Abstract
For maintenance of cytoplasmic protein quality control (PQC), cytoplasmic heat shock proteins (HSPs) negatively control heat shock transcriptional factor (HSF) in a negative feedback loop. However, how mitochondrial protein quality control (mtPQC) is maintained is largely unknown. Here we present evidence that HSF directly monitors mtPQC in the budding yeast Saccharomyces cerevisiae. Mitochondrial HSP70 (Ssc1) negatively regulated HSF activity. Importantly, HSF was localized not only in the nucleus but also on mitochondria. The mitochondrial localization of HSF was increased by heat shock and compromised by SSC1 overexpression. Furthermore, the mitochondrial protein translocation system downregulated HSF activity. Finally, mtPQC modulated the mtHSP genes SSC1 and MDJ1 via HSF, and SSC1 overexpression compromised mitochondrial function. These findings illustrate a model in which HSF directly monitors mtPQC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitochondria have crucial functions in the cell, including ATP generation, iron–sulfur cluster biogenesis, nucleotide biosynthesis, lipid and steroid synthesis and amino acid metabolism. In addition, mitochondria commit apoptosis. Therefore, mitochondrial dysfunction causes a broad spectrum of human diseases, such as myopathies, diabetes, obesity, and liver and renal dysfunctions, age-related neurodegenerative diseases, and aging (Hill and Van Remmen 2014; Hu and Liu 2011; Lasserre et al. 2015). Mitochondria are thought to have evolved from free-living eubacterium endosymbionts, but almost all of the genes encoding mitochondrial proteins have been transferred to the nuclear genome during evolution of eukaryotes (Dyall et al. 2004; Gray 2014). Therefore, mitochondria-to-nucleus signaling is critical for maintenance of mitochondrial functions and thereby cellular homeostasis (Cannino et al. 2007; Eisenberg-Bord and Schuldiner 2017).
Mitochondria have dedicated molecular chaperones and proteases that promote proper protein folding, complex assembly and quality control. In the baker’s yeast Saccharomyces cerevisiae there are many nuclear-encoded mitochondrial heat shock protein (mtHSP) genes, for example: the HSP70/DnaK homolog SSC1, the HSP40/DnaJ homolog MDJ1, and the chaperonin pair HSP60 and HSP10. These mitochondrial proteins are involved in mitochondrial protein import, protein folding and assembly and proteolysis. Loss of these functions causes mitochondrial DNA (mtDNA) loss and a decrease in mitochondrial-dependent metabolism, and SSC1, HSP60 and HSP10 are essential genes (Craig et al. 1989; Hohfeld and Hartl 1994; Ikeda et al. 1994; Laloraya et al. 1994; Reading et al. 1989).
Heat shock causes protein denaturation and HSP has critical roles in refolding and clearance of denatured protein (Saarikangas and Barral 2016). Heat shock transcriptional factor (HSF) mediates aspects of the heat shock response in all eukaryotic cells. In S. cerevisiae, HSF is encoded by a sole HSF1 gene. Upon heat shock, HSF is activated and induces the expression of the genes encoding heat shock proteins (HSPs) resident in various intracellular compartments via binding to the heat shock element (HSE). The perfect minimum consensus HSE sequence is “nGAAnnTTCnnGAA”; where n is any nucleotide (Mager and De Kruijff 1995; Mager and Ferreira 1993). It is assumed that the HSF/HSE system is activated by the accumulation of unfolded proteins in the cytoplasm (Craig and Gross 1991). The cytoplasmic Hsp70 negatively regulates HSF activity (Conn and Qian 2011; Richter et al. 2010). In the endoplasmic reticulum (ER), when the secretion is inhibited, unfolded proteins accumulated in the ER induce expression of ER HSP genes by the different system (Kohno 2007; Schroder and Kaufman 2005). Thus, the protein quality controls (PQCs) in the cytoplasm and the ER are well documented.
