Abstract
Saccharomyces cerevisiae vacuoles are functionally analogous to mammalian lysosomes. Both also serve as physical platforms for Tor Complex 1 (TORC1) signal transduction, the master regulator of cellular growth and proliferation. Hygromycin B is a eukaryotic translation inhibitor. We recently reported on hygromycin B hypersensitive (hhy) mutants that fail to grow at subtranslation inhibitory concentrations of the drug and exhibit vacuolar defects (Banuelos et al. in Curr Genet 56:121–137, 2010). Here, we show that hhy phenotype is not due to increased sensitivity to translation inhibition and establish a super HHY (s-HHY) subgroup of genes comprised of ARF1, CHC1, DRS2, SAC1, VPS1, VPS34, VPS45, VPS52, and VPS54 that function exclusively or inclusively at trans-Golgi and late endosome interface. Live cell imaging of s-hhy mutants revealed that hygromycin B treatment disrupts vacuolar morphology and the localization of late endosome marker Pep12, but not that of late endosome-independent vacuolar SNARE Vam3. This, along with normal post-late endosome trafficking of the vital dye FM4-64, establishes that severe hypersensitivity to hygromycin B correlates specifically with compromised trans-Golgi and late endosome interface. We also show that Tor1p vacuolar localization and TORC1 anabolic functions, including growth promotion and phosphorylation of its direct substrate Sch9, are compromised in s-hhy mutants. Thus, an intact trans-Golgi and late endosome interface is a requisite for efficient Tor1 vacuolar localization and TORC1 function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hygromycin B is an aminoglycoside antibiotic isolated from Streptomyces hygroscopicus that inhibits translation and is cytotoxic to budding yeast at concentrations >100 μg/ml (Eustice and Wilhelm 1984; Cabanas et al. 1978). At lesser concentrations, hygromycin B has been shown to be detrimental to certain yeast mutants. The growth of yeast mutants defective in glycosylation functions is inhibited at 50 µg/ml hygromycin B (Ali et al. 2004). Most interestingly, a small group from over 200 different vacuolar trafficking mutants have been reported to exhibit growth sensitivities at hygromycin B concentrations below 50 µg/ml, including a handful of vacuolar protein sorting (vps) mutants and endosome and vacuole interface 1 (env1) mutant (Conboy and Cyert 2000; Ali et al. 2004; Mukherjee et al. 2006; Takahashi et al. 2008). The vacuole of Saccharomyces cerevisiae is analogous to the mammalian lysosome. Both organelles carry out vital cellular functions, including macromolecule degradation, receptor downregulation, autophagy, and stress survival responses (Bowers and Stevens 2005; Ostrowicz et al. 2008; Veses et al. 2008; Li and Kane 2009; Armstrong 2010; Richards et al. 2012; Hecht et al. 2014; Klionsky and Eskelinen 2014; Feyder et al. 2015). We recently performed a genomic screen of the yeast haploid deletion strain collection of non-essential genes for hypersensitivity to hygromycin B (hhy) mutants; the small collection of hhy mutants is presented in Table 1 and all were shown to have defects in vacuolar trafficking, morphology and/or stress survival function (Banuelos et al. 2010). Furthermore, HHY gene collection is enriched for established gene functions at the Golgi and late endosome trafficking interface.
Many functional proteins of the vacuole enter the endomembrane system in the ER and transit through Golgi. Such membrane and lumenal vacuolar proteins are then delivered from trans-Golgi to the lysosomal vacuole by one of two conserved vesicular mechanisms. The late endosome-dependent pathway delivers vacuolar cargo in a clathrin-dependent manner through the late endosomal compartment (Vida et al. 1993; Payne and Schekman 1985; Seeger and Payne 1992). An alternative late endosome-independent pathway delivers vacuolar cargo in a clathrin-independent manner and utilizes the adaptor protein AP3 (Armstrong 2010; Gautreau et al. 2014; Saftig and Klumperman 2009; Li and Kane 2009; Kummel and Ungermann 2014; Ostrowicz et al. 2008; Richards et al. 2010; Viotti 2014; Wickner 2010). The vacuole membrane also houses conserved proteins and protein complexes that do not enter the endomembrane system, but associate with the vacuole membrane on its cytoplasmic side. These include protein complexes involved in regulation of vacuolar fusion/fission, including Yck3, Env7, and Vac8 (LaGrassa and Ungermann 2005; Manandhar et al. 2013; Manandhar and Gharakhanian 2014; Subramanian et al. 2006). They also include the conserved master regulatory signaling complex involved in regulation of cell proliferation and growth, Tor Complex 1 (TORC1) (reviewed in Loewith and Hall 2011). TORC1 positively regulates several anabolic processes including ribosome biogenesis, protein synthesis, and nutrient uptake; it also negatively regulates catabolic processes, such as autophagy and ubiquitin-mediated proteolysis [Hall 2008; Cybulski and Hall 2009; reviewed in (Reggiori and Klionsky 2013; Delorme-Axford et al. 2015)]. mTOR complex 1 (mTORC1) is known to activate stress-responsive transcription factors (reviewed in Aramburu et al. 2014; Ho 2015). Most recently, TORC1 pathway has been linked to neutral lipid homeostasis in yeast (Madeira et al. 2015) and vacuolar fragmentation (Stauffer and Powers 2016). TORC1 function, in turn, is regulated through nutrient sensing and by the RAGulator or its yeast counterpart, the EGO complex—both of which are also localized to the vacuolar/lysosomal membrane. TORC1 upregulation is associated with disease states, including cancer and diabetes (Chakrabarti et al. 2010). Tor is also involved in regulation of the aging process (McCormick et al. 2011). The TORC1 resident kinase, target of rapamycin (Tor), is a conserved phosphoinositide 3-kinase (PI3K)-related serine/threonine kinase whose direct substrate includes ribosomal kinase S6k in mammals and its homolog Sch9 in yeast (Rohde et al. 2008; Soulard et al. 2009; Neufeld 2010; Noda and Ohsumi 1998; Urban et al. 2007). While higher eukaryotes only have one TOR gene, yeast encodes Tor1 and Tor2 proteins, which can substitute for each other in TORC1. Additional proteins in TORC1 are Lst8, Kog1, and Tco89 (Wullschleger et al. 2006; Sturgill et al. 2008). Recently, it has been discovered that ubiquitin is involved in TORC1 function and regulation through binding with Kog1, a regulatory subunit of TORC1 (Jiang 2016). While mTORC1 is recruited to lysosomes and late endosomes in a nutrient sensitive manner, yeast TORC1 and Tor1 appear to constitutively reside on vacuole membranes (Betz and Hall 2013; Binda et al. 2009). All hhy mutants are also hypersensitive to rapamycin and caffeine (Banuelos et al. 2010)—two drug sensitivities suggestive of compromised TORC1 function (Kuranda et al. 2006; Reinke et al. 2006; reviewed in Aronova et al. 2007). This led us to hypothesize that TORC1 function may be compromised in hhy mutants and to further explore the state of vacuolar trafficking and TORC1 in these mutants.
