Harada Y, Oka T, Matsumoto S, Takegawa K, Furukawa K (2009) Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot Cell 8:1465–1474
Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H et al (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 23:1053–1068
Guo M, Chen Y, Du Y, Dong Y, Guo W, Zhai S et al (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7:e1001302
Guo M, Gao F, Zhu X, Nie X, Pan Y, Gao Z (2015) MoGrr1, a novel F-box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae. Appl Microbiol Biotechnol 99:8075–8088
Guo M, Tan L, Nie X, Zhu X, Pan Y, Gao Z (2016a) The Pmt2p-mediated protein O-mannosylation is required for morphogenesis, adhesive properties, cell wall integrity and full virulence of Magnaporthe oryzae. Front Microbiol 7:630
Guo M, Zhu X, Li H, Tan L, Pan Y (2016b) Development of a novel strategy for fungal transformation based on a mutant locus conferring carboxin-resistance in Magnaporthe oryzae. AMB Express 6:57
Guo M, Tan L, Nie X, Zhang Z (2017) A class-II myosin is required for growth, conidiation, cell wall integrity and pathogenicity of Magnaporthe oryzae. Virulence 8(7):1335–1354
Harries E, Gandia M, Carmona L, Marcos JF (2015) The Penicillium digitatum protein O-mannosyltransferase Pmt2 is required for cell wall integrity, conidiogenesis, virulence and sensitivity to the antifungal peptide PAF26. Mol Plant Pathol 16:748–761
Howard RJ, Valent B (1996) Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu Rev Microbiol 50:491–512
Howard RJ, Ferrari MA, Roach DH, Money NP (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA 88:11281–11284
Jeon J, Goh J, Yoo S, Chi M-H, Choi J, Rho H-S et al (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol Plant Microbe Interact 21:525–534
Le THT, Oki A, Goto M, Shimizu K (2018) Protein O-mannosyltransferases are required for sterigmatocystin production and developmental processes in Aspergillus nidulans. Curr Genet. https://doi.org/10.1007/s00294-00018-00816-x
Lehle L, Strahl S, Fau-Tanner W, Tanner W (2006) Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl 5:6802–6818
Loibl M, Strahl S (2013) Protein O-mannosylation: what we have learned from baker’s yeast. Biochim Biophys Acta 1833:2438–2446
Lommel M, Strahl S (2009) Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19:816–828
Lommel M, Schott A, Jank T, Hofmann V, Strahl S (2011) A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J Biol Chem 286:39768–39775
Matar KAO, Chen X, Chen D, Anjago WM, Norvienyeku J, Lin Y et al (2017) WD40-repeat protein MoCreC is essential for carbon repression and is involved in conidiation, growth and pathogenicity of Magnaporthe oryzae. Curr Genet 63:685–696
Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130
Money NP (1989) Osmotic pressure of aqueous polyethylene glycols: relationship between molecular weight and vapor pressure deficit. Plant Physiol 91:766–769
Mouyna I, Kniemeyer O, Jank T, Loussert C, Mellado E, Aimanianda V et al (2010) Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol Microbiol 76:1205–1221
Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198:249–266
Olson GM, Fox DS, Wang P, Alspaugh JA, Buchanan KL (2007) Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot Cell 6:222–234
Prill SK, Klinkert B, Fau-Timpel C, Timpel C, Fau-Gale CA, Schroppel K, Ernst KF, Ernst JF (2005) PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol Microbiol 55:546–560
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Song W, Dou X, Qi Z, Wang Q, Zhang X, Zhang H et al (2010) R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS One 5:e13193
Strahl-Bolsinger S, Gentzsch M, Tanner W (1999) Protein O-mannosylation. Biochim Biophys Acta 1426:297–307
Talbot NJ (2003) On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57:177–202
Wang Y, He D, Chu Y, Zuo Y-S, Xu X-W, Chen X-L et al (2016) MoCps1 is important for conidiation, conidial morphology and virulence in Magnaporthe oryzae. Curr Genet 62:861–871
Willer T, Valero MC, Tanner W, Cruces J, Strahl S (2003) O-mannosyl glycans: from yeast to novel associations with human disease. Curr Opin Struct Biol 13:621–630
Willer T, Brandl M, Sipiczki M, Strahl S (2005) Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol Microbiol 57:156–170
Xu Y-B, Li H-P, Zhang J-B, Song B, Chen F-F, Duan X-J et al (2010) Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet Biol 47:205–215
Yan X, Li Y, Yue X, Wang C, Que Y, Kong D et al (2011) Two novel transcriptional regulators are essential for infection-related morphogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7:e1002385
Yi M, Chi MH, Khang CH, Park SY, Kang S, Valent B et al (2009) The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21:681–695
Acknowledgements
Thanks are given to Dr. S. Strahl-Bolsinger for providing S. pombe strains and to Dr. Susan Forsburg for providing the plasmid. This work was supported by the National Natural Science Foundations of China (Grant No: 31671976 to MG; Grant No: 31101401 to MG), the Key Grant for Excellent Young Talents of Anhui Higher Education Institutions (gxyqZD2016037 to MG), Natural Science Foundations of Anhui province (Grant No: 1608085QC49 to MG), the Foundation for the Excellent Talents of Anhui Agricultural University (Grant No: RC2015002 to MG), and the Anhui Agricultural University Postgraduate Innovation Foundation (Grant No. 2018yjs-4 to RP). The Zhang laboratory research was supported by the National Science Foundation for Distinguished Young Scholars of China (Grant No. 31325022 to ZZ).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: M. Guo and Z. Zhang. Performed the experiments: Y. Pan, R. Pan, and L. Tan. Analyzed the data: M. Guo and R. Pan. Wrote the paper: M. Guo.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pan, Y., Pan, R., Tan, L. et al. Pleiotropic roles of O-mannosyltransferase MoPmt4 in development and pathogenicity of Magnaporthe oryzae. Curr Genet 65, 223–239 (2019). https://doi.org/10.1007/s00294-018-0864-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0864-2