In contrast, the PQC in mitochondrial (mtPQC) is less understood. Unfold protein accumulation within the mitochondrial matrix induces nuclear genes encoding mtHSPs (Martinus et al. 1996; Zhao et al. 2002). This observation suggests the existence of a mitochondrial PQC (mtPQC). We previously showed that SSC1 and MDJ1 are regulated by the HSF/HSE system in budding yeast (Tachibana et al. 2002). Here we show that Ssc1–Mdj1 conversely downregulates HSF. Furthermore, we found that HSF is resident on mitochondria, suggesting that HSF directly monitors mtPQC.
Results
Ssc1 negatively regulates HSF activity
First, we examined whether the mitochondrial HSP70 Ssc1 downregulates HSF, like cytoplasmic HSP70. Importantly, overexpression of SSC1 drastically compromised heat activation of HSF (Fig. 1a), suggesting that Ssc1 negatively regulates HSF in a negative feedback manner. Consistently, SSC1-overexpressing cells were hypersensitive to heat shock: SSC1-overexpressing cells were lethal, when transferred from 20 °C to 37 °C, but not from 30 °C to 37 °C (Fig. 1b).
Ssc1 negatively regulates HSF. a Exponentially growing cells of the wild-type strain (KMY1005) harboring a plasmid pSCU69 (p424GPD-SSC1) or an empty vector pSCU60 (p424GPD) were preincubated at 23 °C (white bars) and then transferred to 39 °C for one hour (black bars). Cells possessed a plasmid pSCU23 (pHSE-CYC1-lacZ) and HSF activity was monitored by β-galactosidase activity. b The same cells used in panel (a) were preincubated at 20 °C or 30 °C and then incubated on agar plates at 37 °C for 2 days. c Cells of (1) the wild-type (W303·), (2) SCU27 (mdj1∆ ρ0), and (3) SCU29 (ρ0) harboring pSCU69 (p424GPD-SSC1) were preincubated in the liquid media at 20 °C and then incubated on agar plates at 37 °C for 3 days. d Cells of the wild-type strain (KMY1005) were preincubated at 30 °C in the liquid media and then incubated on agar plates at 37 °C for 3 days. Cells possess the empty vectors pSCU60 (p424GPD) and pSCU66 (p423GPD) (Vector; 1), pSCU69 (p424GPD-SSC1) and p423GPD (SSC1; 2), p424GPD and pSCU96 (p423GPD-MDJ1) (MDJ1; 3), or p424GPD-SSC1 and p423GPD-MDJ1 (SSC1 MDJ1; 4)
To assess whether SSC1 overexpression-mediated heat shock lethality is related to the role of Ssc1 as mitochondrial HSP70, we examined effects of deletion of the co-chaperon gene MDJ1 cancels on the lethality. Loss of Mdj1 produces ρ0 cells without mtDNA (Rowley et al. 1994). SSC1 overexpression failed to confer heat shock lethality to ρ0 cells lacking MDJ1, but it again rendered ρ0 cells heat shock hypersensitive when Mdj1 was added back (Fig. 1c). These findings indicated that SSC1 overexpression-mediated heat shock lethality is dependent on the role of Ssc1 as mitochondrial HSP70.
Conversely, co-overexpression of MDJ1 exaggerated heat shock hypersensitivity in SSC1-overexpressing cells (Fig. 1d), while a sole overexpression of MDJ1 compromised neither HSF activity nor heat shock sensitivity (Fig. 1d and data not shown). These indicated that enhancement in a cooperative action of Ssc1 and Mdj1 caused heat shock sensitivity. Taken together, after heat shock an excessive mtHSP activity compromised heat shock response via HSF activation.
HSF is colocalized with mitochondria
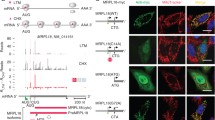
Interestingly, it was reported that HSF was co-purified with isolated mitochondria (Sickmann et al. 2003). This suggests that HSF is associated to the mitochondrial outer membrane. In the case of budding yeast, HSF (monitored by Hsf1–GFP) was predominantly localized in the nucleus even in normal conditions (Haitani and Takagi 2008) (Fig. 2a). However, we found that a small portion of HSF was also colocalized with mitochondria (monitored by mitochondrial RFP) (Fig. 2b). It is most likely that HSF is peripherally associated to the cytoplasmic side of the mitochondrial outer membrane.