Here, we report further studies of hhy mutants that establish a super hygromycin B hypersensitive subgroup (s-hhy mutants) and genetically, as well as phenotypically, point to a compromised trans-Golgi and late endosome interface in those mutants. We also show that Tor1p vacuolar localization and several parameters of TORC1 function are defective in s-hhy mutants. Taken together, our results implicate intact Golgi and late endosome interface in efficient Tor1p vacuolar localization and TORC1 anabolic functions. Our studies also support a therapeutic potential for subtranslation inhibitory levels of hygromycin B in regulation of TORC1 activity, as well as TORC1 hyperactivity in disease.
Materials and methods
Yeast strains and plasmids
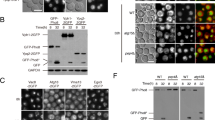
Saccharomyces cerevisiae deletion strains used in these studies were from the MAT-α haploid deletion strain library of non-essential genes derived from the parental strain BY4742 (Genotype: MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0) and were gifts from Dr. Gregory Payne, University of California, Los Angeles (UCLA). All yeast strains used in this study are listed in Table 2. Each mutant contains a PCR-based disruption of its open reading frame by integration of a KanMX4 module through homologous recombination (Wach et al. 1994). TOR1-3XGFP expressing strain MP52-2A (Genotype: [YL516] MATa; TOR1-D330-3xGFP) was a gift from Dr. Claudio De Virgilio and has been described previously (Binda et al. 2009). A PCR-based approach was used to obtain hhy∆ and vps16∆ mutants in a TOR1-3xGFP background (TOR1-GFP). Primers were designed approximately 50–100 bases upstream and downstream of each target gene (see Table 3 for primers used in this study). PCR amplification was performed with Phusion polymerase (NEB) using genomic DNA extracted from each respective deletion strain. Amplicons containing the kanamycin resistance cassette (KanMX4) flanked by gene-specific upstream and downstream sequences were subjected to agarose gel electrophoresis and purification using Zymoclean Gel DNA Recovery Kit (Zymo Research; Orange, CA) prior to separate transformations into TOR1-3XGFP background. Transformants were selected on YPD plates supplemented with 200-µg/ml geneticin (G418), a functional analog of kanamycin; correct kanamycin cassette insertion and gene deletion were confirmed by PCR using KanMX4 and locus-specific primer pairs. Plasmid p1462 encoding Sch9-HA was a gift from Dr. Claudio De Virgilio and has been described before in Urban et al. (2007). PEP12-RFP and VAM3-RFP expressing plasmids were gifts from Dr. Christian Ungermann (University of Osnabrück) and have been described before (Markgraf et al. 2009). Their auxotrophic markers were changed from Trp to Leu to be utilized in our strains. Yeast transformations were performed by the lithium acetate (PLATE) method.
Media and growth analyses
Cells were routinely grown (Sherman 2002) in YPD (2% peptone, 1% yeast extract and 2% dextrose) or supplemented selective (SM) medium with or without agar at 30 °C unless stated otherwise. Deletion strains with a KanMX4 cassette were selected for as described above. For liquid growth kinetics studies, overnight cultures were diluted to an OD600 = 0.2 in YPD and transferred to a 96-well microtiter plate. Growth of each strain was assessed in the presence of hygromycin B or cycloheximide. Sub-inhibitory concentrations for each drug were experimentally determined by growth assays of the parental wild type (BY4742α). Final concentrations for growth assessment in hygromycin B were 0, 10, 20, and 40 μg/ml. Final concentrations for growth assessment in cycloheximide were 0, 0.025, 0.05, and 0.1 μg/ml. PBS and DMSO served as vehicles for hygromycin B and cycloheximide treatments, respectively. The microtiter plates were incubated at 30 °C with aeration (220 rpm), and depending on the instrument used, OD600 or OD595 was determined at specified intervals as stated in legends of Figs. 1 and 2. Growths for each treatment condition were carried out in triplicates, and the average absorbance was used to make each graph. Each strain was assessed in at least three separate experiments with reproducible results. Growth assays were also performed for hhy mutants in the TOR1-GFP background and showed similar profiles to those in the parental background (data not presented).
hhy mutants exhibit a dichotomy of growth kinetics in the presence of hygromycin B. Wild-type and hhy∆ strains were grown in the presence of 0, 10, 20, and 40 μg/ml of hygromycin B and their OD595 were read every 2 h for 12 h and then at 24 h. hhy mutants were subsequently divided into two groups: those that did not show growth at any of the tested hygromycin B concentrations (super-hhy = s-hhy), and those that showed dose-dependent growth (d-hhy); they are listed along with their known gene functions in Table 1
Microscopic localization and vacuole morphology studies
To preserve fluorescence of fusion proteins, cells were grown and manipulated in amber conical tubes and microtubes. OD600 = 0.8 of cells were divided into hygromycin B-treated (10 μg/ml) and mock-treated samples. Total hygromycin B treatment time for all samples was 4 h in YPD at 30 °C after which cells were washed three times in PBS. 1-µl oxyrase was added for each 25 µl of cell suspension to preserve fluorescence, and samples were kept on ice until addition to concanavalin A-treated glass slides to improve cell adherence. In experiments where cells were also labeled with the endocytic and vacuolar marker dye FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridium dibromide] (Molecular Probes Inc., Eugene, OR), staining was carried out as described by Vida and Emr (1995) after the first hour of hygromycin B treatment or mock-treatment. All strains were viewed with an Olympus Fluoview 1000 confocal laser scanning system mounted on an inverted microscope (Olympus IX-81) and a 60× oil immersion UPLSAPO (NA 1.35, WD 0.15 mm) objective. The argon ion (488 nm) and blue/red diode (405 nm/635 nm) lasers were used for image capturing. Fluorescent and Nomarski optics images were captured from at least ten random fields for each strain and from three separate experiments; shown images represent the overall consensus of localization patterns visualized. Images were captured using Olympus’s FV-10-ASW 3.0 software, were transferred as JPG files to Photoshop CS2 for cropping, and were then merged to visualize co-localization. Vacuolar morphology for each strain was assessed by scoring at least 200 cells into four categories: Wild-type, fragmented, enlarged or absent. TOR1-GFP vacuolar localization was assessed in at least 200 cells, and the p value was calculated using a Chi-squared test.
Cell viability and recovery assays
Strains were grown overnight, diluted to OD600 = 0.2 in YPD, and divided into two tubes of untreated and treated with hygromycin B to a final concentration of 10 µg/ml. Cultures were then incubated at 30 °C with aeration for 4 h. At 4 h, 1.1 ml of cell culture was removed; one ml was stained with propidium iodide (Molecular Probes Inc., Eugene, OR) and 0.1 ml was used to measure OD600. Hygromycin B was removed from the remaining treated culture by washing 3 times in 3 ml YPD. Washed cells were then incubated for an additional 24 h at 30 °C with aeration, after which OD600 was assessed and an additional propidium iodide staining was performed. Viability was assessed by scoring at least 300 cells from three separate experiments. For plate assays of recovery, s-hhy cultures were treated with 10-µg/ml hygromycin B for 4 h, then washed and quantitatively spotted as tenfold serial dilution on YPD plates using a multichannel micropipetter as performed by Binda et al. (2009). For viability staining, 1 µl of 1 µg/ml propidium iodide was added to 1 ml of fresh cell culture in amber tubes, and cells were incubated at 30 °C for 30 min with aeration. Cells were collected by centrifugation, resuspended in PBS after three washes, and prepared for microscopy as above. Nomarski optics and fluorescence images were captured from random fields and used to count dead (fluorescing) and total cells. Heat killed cells served as positive staining control. For rapamycin recovery assays, experiments were performed as detailed for hygromycin B; cells were assessed immediately and 4 h after treatment, and then 24 and 48 h after removal of drug. All growth recovery assays were performed in triplicates and for at least two separate experiments, and standard error was calculated.