HSF is colocalized with mitochondria. a Cells of strain SCU2544 (HSF1–GFP) were incubated at 23 °C. Cells were fixed with ethanol and stained with DAPI. GFP, DAPI and cell images were captured. b Cells of strain SCU2544 (HSF1–GFP) harboring a plasmid pSUC1355 (p416GPD-mtRFP) were incubated at 23 °C. Cells were not fixed to prevent reduction in weak Hsf1–GFP signals. GFP, RFP and cell images were captured. White arrowheads indicate Hsf1–GFP colocalized on mitochondria
Mitochondrial localization of HSF is regulated by heat shock and Ssc1
We detected mitochondrial localization of HSF in about 60% cells under non-stressed conditions (Fig. 3a, b, vector). Interestingly, heat shock promoted mitochondrial localization of HSF. This suggested that mitochondrial HSF is implicated in heat shock response. Furthermore, SSC1 overexpression reduced mitochondrial localization of HSF in non-stressed and heat-treated cells (Fig. 3a, b, SSC1). This demonstrated that Ssc1 negatively regulated mitochondrial localization of HSF. Collectively, heat shock recruited HSF to mitochondria, but the excessive Ssc1 repressed the recruitment. We noted that SSC1 overexpressing caused delocalization of mitochondria to plasma membrane-proximal regions and increase in cell size (Fig. 3b). These findings suggested that the excessive Ssc1 affected mitochondrial functions and cell growth.
Mitochondrial localization of HSF is regulated by heat shock and SSC1 overexpression. a, b Cells of strain SCU2544 (HSF1–GFP) harboring a plasmid pSUC1355 (p416GPD-mtRFP) in combination with the empty vector (pSCU60) or pSCU69 (p424GPD-SSC1) were preincubated at 23 °C and then exposed to 39 °C for 10 min. The percentage of cells with mitochondria colocalized on mitochondria is shown in (b). More than 100 cells were analyzed, and experiments were repeated at least two times to confirm reproducibility of the results. p values (two tailed) were obtained by Fisher’s exact test. *p < 0.001. ns not significant
The mitochondrial protein transporter mediates the mtHSP–HSF signaling
Because Ssc1 localized at the inside of mitochondria affect HSF located at the outside of mitochondria, Ssc1 should regulate HSF indirectly. Ssc1 is involved not only in folding of unfolded proteins but also in import of mitochondrial proteins, and hence the status of Ssc1 function affects mitochondrial protein import activity (Herrmann and Neupert 2000; Voos et al. 1999). We suspected whether the excessive Ssc1 repressed HSF by modulating mitochondrial protein import. When translocation of mitochondrial preproteins is retarded on the mitochondrial outer membranes, where HSF might sense this unwanted event.
To assess this idea, we modulated activity of the mitochondrial protein transport. Tom40 is an essential component of the mitochondrial protein translocase of the outer membrane (TOM) complex and is directly associated with mitochondrial preproteins (Endo and Kohda 2002; Pfanner and Chacinska 2002; Shiota et al. 2015). Overexpression of TOM40 again impaired heat activation of HSF (Fig. 4a). Conversely, loss of Tom40 activated HSF even in non-stressed conditions (Fig. 4b). These findings indicated that Tom40 negatively regulated HSF activity, like Ssc1.