Sch9 phosphorylation analyses
p1462 plasmid expressing Sch9-HA has been previously described (Urban et al. 2007). Wild-type and s-hhy mutants were transformed with p1462, and Sch9-HA phosphorylation was analyzed in westerns with anti-HA monoclonal antibody (Cell Signaling Technology, Beverly, MA) and chemiluminescent approach (Thermo Scientific, Pittsburg, PA) using the Sch9 chemical fragmentation analysis described by Urban et al. (2007) with the following modifications. In addition to the previously described treatments, cells were treated with hygromycin B (10 µg/ml, 4 h) or in combination with cycloheximide (25 µg/ml, 30 min) where cycloheximide was added during the last 30 min of the hygromycin B treatment. Each mutant was analyzed at least 2 times and from two independent cell preparations. Lower exposure bands were quantified densitometrically using image J. To compare extent of Sch9 phosphorylation, mid-region of the fragmentation profile was used as it was consistently visualized in all mutant samples. Extent of Sch9 phosphorylation was calculated by expressing the densitometry value of the slowest migrating band (most highly phosphorylated) divided by that of the total values of the slowest and fastest migrating (non-phosphorylated) bands for each mutant and all treatments of the mutants. For comparison, this ratio was set to 100% for wild-type (WT) cells without treatment, and Sch9 phosphorylation of untreated mutants was expressed as percent of WT in parallel experiments.
Results
Deletion of HHY genes that function at trans-Golgi and late endosome interface leads to super-hypersensitivity to hygromycin B
hhy mutants are unable to grow in the presence of 25 μg/ml of hygromycin B (Banuelos et al. 2010). To determine the minimum inhibitory concentration of hygromycin B and study the growth kinetics of hhy mutants in a range of hygromycin B concentrations, we conducted 24-h growth assays in liquid media in the presence of 0, 10, 20, and 40 μg/ml hygromycin B (Fig. 1). As expected, hygromycin B did not affect growth of the parental wild-type strain at any of the tested concentrations. hhy mutants, however, exhibited a dichotomy of growth phentoypes in the presence of hygromycin B. arf1∆, chc1∆, drs2∆, sac1∆, vps1∆, vps34∆, vps45∆, vps52∆, and vps54∆ lacked significant growth under all three tested hygromycin B concentrations; this subset of hhy mutants will be referred to as super-hypersensitive to hygromycin B (s-hhy). bud32∆, dhh1∆, paf1∆, and tpd3∆ exhibited increased growth inhibition with increasing hygromycin B concentration and will be referred to as dose-dependent hypersensitivity to hygromycin B (d-hhy) mutants. s-hhy mutant arf1∆ repeatedly exhibited a slight dose-dependent growth at the lowest tested hygromycin B concentration. Consistent with previous reports, bud32∆, chc1∆, dhh1∆, drs2∆, paf1∆, tpd3∆, and vps34∆ strains showed compromised fitness relative to wild type in the absence of hygromycin B. In the course of these studies, two of the original hhy mutants were dismissed as their phenotypes could not be confirmed—yelo59w∆ and sbh2∆. In addition, several deletion mutants of genes involved in vesicular trafficking were re-evaluated at the original screen conditions due to their reported genetic and/or physical interactions with HHY genes. Of these, only vps1∆ lacked significant growth, was assessed to be a s-hhy mutant, and was included as such in subsequent studies.
The s-HHY and d-HHY genes and summaries of their known molecular functions (as gleaned from yeastgenome.org) are presented in Table 1.
The dichotomy of hygromycin B sensitivities of s-hhy and d-hhy deletion mutants corresponds to the functional dichotomy of the deleted genes. s-HHY genes are directly involved in vesicular trafficking functions, while d-HHY genes are not. Furthermore, the common denominator in the vesicular trafficking function of s-HHY gene products is their role, either exclusively or inclusively, at the trans-Golgi and late endosome interface. Thus, super-hypersensitivity to hygromycin B in hhy mutants correlates with compromised trans-Golgi and late endosome interface.
Alternatively, the observed s-hhy phenotype may have been due to increased sensitivity to the translation inhibitory function of hygromycin B. This possibility was probed via growth assays of wild type and hhy mutant strains using another potent translation inhibitor, cycloheximide. Cycloheximide concentrations sub- to minimally inhibitory to growth of the wild-type parental strain were experimentally determined to range from 0.025 to 0.10 µg/ml (data not presented). 24-h growth assays were performed with wild-type parental and hhy mutant strains in the presence of 0, 0.025, 0.05, and 0.1 µg/ml cycloheximide. Most hhy mutants did not show significantly increased growth sensitivities to cycloheximide relative to that observed with wild type, nor was there a dichotomy of sensitivities based on gene function as seen with hygromycin B treatments (Fig. 2). Thus, the unique hygromycin B hypersensitivity of hhy mutants does not appear to be due to increased vulnerabilities to translation inhibition. Taken together, these results suggest that the compromised trans-Golgi and late endosome interface of s-hhy mutants correlates with their super-hypersensitivity to hygromycin B.
Hygromycin B treatment of s-hhy mutants disrupts late endosome-dependent but not late endosome-independent post-Golgi trafficking
The vacuolar defects of hhy mutants, the absence of increased growth sensitivity to another potent translation inhibitor, and the common function of s-hhy mutants at the trans-Golgi and late endosome interface led us to hypothesize that a compromised trans-Golgi and late endosome interface is highly susceptible to hygromycin B in s-hhy mutants. To test this, we investigated the state of both late endosome-dependent and late endosome-independent trafficking in the s-hhy mutants. s-hhy mutants have been shown biochemically to have severe defects in correct trafficking and proteolytic processing of the common endosome-dependent vacuolar marker carboxypeptidase Y, resulting in secretion of significant levels of the enzyme (Bankaitis et al. 1986; Banta et al. 1988; Robinson et al. 1988; Rothman et al. 1989; Bonangelino et al. 2002). Therefore, we instead explored localization of Vam3 and Pep12 as respective markers of the two trafficking pathways by live cell imaging. Pep12-RFP and Vam3-RFP are high copy plasmids with two copies of DsRed attached to the N terminus as described in Markgraf et al. (2009). Vam3 is a Q-SNARE that resides on the vacuole membrane and is transported there by the endosome-independent pathway (reviewed in Ostrowicz, et al. 2008). Pep12 is a late-endosome Q-SNARE that is delivered to that compartment in a clathrin-dependent manner and serves as a late endosome marker (Becherer et al. 1996). WT and s-hhy mutant strains were transformed with plasmids constitutively expressing either Vam3-RFP or Pep12-RFP, and localization patterns of the two proteins were assessed in live cells mock-treated or treated for 4 h with 10 µg/ml hygromycin B, which is the lowest tested inhibitory dose for s-hhy mutants (Fig. 3). The 4-h treatment time was experimentally determined to be the shortest incubation that resulted in visible changes in slow-growing s-hhy mutants. As expected, mock-treated wild-type strain exhibited localization of Vam3-RFP to vacuolar membranes, which are marked as the crater boundaries in Nomarski optics imaging, and localization of Pep12-RFP to 1–3 punctate structures characteristic of the late endosome. In contrast, while the punctate staining pattern of Pep12-RFP was slightly compromised in mock-treated s-hhy mutants, the vacuolar staining of Vam3-RFP was not. Upon hygromycin B treatment, no changes were observed in Vam3-RFP nor Pep12-RFP localizations in wild-type strain. In contrast, within 4 h of hygromycin B treatment, Pep12-RFP punctate staining patterns were abrogated in the s-hhy mutants, while Vam3-RFP vacuolar localization remained undisturbed. These results indicate that in s-hhy mutants, Golgi and late-endosome interface is compromised and further disrupted at subtranslation inhibitory levels of hygromycin B, while late endosome-independent post-Golgi trafficking is not.