The mitochondrial translocase Tom40 negatively regulates HSF. a Cells of the wild-type strain (KMY1005) harboring a plasmid pSCU95 (p424GAL1-TOM40) or an empty vector pSCU62 (p424GAL1) were incubated in the galactose-based medium at 23 °C and exposed to 39 °C for 40 min. Cells possessed a plasmid pSCU23 (pHSE-CYC1-lacZ) and HSF activity was monitored by β-galactosidase activity. b Cells of a strain SCU59 (GAL1-TOM40) harboring a plasmid pSCU23 (pHSE-CYC1-lacZ) were incubated in the glucose-based medium for shutoff of expression of TOM40 for 24 h. Culture was done at 30 °C throughout. HSF activity was monitored by β-galactosidase activity. Cell growth was arrested after 24 h, indicating that Tom40 are efficiently depleted. Cells of the wild-type strain (W303·) treated with the same procedures were used as the control. c Cells of SCU19 (SSC1) and SCU20 (ssc1-ts) strains harboring a plasmid pSCU23 (pHSE-CYC1-lacZ) were cultured at 23 °C and 30 °C. HSF activity was monitored by β-galactosidase activity. d Cells of strains SCU20 (ssc1-ts) and SCU19 (SSC1) harboring pSCU225 (p423GAL1-TOM40) or pSCU67 (p423GAL1) were incubated in the galactose-based medium at 30 °C. Cells possessed a plasmid pSCU23 (pHSE-CYC1-lacZ) and HSF activity was monitored by β-galactosidase activity
Similarly, Ssc1 dysfunction activated HSF even in non-stressed conditions (Fig. 4c), like depletion of Tom40. This supported that Ssc1 negatively regulates HSF. Furthermore, TOM40 overexpression cancelled this HSF activation induced by deficient Ssc1 (Fig. 4d). This indicated that Ssc1 negatively regulated HSF via the TOM translocase and supported the idea that the mitochondrial protein transport system mediates the mtHSP–HSF signaling.
Ssc1 preferentially impacts on SSC1 and MDJ1
Because Ssc1 and Mdj1 are transcriptionally regulated by HSF, the fact that the Ssc1–Mdj1 activity controls HSF indicates that Ssc1–Mdj1 activity should be autoregulated via HSF. In addition, Ssc1–Mdj1 activity might also affect other cellular HSPs via HSF. We assessed this idea. Consistent with the fact that HSF activity was repressed by SSC1 overexpression, heat induction of both mtHSP genes, SSC1 and MDJ1 were severely compromised by SSC1 overexpression (Fig. 5). However, cytoplasmic HSP genes (SSA3 and HSP12) were mildly impaired by SSC1 overexpression (Fig. 5). This indicated that the mtPQC signaling preferentially tuned expression of mtHSP via HSF. In the budding yeast S. cerevisiae Hsf1 is a sole HSF, and therefore, Hsf1 must take care of diverse HSP genes. HSF finely tunes each HSP gene according to different HSE contexts. SSC1 and MDJ1 might have specific HSEs in their promoter regions (see “Discussion”).
SSC1 overexpression represses HSF gene expression. Cells of the wild-type strain (KMY1005) harboring pSCU69 (p424GPD-SSC1) or the empty vector pSCU69 (p424GPD) were preincubated at 23 °C and then transferred to 37 °C for one hour. Cells possessed a plasmid pSCU34 (pSSC1-lacZ), pSCU35 (pMDJ1-lacZ), pSCU33 (pSSA3-lacZ) or pSCU64 (pHSP12-lacZ), and the promoter activity of each gene is were monitored by the β-galactosidase assay
SSC1 overexpression compromises mitochondrial function
Defects in mtHSP functions cause mtDNA loss and a decrease in mitochondrial metabolism (Craig et al. 1989; Hohfeld and Hartl 1994; Ikeda et al. 1994; Laloraya et al. 1994; Reading et al. 1989). Because SSC1 overexpression perturbed induction of mtHSP gene after heat shock (Fig. 5), we suspected whether SSC1 overexpression causes mtDNA loss and defects in mitochondrial metabolisms at high temperature. After the control cells harboring an empty vector were incubated at 30 or 37 °C, these cells possessed mtDNA stained by DAPI (Fig. 6a). However, SSC1-overexpressing showed no clear mtDNA signals, if they were precultured at 37 °C (Fig. 6a). Although mtDNA signals were still found in SSC1-overexpressing cells, even if these cells were precultured at 30 °C (Fig. 6a), these cells failed to grow on media containing a non-fermentable carbon source glycerol (Fig. 6b). This indicated that SSC1 overexpression diminished mitochondrial respiration even at normal temperature. We confirmed that SSC1-overexpressing cells precultured at 37 °C similarly showed no growth on the glycerol-based media (data not shown). Thus, SSC1 overexpression compromises mitochondrial function.
SSC1 overexpression compromises mitochondrial function. a Exponentially growing cells of the wild-type strain (KMY1005) harboring a plasmid pSCU69 (p424GPD-SSC1) or an empty vector pSCU60 (p424GPD) were preincubated on glucose-containing SD medium at 30 °C or 37 °C for two days. Thereafter, cells were incubated in YPAD medium and exponentially growing cells were stained with DAPI. DAPI and cell images were captured using a microscope. b The 30°C-precultured cells used in panel (a) were incubated in YPAD at 30 °C and then serially fivefold diluted cells were spotted from left to right on YPAD (glucose) or YPAGly (glycerol) plates. YPAD and YPAGly plates were kept at 30 °C for 1 days or 3 days, respectively
Discussion
This study showed that mtHSPs negatively regulates HSF. The HSF/HSE system is adopted to maintain mtPQC in yeast cells. It might be expected that the mtHSP abnormality caused respiration or metabolisms in mitochondria, stimulating HSF activity. However, inhibition of respiration by NaN3, and inhibition of mitochondrial ribosome-mediated translation by chloramphenicol and streptomycin led to no HSF activation (data not shown). In addition, the ρ0 strain showed no HSF activation (data not shown). These suggested that mere dysfunction of mitochondrial metabolism or respiration did not stimulate HSF activity.
We showed evidence that the mtHSP-translocase axis modulated HSF. Based on the results of this study, we propose a model as follows. In normal (non-stressed) conditions, Ssc1 is involved both in import of mitochondrial proteins and in folding of client proteins in mitochondria (Fig. 7a). However, when denatured proteins are increased in mitochondria (e.g., upon heat shock), a large portion of Ssc1 proteins should be recruited to the denatured client proteins to refold them (Fig. 7b). Subsequently, preproteins should be accumulated on TOM complex on the mitochondrial outer membrane, which might be sensed by mitochondria-associated HSF, causing HSF activation. By contrast, when excessive Ssc1 molecules are present, Ssc1-mediated protein transport is not impaired even after unfolded proteins accumulation in mitochondria, causing no proper HSF activation (Fig. 7c). Proper activities of mtHSP and the translocase should be critical for surveillance of mtPQC and accurate HSF activation to maintain mtPQC. Our model is different from recently proposed one: defective mitochondrial protein translocation system causes accumulation of mistargeted mitochondrial preproteins in the cytoplasm, evoking the unfolded protein response (Wrobel et al. 2015). It is most likely that these two distinct systems cooperatively and complementarily sense and maintain mtPQC.
Model in which HSF activity is regulated by the Ssc1-Tom40 axis in a negative feedback manner. a Ssc1 is involved in folding of unfolded proteins and mitochondrial protein import in normal conditions. Mitochondrial preproteins are properly incorporated into the mitochondria in these conditions. b After heat shock, most of Ssc1 molecules are devoted to refolding of the unfolded proteins accumulated in mitochondria, impairing Ssc1-mediated protein transport. Subsequently, preproteins are accumulated on the outside of the mitochondrial outer membrane. This is sensed by mitochondria-associated HSF, causing HSF activation. c When excessive Ssc1 molecules are present in mitochondria, Ssc1-mediated protein transport is not impaired after heat shock, causing insufficient activation of HSF
How does HSF on mitochondrial outer membrane senses preprotein status, directly or indirectly? We suspected that the yeast Hsp90 homolog Hsc82 might be involved in the mtPQC signaling to HSF on mitochondrial outer membrane, because HSP90 escorts the mitochondrial protein precursor to the TOM complex, in conjunction with cytoplasmic HSP70, and Hsc82 is co-purified with mitochondria and represses Hsf1 activity by unknown mechanisms (Duina et al. 1998; Sickmann et al. 2003). In mammalian cells, HSP90 binds and inactivates monomeric HSF in the absence of stress (Fan et al. 2006; Zou et al. 1998). These findings suggest a possibility that Hsc82 is located near the TOM complex and binds to HSF to transmit the mtPQC signal to HSF. However, we found no physical interaction of Hsc82 and its homolog Hsp82 with Hsf1 by pull-down assay (our unpublished data). In addition, GFP-tagged Hsp82 and Hsc82 showed a broad intracellular localization including the nucleus, but not a specific mitochondrial location (data not shown). Thus, we could not obtain evidence showing that Hsp82/Hsc82 directly represses Hsf1 on mitochondria. It is an important future work to reveal molecular features of mitochondrial HSF.