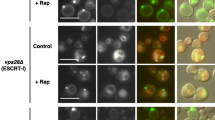
Hygromycin B treatment alters localization of late endosomal marker Pep12-RFP but not that of the late endosome-independent vacuolar SNARE Vam3-RFP. Localization of Vam3-RFP and Pep12-RFP was assessed in s-hhy∆ strains using confocal and Nomarski optics microscopy after 4 h of growth in the presence or absence of 10-µg/ml hygromycin B. Scale bar 5 μm for all panels
Vacuolar morphology and Tor1-GFP localization are disrupted in hygromycin B-treated s-hhy mutants
We have previously shown that hhy mutants are sensitive to caffeine and rapamycin suggestive of defects in TORC-1 signaling pathway (Banuelos et al. 2010). Caffeine is a phosphodiesterase inhibitor that has been reported to target yeast TOR Complex1 (TORC1) and TOR Complex2 (TORC2) (Guan et al. 2009; Reinke et al. 2006; Wanke et al. 2008). Rapamycin is an anti-fungal agent that directly targets TORC1 (Park et al. 2005; Kuranda et al. 2006; reviewed in Levin 2005; Aronova et al. 2007). Since TORC1 localizes to the vacuole (Betz and Hall 2013; Binda et al. 2009), and all HHY genes have been implicated in vacuolar function and/or trafficking, a defective TORC1 signaling due to vacuolar trafficking defects is plausible. We hypothesized that TORC1 vacuolar localization is defective and hypersensitive to hygromycin B in hhy mutants and investigated the localization of the TORC1 kinase Tor1. Tor1 and Tor2 are the interchangeable kinase subunits of TORC1 and, unlike TOR2, TOR1 is not an essential gene. Thus, we confirmed that tor1Δ mutants are not hypersensitive to hygromycin B in the wild type background (Fig. 1). In addition, deletion of TCO89, another non-essential component of TORC1, did not lead to hygromycin B hypersensitivity (Fig. 1). Together, these controls indicate that the absence of Tor1 (and its presumable replacement by Tor2) or a structurally compromised TORC1 is not sufficient for hygromycin B hypersensitivity. For the localization studies, we utilized a strain in which TOR1 gene is endogenously tagged with 3x-GFP (Tor1-GFP) in a central region, and the fusion protein is active and localizes to the vacuole membrane (Binda et al. 2009). HHY genes were individually deleted in that background, and Tor1-GFP localization was assessed by confocal microscopic live cell imaging after 4 h of either mock-treatment or treatment with 10-µg/ml hygromycin B. Cells were also stained with the vital endocytic vacuolar membrane marker FM4-64 to assess the endocytic pathway, mark the vacuolar membrane and quantitatively score for vacuolar morphology for each mutant. FM4-64 is a lipophilic styryl vital dye that localizes to the vacuolar membrane through the endocytic pathway and moves from plasma membrane through early endosomes and late endosomes to the vacuole (Vida and Emr 1995). Representative images from treated and mock-treated strains are shown in Fig. 4, and the quantified results of scored cells for Tor1-GFP localization and vacuolar morphology are summarized in Table 4.
Tor1-GFP localization and vacuolar morphology are altered and sensitive to hygromycin B treatment in s-hhy mutants. HHY genes were individually deleted in a strain in which endogenous TOR1 was tagged with 3x-GFP (TOR1-GFP). Tor1-GFP localization was then assessed by confocal microscopy and Nomarski optics after 4 h of treatment with 10 µg/ml hygromycin B. To assess vacuolar morphology and Tor1-GFP vacuolar co-localization, cells were stained with the endocytic vacuolar membrane marker FM4-64. Scale bar 5 μm for all panels. >150 cells were scored for each mutant from multiple trials, and shown in Table 4. a s-hhy∆ strains and b d-hhy∆ strains
In wild-type cells, in the presence or absence of hygromycin B, vacuole morphology is unchanged and both Tor1-GFP and FM4-64 localize to vacuole membranes. hhy mutants, however, exhibited two distinct Tor1-GFP localization and vacuolar morphology phenotypes upon hygromycin B treatment; this dichotomy again corresponds to their s-hhy and d-hhy classifications. In most s-hhy mutants, Tor1-GFP vacuolar localization was slightly compromised in untreated cells and further perturbed in the presence of hygromycin B. s-hhy mutants also exhibited their previously reported aberrant vacuolar morphologies in the absence of hygromycin B, which were then exaggerated upon treatment. As in s-hhy mutants, Tor1-GFP vacuolar localization was slightly compromised in untreated d-hhy cells. However, in contrast to s-hhy mutants, both Tor1-GFP localization and vacuolar morphology were not further compromised in hygromycin B-treated d-hhy strains. Studies of Vam3-RFP or Pep12-RFP localization in TOR1-GFP background confirmed that hygromycin B treatment (10 µg/ml for 4 h) disrupts Tor1-GFP and Pep12-RFP localizations but not that of Vam3-RFP (data not presented).
The onset of severe vacuolar morphology defects upon hygromycin B treatment supports a general block in endosome-dependent vacuolar trafficking that includes, but may not be limited to, localization of Pep12 and Tor1. The undisturbed pattern of FM4-64 localization to the vacuole in hygromycin B-treated hhy strains is indicative of undisturbed bulk endocytosis which includes late endosome to vacuole stage of trafficking. Taken together, these results further pinpoint the trans-Golgi and late endosome interface specifically as the compromised stage of vacuolar trafficking in s-hhy mutants and as correlative with hygromycin B super-hypersensitivity. Furthermore, as Tor1 trafficking in s-hhy mutants is defective and hypersensitive to hygromycin B, our results also implicate intact trans-Golgi and late endosome interface as a requisite for efficient Tor1 vacuolar localization.
Hygromycin B-treated s-hhy mutants are viable but exhibit growth arrest and defects in recovery from growth arrest
Since our results established compromised Tor1 vacuolar localization, we next probed whether TORC1 function was compromised and/or hypersensitive to hygromycin B in hhy mutants. First, we assessed whether the observed lack of growth of mutants in 10-µg/ml hygromycin B is due to growth arrest indicative of defective TORC1 signaling or whether it is due to cell death. To assess cell death, a viability assay was performed using propidium iodide, which only penetrates dead cells due to their leaky membranes. Cells were stained with propidium iodide after the routinely used 4-h hygromycin B treatment and were subsequently analyzed by epifluorescence and stained cells were quantified by scoring (Fig. 5a). Hygromycin B-treated hhy mutants showed slight or no drop in cell viability relative to their mock-treated counterparts, implicating growth arrest in the presence of hygromycin B.
Hygromycin B-treated s-hhy strains are viable and defective in recovery from hygromycin B-induced growth arrest. a Viability of hhy mutants after 4 h of treatment with 10 µg/ml hygromycin B was assessed microscopically using propidium iodide (PI) staining. Percent viability is the number of live (unstained) cells over total (stained + unstained) cells counted and is shown for treated (black bars) and untreated (white bars) cells. b Ability of hhy∆ strains to recover from hygromycin B-induced growth arrest was assessed by determining cell density after 4 h of treatment or mock-treatment with 10-µg/ml hygromycin B (white bars) and 24 h after removal of hygromycin B (black bars). c s-hhy cultures were treated with 10 µg/ml hygromycin B for 4 h, then washed and spotted as tenfold serial dilutions on YPD plates. d Ability of s-hhy∆ strains to recover from rapamycin-induced growth arrest was assessed by determining cell density immediately and after 4 h of 200 ng/ml rapamycin treatment and also 24 and 48 h after removal of the drug. Parallel untreated samples are denoted as NT. vps16∆ strain served as a control growth recovery defective mutant
Deletion of individual components of TORC1, or those of the EGO complex which regulates TORC1, results in inability to recover from rapamycin-induced growth arrest (Dubouloz et al. 2005). More recently, EGO complex–TORC1 interactions were shown to be perturbed in vacuolar trafficking Vps-C mutants as assessed by their defect in recovery from rapamycin-induced growth arrest (Kingsbury et al. 2014). To evaluate the state of EGO complex–TORC1 interactions in hhy mutants, we assessed the ability of hhy mutants to recover from both hygromycin B-induced and rapamycin-induced growth arrest. Wild-type and mutant strains were either mock-treated or treated with 10-μg/ml hygromycin B for 4 h, the drug was removed by multiple washes, and cells were resuspended in YPD without hygromycin B and incubated with aeration for another 24 h. The OD600 of each individual culture was read immediately following 4-h incubation and again after 24-h post-treatment (Fig. 5b). All strains remained viable 24-h post-hygromycin B treatment as assessed by propidium iodide staining (data not presented). In wild-type cells, both treated and mock-treated cells showed equivalent growth 24-h post-hygromycin B treatment. Interestingly, hhy mutants once again exhibited a dichotomy in cell cycle arrest recovery that corresponded to that seen for hygromycin B hypersensitivity and Tor1-GFP localization. Hygromycin B-treated s-hhy mutants showed strong inhibition of growth recovery following hygromycin B treatment relative to their mock-treated counterparts, while d-hhy mutants did not. Growth recovery defects of s-hhy mutants were confirmed by quantitative growth assays of serially diluted strains on rich media following mock or hygromycin B treatment (Fig. 5c). In both liquid and solid media growth assays, s-hhy mutant arf1Δ exhibited an intermediate growth defect relative to other s-hhy’s. To assess whether the growth recovery defect of s-hhy mutants is unique to hygromycin B treatment, we also probed their growth recovery following rapamycin treatment (Fig. 5d). With the exception of chc1∆ and vps34∆ which exhibited full inhibition of growth recovery comparable to that observed in the control class C vps16 mutant, s-hhy mutants exhibited a wide range of partial growth recovery defects following rapamycin treatment.
The viability and recovery results implicate compromised cell cycle progression and growth arrest recovery in s-hhy mutants, both of which are hallmarks of compromised TORC1 function. s-hhy mutants exhibited a wide range of growth recovery defect following rapamycin treatment and uniform severe recovery defect following hygromycin B treatment, suggesting that the observed super-hypersensitivity is unique to hygromycin B.
TORC1 kinase function is compromised in s-hhy mutants
Since our results suggest TORC1 signaling defects, we next assessed kinase activity of TORC1 directly and analyzed the phosphorylation of Sch9, a direct substrate of TORC1 (Urban et al. 2007). Sch9 is the yeast equivalent of mTORC1 substrate S6 kinase and a regulator of translational initiation and sphingolipid metabolism in S. cerevisiae (Urban et al. 2007; Swinnen et al. 2014). We followed the Sch9 mobility upshift assay approach reported by Urban et al. (2007), where due to the large size of Sch9, partial proteolytic cleavage products of exogenously expressed Sch9-HA were generated by 2-nitro-5-thiocyanobenzoic acid (NTCB), and fragments were assessed for phosphorylation-dependent upshift in SDS–PAGE. Figure 6a shows the migration profile of full length and partial degradation products of Sch9 that retain the HA-tagged c-terminus under various drug treatments for the wild-type parental strain. Consistent with previous reports, phosphorylation of both full length and c-terminal partial degradation products of Sch9-HA was severely inhibited in the presence of rapamycin. Also consistent with previous reports, Sch9-HA phosphorylation was maximized in the presence of the translation inhibitor cycloheximide (Fig. 6a), presumably due to upregulation of TORC1 as free intracellular amino acid levels rise during translation inhibition, as suggested by Urban et al. (2007). As expected, Sch9 phosphorylation was not sensitive to 10 µg/ml hygromycin B in wild type, and when wild-type cells were treated with hygromycin B and cycloheximide simultaneously, there was no significant difference in Sch9 phosphorylation compared to cycloheximide-only treatment (Fig. 6a).
Tor1 kinase function is compromised in s-hhy mutants. Wild type and s-hhy strains expressing a plasmid-based copy of Sch9-HA tag were grown exponentially and treated for 30 min with rapamycin (200 ng/ml), 30 min with cycloheximide (25 µg/ml), or 4 h with hygromycin B (10 µg/ml) with cycloheximide (25 µg/ml) added for the final 30 min. Proteins were extracted as described by Urban et al. (2007) and treated or mock-treated with NTCB for cleavage. NTCB samples were analyzed by SDS–PAGE, and immunoblots were probed with anti-HA antibody. a Full migration profile of Sch9 in wild-type cells under the various treatments. Left panel contains NTCB mock-treated samples. Right panel contains samples treated with NTCB; the boxed mid-region was consistently visualized in all mutant strains and was used for quantifications. b The region corresponding to the boxed area in 6A is presented. Extent of phosphorylation was quantified by densitometric analysis of this region using Image J software as described in “Materials and methods”. 0 No treatment, R rapamycin, C cycloheximide, CH cycloheximide plus hygromycin B. c For comparison of phosphorylation among untreated samples, extent of phosphorylation was set to 100% for untreated WT cells and phosphorylation of mutants was expressed relative to WT in parallel experiments. Data include two independent trials per mutant
Next, s-hhy mutants were assessed similarly in multiple replicates. Different s-hhy mutants vary in their growth rates and in protein yields from wild-type samples processed in parallel. Therefore, we used the mid-range of partial degradation products highlighted in the box in Fig. 6a to compare extent of Sch9 phosphorylation, since they were consistently visible in mutant strains. Low exposure autoradiographs were subjected to densitometry, and percent full phosphorylation was calculated as the top band of the boxed region over the sum of top plus bottom bands. A typical experimental set is shown in Fig. 6b and percent full phosphorylation is expressed under each treatment lane. For untreated samples, extent of full phosphorylation from at least two different experimental sets was normalized to that of untreated wild-type Sch9 and expressed in Fig. 6c. These results show that Sch9 phosphorylation is significantly compromised in s-hhy mutants even in the absence of hygromycin B treatment. sac1Δ, vps1Δ, vps34Δ, and vps52Δ showed the largest defect in Sch9 phosphorylation levels at consistently <40% of wild type. As in wild-type cells, phosphorylation of Sch9 was lost in s-hhy mutants in the presence of the TORC1 inhibitor rapamycin and was significantly increased in the presence of cycloheximide, indicating that these parameters of TORC1 regulation are not disturbed in s-hhy mutants. We were not able to see significant or consistent changes in Sch9 phosphorylation in s-hhy mutants upon the same duration and extent of treatment with hygromycin B alone (data not presented), presumably due to their already compromised TORC1 activity shown in Fig. 6c. However, when s-hhy mutants were treated with hygromycin B while simultaneously stimulating their TORC1 activity with cycloheximide, their Sch9 phosphorylation was further compromised relative to levels seen in cycloheximide treatment alone. The exceptions were arf1Δ strain which uniquely showed increased phosphorylation and drs2Δ whose Sch9 phosphorylation did not show statistically significant changes. The antagonistic effects of the two translation inhibitors hygromycin B and cycloheximide on almost all s-hhy mutants further support a translation-independent target for hygromycin B in these mutants.