When unfolded proteins are accumulated specifically in mitochondria in some conditions, the mtPQC-HSF signaling should induce not only mtHSPs but also HSPs localized outside of mitochondria. This may rather be a desirable response, because unfolded protein accumulation in mitochondria impedes mitochondrial protein import activity, leading to accumulation of preproteins in the outside of mitochondria. Such accumulated preproteins in the cytoplasm may titrate out cytoplasmic HSP70. Thus, mtPQC signaling system might be important the cytoplasmic PQC, in addition mtPQC.
The excessive Ssc1 severely repressed SSC1 and MDJ1, as compared with SSA3 and HSP12 (Fig. 5), suggesting that mtPQC preferentially regulates HSF-mediated induction of mtHSP genes, although these HSPs are all controlled by HSF. We suspect whether SSC1 and MDJ1 might have specific HSEs (sequences and/or positions) to respond to the mtPQC, although these four genes similarly possess step-type HSEs, which contains two gaps (nTTCn(5 bp)nTTCn(5 bp)nTTCn) (Sakurai and Takemori 2007). Alternatively, another unidentified monitoring system may cooperatively induce mtHSP genes in response to the mtPQC.
It has been reported that unfold protein accumulation within the mitochondrial matrix induces mtHSP genes, but not HSP genes encoding stress proteins resident in the ER in mammalian cells (Martinus et al. 1996; Zhao et al. 2002). The bZIP transcription factor CHOP is responsible for the mammalian mtPQC (Zhao et al. 2002). In the case of C. elegans, mtPQC stress evokes the bZIP transcriptional factor ZC376.7-mediated gene induction of mitochondrial Hsp70 and Hsp60 and requires the transcription factor DVE-1, the small ubiquitin-like protein UBL-5, the mitochondrial matrix protease ClpP and the mitochondrial matrix peptide exporter haf-1 (Haynes et al. 2007; Haynes and Ron 2010). The authors presented a model whereby perturbation of the protein-folding environment in the mitochondrial matrix promotes ClpP-mediated generation of peptides whose haf-1-dependent export from the matrix contributes to mtPQC signaling across the mitochondrial membrane. We cannot exclude a possibility that there is an additional pathway for the mtPQC signal in yeast. Computational analyses predict that expression of SSC1 may be regulated by stress-responsible transcriptional activators, Gcr1 (required for expression of glycolytic genes), Rpn4 (required for expression of proteasome subunit genes), Yap1 (required for expression of antioxidative genes), and Zap1 (required for expression of genes involved in zinc uptake) (Qian et al. 2003). This prediction suggests that these transcriptional activators might also be involved in the mtPQC. Conversely, it is the most likely that higher eukaryotic cells might additionally possess HSF/HSE-dependent mtPQC signaling system, since the molecular mechanisms of the HSF/HSE system and mitochondrial protein import are highly conserved among eukaryotic cells. We believe that our study could open the door to evaluate evolutional conservation and diversity of mtPQC sensing systems.