Thus, Tor1 kinase function, assayed by phosphorylation of its direct substrate, is compromised in s-hhy mutants. These results indicate that intact Golgi and late endosome interface is necessary for wild-type levels of TORC1 kinase activity. Taken together with defects in Tor1-GFP localization, cell proliferation, and growth recovery in s-hhy mutants, our results suggest that efficient Tor1 vacuolar localization is necessary for wild-type levels of growth promoting functions of TORC1.
Discussion
We previously reported a genomic screen for hypersensitivity to hygromycin B (hhy) mutants and established that hhy mutant collection is defective in vacuolar events; the collection is also hypersensitive to rapamycin and caffeine suggestive of compromised TORC1 signaling (Banuelos et al. 2010). Here, we define a super-hypersensitive (s-HHY) subgroup of genes that inclusively or exclusively function at the trans-Golgi and late-endosome interface. We also show that Tor1 vacuolar localization, as well as TORC1 growth promoting functions, are compromised and hypersensitive to hygromycin B in s-hhy mutants. Together, our results establish correlation between compromised Golgi and late endosome interface and super-hypersensitivity to hygromycin B. They also implicate an intact trans-Golgi and late endosome trafficking interface as a requisite for efficient Tor1 vacuolar localization and TORC1 anabolic functions.
Hygromycin B vulnerability and compromised trans-Golgi and late endosome interface
The s-HHY genes are ARF1, CHC1, DRS2, SAC1, VPS1, VPS34, VPS45, VPS52, and VPS54; each has a known role in vesicular trafficking at the trans-Golgi and late endosome interface. Thus, a compromised trans-Golgi and late endosome interface directly correlates with severe hygromycin B hypersensitivity. Undisturbed endosome-independent vacuolar localization of Vam3-RFP in s-hhy mutants in the presence or absence of hygromycin B indicates that direct Golgi to vacuole trafficking is neither compromised nor hypersensitive to the drug. Similarly, undisturbed FM4-64 endocytosis and localization to the vacuole membrane in hhy mutants indicates that late endosome to vacuole trafficking is also neither compromised nor hypersensitive to hygromycin in these mutants. Exclusion of all class E and class C vps mutants from the uncovered hhy collection indicates that compromised multivesicular body formation or vacuolar biogenesis, respectively, are not associated with hypersensitivity to hygromycin B. Last, unique hypersensitivity of hhy mutants, specifically s-hhy mutants, to hygromycin B but not to another translation inhibitor suggests that hygromycin B hypersensitivity is independent of the state of translation machinery; a similar conclusion was reached by Conboy and Cyert (2000) whose mutants were sensitive to hygromycin B but not to additional translation inhibitors. These results, together with the extensive vacuolar morphology defects as well as localization disturbances of Tor1 and the endosomal marker Pep12 in s-hhy mutants, support a general trafficking disturbance specifically at Golgi and late endosome interface. Among s-hhy mutants, arf1Δ is unique in its range of milder or no hygromycin super-hypersensitive phenotypes assayed; this is most likely due to partial functional compensation by Arf2.
What may be the molecular target of hygromycin B in s-hhy mutants? While all s-HHY gene products function at the trans-Golgi and late endosome interface, they are not part of one molecular complex. Arf1, Chc1, and Vps1 are involved in clathrin-coated vesicle formation and budding at trans-Golgi; Vps45 is involved in vesicle docking at late endosome; and Vps52 and Vps54 are two members of the Golgi-associated retrograde protein (GARP) complex involved in fusion of late endosome derived retrograde vesicles with Golgi (Bonifacino and Hierro 2011; Conibear and Stevens 2000; Bryant and James 2001). What these gene products have in common is association with membranes during either budding or fusion events at Golgi and late endosome interface. Interestingly, the other two members of GARP complex, Vps51 and Vps53, were not picked up in the screen and did not show hhy phenotype upon reexamination (data not presented), suggesting that only GARP complexes missing Vps52 or Vps54 may be compromised sufficiently to be associated with hygromycin B hypersensitivity. The remainder of s-HHY gene products function directly in membrane lipid events. Drs2 is the trans-Golgi flippase associated with promoting microdomains and membrane curvature (Graham 2004; Hua et al. 2002), Sac1p is the phosphoinositide phosphatase involved in generation of PI(3)P, and Vps34p is the kinase that phosphorylates phosphatidylinositol to produce PI(3)P. Interestingly, PI(3)P is the phosphoinositide species enriched in endosomal membranes and implicated in vesicular trafficking (Bhandari et al. 2007; De Camilli et al. 1996); it has also been implicated in TORC1 regulation (Bridges et al. 2012). Another phosphoinositide, PI(3, 5)P2, has recently been reported to play a role in nutrient sensing by TORC1 (Jin et al. 2014). It is tempting to speculate, therefore, that hygromycin B may be targeting membrane microdomains engaged in fusion and fission events, and that such microdomains may also be important in recruitment and/or trafficking of Tor1. As such, compromised or PI3P-deficient microdomains predicted in drs2Δ, sac1Δ, and vps34Δ, or compromised budding/fusion complexes predicted in the remainder of s-hhy mutants may be especially vulnerable to hygromycin B. In fact, aminoglycoside drugs have been reported to interfere with coatomer formation and secretion in mammalian cells (Hudson and Draper 1997; Hu et al. 1999). While translation inhibition function of hygromycin B is a cytoplasmic event, the pathway of hygromycin B entry into cells remains unknown. Prior research has suggested that in mammalian cells, aminoglycoside antibiotics are taken up by receptor-mediated endocytosis (Hashino and Shero 1995; Hashino et al. 1997), which would not be inconsistent with direct endomembrane interactions at late endosome and trans-Golgi interface. Alternatively, hygromycin B may be indirectly disruptive to a compromised trans-Golgi and late endosome interface.
Late endosome-dependent vacuolar trafficking as a requisite for wild type levels of Tor1 localization and TORC1 function
Our results indicate that late endosome-dependent vacuolar trafficking is compromised in s-hhy mutants and necessary for wild-type levels of Tor1 vacuolar localization and TORC1 anabolic functions. Since TORC1 functions are compromised in hygromycin B-treated s-hhy mutants, localization of Tor2, which can functionally substitute for Tor1, is presumably blocked as well. Since our results suggest an intact late endosome-independent trafficking pathway to vacuoles, late endosome-dependent trafficking may be the main pathway utilized by Tor1 and Tor2 for their vacuolar localization. Consistent with that hypothesis, both proteins lack the only two known late endosome-independent and AP-3-dependent trafficking consensus sequences. Alternatively, late endosome-dependent trafficking of other molecules involved in recruitment of Tor1/2 and TORC1 activation may be compromised in s-hhy mutants.
s-hhy strains have compromised Tor1-GFP vacuolar localization and Sch9 phosphorylation even in the absence of hygromycin B. This is not surprising considering the direct roles of s-HHY gene products in vacuolar trafficking. Furthermore, Vps34p has been shown to directly interact with Tor1p and may be an upstream regulator of TORC1 (Aronova et al. 2007; Jacinto 2008), and physical interactions have been reported between Vps54p and Tor1p (Krogan et al. 2006). Several studies to date have suggested a link between compromised TORC1 function and defects in vacuolar trafficking and function (Zurita-Martinez et al. 2007; Brown et al. 2010; Flinn and Backer 2010). Most recently, Kingsbury et al. (2014) reported that mutations in members of HOPS and CORVET complexes (required for homotypic and heterotypic vacuolar fusion events, endosomal trafficking, and retrograde trafficking from the vacuole) cause defects in TORC1 signaling. While HHY gene products and HOPS/CORVET components do not overlap, these results together further implicate intact endomembrane trafficking to the vacuole as a requisite for TORC1 signaling. While mTORC1 is recruited to the lysosomal membrane and activated based on nutrient signaling, yeast TORC1 localization appears to be constitutively at the vacuole (Betz and Hall 2013; Binda et al. 2009). As Tor1 localization remains punctate following treatment in most s-hhy mutants, our results support association of Tor1p with the endomembrane at pre-vacuolar compartments, and its subsequent delivery to the vacuole through the late endosome-dependent pathway. This is consistent with a recent report where during Ras-induced senescence, mTOR is recruited to trans-Golgi before transport to a novel lysosomal derived TOR-autophagy spatial coupling compartment (TASCC) (Narita et al. 2011). Interestingly, we do not observe upregulated autophagy in hygromycin B-treated s-hhy mutants; in fact, they appear to be defective in autophagy under starvation and rapamycin treatment (Ejzykowicz, Ruiz and Gharakhanian, preliminary results). This suggests that vacuolar localization may be required for both anabolic and catabolic functions of TORC1.
Implications
Rapamycin and its analogs (rapalogs) are the main pharmacological agents currently explored to control TORC1 activity as well as mTORC1 hyperactivity in cancer and diabetes (reviewed in Seto 2012; Cornu et al. 2013). However, effective concentrations are highly toxic, and cancer cells rapidly develop resistance to single drug approaches. The search for additional inhibitors of TORC1 is ongoing, and the small molecule TORC1 inhibitor CID 3528206 has been reported from a systemic screen (Chen et al. 2012). An interesting implication of our studies is that subtranslation inhibitory levels of hygromycin B can be a potent negative regulator of TORC1. Low doses of hygromycin B in combination with rapamycin or with other drugs that either disturb late endosome-dependent vesicular trafficking or inhibit a particular s-HHY gene product may be effective in TORC1 downregulation. We are currently exploring such drug combinations and our preliminary results indicate a synergistic drug combination effect between hygromycin B and rapamycin (Priscilla Bravo and E. Gharakhanian, unpublished results). Controlling TORC1 activity by low-dose drug combinations with hygromycin B offers alternatives for regulating TORC1 in functional studies as well as translational potential in regulating mTORC1 hyperactivity in disease.
References
Ali R, Brett CL, Mukherjee S, Rao R (2004) Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffick in yeast. J Biol Chem 279:4498–4506
Aramburu J, Ortells MC, Tejedor S, Buxade M, Lopez-Rodriguez C (2014) Transcriptional regulation of the stress response by mTOR. Sci Signal 7:re2
Armstrong J (2010) Yeast vacuoles: more than a model lysosome. Trends Cell Biol 20:580–585
Aronova S, Wedaman K, Anderson S, Yates J, Powers T (2007) Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell 18:2779–2794
Bankaitis VA, Johnson LM, Emr SD (1986) Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA 83:9075–9079
Banta LM, Robinson JS, Klionsky DJ, Emr SD (1988) Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol 107:1369–1383
Banuelos MG, Moreno DE, Olson DK, Nguyen Q, Ricarte F, Aguilera-Sandoval CR, Gharakhanian E (2010) Genomic analysis of severe hypersensitivity to hygromycin B reveals linkage to vacuolar defects and new vacuolar gene functions in Saccharomyces cerevisiae. Curr Genet 56:121–137
Becherer KA, Rieder SE, Emr SD, Jones EW (1996) Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell 7:579–594
Betz C, Hall MN (2013) Where is mTOR and what is it doing there? J Cell Biol 203:563–574
Bhandari R, Chakraborty A, Snyder SH (2007) Inositol Pyrophosphate Pyrotech. Cell Metab 5:321–323
Binda M, Peli-Gulli M, Bonfils G, Panchaud N, Urban J, Sturgill T, Loewith R, De Virgilio C (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35:563–573
Stauffer B, Powers T (2016) Target of rapamycin signaling mediates vacuolar fragmentation. Curr Genet. doi:10.1007/s00294-016-0616-0
Bonangelino CJ, Chavez EM, Bonifacino JS (2002) Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell 13:2486–2501
Bonifacino JS, Hierro A (2011) Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol 21:159–167
Bowers K, Stevens TH (2005) Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1744:438–454
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2):115–132
Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, Saltiel AR (2012) Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J Biol Chem 287:20913–20921
Brown CR, Hung GC, Dunton D, Chiang HL (2010) The TOR Complex 1 is distributed in endosomes and in retrograde vesicles that form from the vacuole membrane and plays an important role in the vacuole import and degradation pathway. J Biol Chem 285:23359–23370
Bryant NJ, James DE (2001) Vps45 stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO 20:3380–3388
Cabanas MJ, Vazquez D, Modolell J (1978) Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur J Biochem 87:21–27
Chakrabarti P, English T, Shi J, Smas CM, Kandror KV (2010) Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes 59:775–781
Chen J, Young SM, Allen C, Seeber A, Péli-Gulli MP, Panchaud N, Waller A, Ursu O, Yao T, Golden JE, Strouse JJ, Carter MB, Kang H, Bologa CG, Foutz TD, Edwards BS, Peterson BR, Aubé J, Werner-Washburne M, Loewith RJ, De Virgilio C (2012) Sklar LA (2012) Identification of a small molecule yeast TORC1 inhibitor with a multiplex screen based on flow cytometry. ACS Chem Biol 4:715–722
Conboy M, Cyert MS (2000) Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol Biol Cell 11:2429–2443
Conibear E, Stevens TH (2000) Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11:305–323
Cornu M, Albert V, Hall MN (2013) mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev 23:53–62