Materials and Methods
Strains, plasmids and culture media
Strains and plasmids used are listed in Tables 1 and 2, respectively. For assay of promoter activity, the promoter region of HSP12 (− 560 to + 3, translation start site is + 1) was amplified by PCR using an Expand High-Fidelity PCR System (Roche). The fragment was inserted into a reporter plasmid pSEY101 (Mori et al. 1996) to fuse in frame to lacZ. Plasmid pHSE2BGY contains an HSE-CYC1-lacZ (Sorger and Pelham 1988). For overexpression of SSC1 and MDJ1, their open reading frame (ORF) regions were amplified by PCR. Each fragment was inserted downstream of GPD promoter of a plasmid p424GPD and p423GPD (Mumberg et al. 1995), or GAL1 promoter of a plasmid p423GAL1 (Mumberg et al. 1994). Glucose-based synthetic medium (SD), and galactose-based synthetic medium (SGalR) were prepared as described previously (Tachibana et al. 2002). Glucose-containing YPAD medium (YPD containing 0.01% adenine) was prepared using a standard method. For assessment of mitochondrial respiratory activity of cells, glycerol (5%) was used instead of glucose. YPAGly contained 5% glycerol instead of glucose.
Microscopic assay
Exponentially growing cells expressing green fluorescent protein (GFP)- and red fluorescent protein (RFP)-tagged proteins were used. For staining with 4′,6-diamidino-2-phenylindole (DAPI; Dojin #D212, Tokyo, Japan), the cells were fixed with 70% ethanol for 10 s. After washing with distilled water, cells were stained with DAPI at 1 µg/ml for 10 s. Cells were viewed a Carl Zeiss Axio Imager M1 microscope (100× objective) and a cooled CCD camera (Carl Zeiss AxioCam MRm, Jena, Germany). For obtain Fig. 3b, more than 100 cells were analyzed, and experiments were repeated at least two times to confirm reproducibility of the results. We used Fisher’s exact test.
β-Galactosidase assay
Cells at mid-log phase were harvested. Assays of β-galactosidase activity in extracts of yeast cells were measured and expressed in Miller units as described (Tachibana et al. 2002). Values are expressed as the mean ± standard deviation (SD) of duplicate determinations of three independent yeast transformants.
References
Cannino G, Di Liegro CM, Rinaldi AM (2007) Nuclear-mitochondrial interaction. Mitochondrion 7:359–366
Conn CS, Qian SB (2011) mTOR signaling in protein homeostasis: less is more? Cell Cycle 10:1940–1947
Craig EA, Gross CA (1991) Is hsp70 the cellular thermometer?. Trends Biochem Sci 16:135–140
Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM (1989) SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol 9:3000–3008
Duina AA, Kalton HM, Gaber RF (1998) Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem 273:18974–18978
Dyall SD, Brown MT, Johnson PJ (2004) Ancient invasions: from endosymbionts to organelles. Science 304:253–257
Eisenberg-Bord M, Schuldiner M (2017) Ground control to major TOM: mitochondria-nucleus communication. FEBS J 284:196–210
Endo T, Kohda D (2002) Functions of outer membrane receptors in mitochondrial protein import. Biochim Biophys Acta 1592:3–14
Fan AC, Bhangoo MK, Young JC (2006) Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J Biol Chem 281:33313–33324
Gray MW (2014) The pre-endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harb Perspect Biol 6:a016097
Haitani Y, Takagi H (2008) Rsp5 is required for the nuclear export of mRNA of HSF1 and MSN2/4 under stress conditions in Saccharomyces cerevisiae. Genes Cells 13:105–116
Haynes CM, Ron D (2010) The mitochondrial UPR—protecting organelle protein homeostasis. J Cell Sci 123:3849–3855
Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D (2007) ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 13:467–480
Herrmann JM, Neupert W (2000) Protein transport into mitochondria. Curr Opin Microbiol 3:210–214
Hill S, Van Remmen H (2014) Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol 2:936–944
Hohfeld J, Hartl FU (1994) Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J Cell Biol 126:305–315
Hu F, Liu F (2011) Mitochondrial stress: a bridge between mitochondrial dysfunction and metabolic diseases? Cell Signal 23:1528–1533
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691
Ikeda E, Yoshida S, Mitsuzawa H, Uno I, Toh-e A (1994) YGE1 is a yeast homologue of Escherichia coli grpE and is required for maintenance of mitochondrial functions. FEBS Lett 339:265–268
Kohno K (2007) How transmembrane proteins sense endoplasmic reticulum stress. Antioxid Redox Signal 9:2295–2303
Laloraya S, Gambill BD, Craig EA (1994) A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc Natl Acad Sci USA 91:6481–6485
Lasserre JP, Dautant A, Aiyar RS, Kucharczyk R, Glatigny A, Tribouillard-Tanvier D, Rytka J, Blondel M, Skoczen N, Reynier P, Pitayu L, Rotig A, Delahodde A, Steinmetz LM, Dujardin G, Procaccio V, di Rago JP (2015) Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis Model Mech 8:509–526
Mager WH, De Kruijff AJ (1995) Stress-induced transcriptional activation. Microbiol Rev 59:506–531
Mager WH, Ferreira PM (1993) Stress response of yeast. Biochem J 290:1–13
Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ (1996) Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem 240:98–103
Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T (1996) Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1:803–817
Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768
Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122
Nakai M, Kato Y, Ikeda E, Toh-e A, Endo T (1994) Yge1p, a eukaryotic Grp-E homolog, is localized in the mitochondrial matrix and interacts with mitochondrial Hsp70. Biochem Biophys Res Commun 200:435–442
Okamoto K, Kondo-Okamoto N, Ohsumi Y (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17:87–97
Pfanner N, Chacinska A (2002) The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim Biophys Acta 1592:15–24
Qian J, Lin J, Luscombe NM, Yu H, Gerstein M (2003) Prediction of regulatory networks: genome-wide identification of transcription factor targets from gene expression data. Bioinformatics 19:1917–1926
Reading DS, Hallberg RL, Myers AM (1989) Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature 337:655–659
Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40:253–266
Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77:249–259
Saarikangas J, Barral Y (2016) Protein aggregation as a mechanism of adaptive cellular responses. Curr Genet 62:711–724
Sakurai H, Takemori Y (2007) Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: binding specificities of yeast HSFs and human HSF1. J Biol Chem 282:13334–13341
Schroder M, Kaufman RJ (2005) ER stress and the unfolded protein response. Mutat Res 569:29–63
Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, Elofsson A, Tomii K, Horton P, Wiedemann N, Pfanner N, Lithgow T, Endo T (2015) Molecular architecture of the active mitochondrial protein gate. Science 349:1544–1548
Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100:13207–13212
Sorger PK, Pelham HR (1988) Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855–864
Tachibana T, Astumi S, Shioda R, Ueno M, Uritani M, Ushimaru T (2002) A novel non-conventional heat shock element regulates expression of MDJ1 encoding a DnaJ homolog in Saccharomyces cerevisiae. J Biol Chem 8:22140–22146
Voos W, Martin H, Krimmer T, Pfanner N (1999) Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta 1422:235–254
Westermann B, Prip-Buus C, Neupert W, Schwarz E (1995) The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J 14:3452–3460
Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, Januszewicz E, Dziembowski A, Koblowska M, Warscheid B, Chacinska A (2015) Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524:485–488
Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ (2002) A mitochondrial specific stress response in mammalian cells. EMBO J 21:4411–4419
Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94:471–480
Acknowledgements
We thank Toshiya Endo, Walter Neupert, Sabine Rospert, Bent Jakobsen, Dennis Winge, Kazutoshi Mori, Shuh-ichi Nishikawa, Takehiko Shibata, Markus Proft, Ramón Serrano, and Koji Okamoto for generous gifts of materials. We especially thank Tomohusa Tachibana and Yoshihiro Kato for execution of preliminary experiments. We give special thanks to Toshiya Endo and Shuh-ichi Nishikawa, and laboratory members of TU for helpful discussions. BY4741 was provided by the National Bio-Resource Project (NBRP) of the MEXT, Japan.
Author information
Authors and Affiliations
Contributions
TU conceived and initiated the project. TU designed the experiments. NK, YH, and TU executed the experiments and analyzed the data. NK, YH, and TU discussed the data. TU wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Koike, N., Hatano, Y. & Ushimaru, T. Heat shock transcriptional factor mediates mitochondrial unfolded protein response. Curr Genet 64, 907–917 (2018). https://doi.org/10.1007/s00294-018-0809-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0809-9