Cybulski N, Hall MN (2009) TOR complex 2: a signaling pathway of its own. Cell 34:620–627
De Camilli P, Emr SD, McPherson PS, Novick P (1996) Phosphoinositides as regulators in membrane traffic. Science 271:1533–1539
Delorme-Axford E, Guimaraes RS, Reggiori F, Klionsky DJ (2015) The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression. Methods 75:3–12
Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C (2005) The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19:15–26
Eustice DC, Wilhelm JM (1984) Fidelity of the eukaryotic codon-anticodon interaction: interference by aminoglycoside antibiotics. Biochemistry 23:1462–1467
Feyder S, De Craene JO, Bar S, Bertazzi DL, Friant S (2015) Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci 16:1509–1525
Flinn RJ, Backer JM (2010) mTORC1 signals from late endosomes: taking a TOR of the endocytic system. Cell Cycle 9:1869–1870
Gautreau A, Oguievetskaia K, Ungermann C (2014) Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect Biol 6:a016832
Graham TR (2004) Flippases and vesicle-mediated protein transport. Trends Cell Biol 14:670–677
Guan XL et al (2009) Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 20:2083–2095
Hall MN (2008) mTOR—what does it do? Trans Proc 40:S5–S8
Hashino E, Shero M (1995) Endocytosis of aminoglycoside antibiotics in sensory hair cells. Brain Res 704:135–140
Hashino E, Shero M, Salvi RJ (1997) Lysosomal targrtting and accumulation of aminoglycoside antibiotics in sensory hair cells. Brain Res 777:75–85
Hecht KA, O’Donnell AF, Brodsky JL (2014) The proteolytic landscape of the yeast vacuole. Cell Logist 4:e28023
Ho YH (2015) Gasch AP (2015) Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr Genet 61:503–511. doi:10.1007/s00294-015-0491-0
Hu T, Kao CY, Hudson RT, Chen A, Draper RK (1999) Inhibition of secretion by 1,3-Cyclohexanebis(methylamine), a dibasic compound that interferes with coatomer function. Mol Biol Cell 10:921–933
Hua Z et al (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell 13:3162–3177
Hudson RT, Draper RK (1997) Interaction of coatomer with aminoglycoside antibiotics: evidence that coatomer has at least two dilysine binding sites. Mol Biol Cell 8:1901–1910
Jacinto E (2008) What controls TOR? IUBMB Life 60:483–496
Jiang Yu (2016) Regulation of TORC1 by ubiquitin through non-covalent binding. Curr Genet 62:553–555. doi:10.1007/s00294-016-0581-7
Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ et al (2014) Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell 25:1171–1185
Kingsbury JM, Sen ND, Maeda T, Heitman J, Cardenas ME (2014) Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics 196:1077–1089
Klionsky DJ, Eskelinen EL (2014) The vacuole versus the lysosome: when size matters. Autophagy 10:185–187
Krogan NJ et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643
Kummel D, Ungermann C (2014) Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr Opin Cell Biol 29:61–66
Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J (2006) Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signaling pathways. Mol Microbiol 61:1147–1166
LaGrassa TJ, Ungermann C (2005) The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol 168:401–414
Levin DE (2005) Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 69:262–291
Li SC, Kane PM (2009) The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta 1793:650–663
Loewith R, Hall MN (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189:1177–1201
Madeira JB, Masuda CA, Maya-Monteiro CM, Matos GS, Montero-Lomeli M, Bozaquel-Morais BL (2015) TORC1 inhibition induces lipid droplet replenishment in yeast. Mol Cell Biol 35:737–746
Manandhar SP, Gharakhanian E (2014) ENV7 and YCK3, which encode vacuolar membrane protein kinases, genetically interact to impact cell fitness and vacuole morphology. FEMS Yeast Res 14:472–480
Manandhar SP, Ricarte F, Cocca SM, Gharakhanian E (2013) Saccharomyces cerevisiae Env7 is a novel serine/threonine kinase 16-related protein kinase and negatively regulates organelle fusion at the lysosomal vacuole. Mol Cell Biol 33:526–542
Markgraf DF, Ahnert F, Arlt H, Mari M, Peplowska KN, Griffith J, Reggiori F, Ungermann C (2009) The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell 20:5276–5289
McCormick MA, Tsai SY, Kennedy BK (2011) TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci 366:17–27
Mukherjee S, Kallay L, Brett CL, Rao R (2006) Mutational analysis of the intramembranous H10 loop of yeast Nhx1 reveals a critical role in ion homoeostasis and vesicle trafficking. Biochem J 398:97–105
Narita M, Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavaré S, Inoki K, Shimizu S, Narita M (2011) Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332(6032):966–970
Neufeld TP (2010) Tor-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 22:157–168
Noda T, Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273:3963–3966
Ostrowicz CW, Meiringer CTA, Ungermann C (2008) Yeast vacuole fusion: a model system for eukaryotic endomembrane dynamics. Autophagy 4:5–19
Park I, Erbay E, Nuzzi P, Chen J (2005) Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp Cell Res 309:211–219
Payne GS, Schekman R (1985) A test of clathrin function in protein secretion and cell growth. Science 230:1009–1014
Reggiori F, Klionsky DJ (2013) Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194:341–361
Reinke A, Chen JC, Aronova S, Powers T (2006) Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem 281:31616–31626
Richards A, Veses V, Gow NAR (2010) Vacuole dynamics in fungi. Fungal Biol Rev 24:93–105
Richards A, Gow NA, Veses V (2012) Identification of vacuole defects in fungi. J Microbiol Methods 91:155–163
Robinson JS, Klionsky DJ, Banta LM, Emr SD (1988) Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 8:4936–4948
Rohde JR, Bastidas R, Puria R, Cardenas ME (2008) Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol 11:153–160
Rothman JH, Howald I, Stevens TH (1989) Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO 8:2057–2065
Saftig P, Klumperman J (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10:623–635
Seeger M, Payne G (1992) A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J 11:2811–2818
Seto B (2012) Rapamycin and mTOR: a serendipitous discovery and implications for breast cancer. Clin Transl Med 1:29
Sherman F (2002) Getting started with yeast. Methods Enzymol 350:3–41
Soulard A, Cohen A, Hall MN (2009) TOR signaling in invertebrates. Curr Opin Cell Biol 21:825–836
Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN (2008) TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 7:1819–1830
Subramanian K, Dietrich LE, Hou H, LaGrassa TJ, Meiringer CT, Ungermann C (2006) Palmitoylation determines the function of Vac8 at the yeast vacuole. J Cell Sci 119:2477–2485
Swinnen E et al (2014) The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol Biol Cell 25:196–211
Takahashi MK, Frost C, Oyadomari K, Pinho M, Sao D, Chima-Okereke O, Gharakhanian E (2008) A novel immunodetection screen for vacuolar defects identifies a unique allele of VPS35 in S. cerevisiae. Mol Cell Biochem 311:121–136
Urban J et al (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26:663–674
Veses V, Richards A, Gow NA (2008) Vacuoles and fungal biology. Curr Opin Microbiol 11:503–510
Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792
Vida TA, Huyer G, Emr SD (1993) Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol 121:1245–1256
Viotti C (2014) ER and vacuoles: never been closer. Front Plant Sci 5:20
Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808
Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, Virgilio CD (2008) Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol 69:277–285
Wickner W (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 26:115–136
Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124:471–484
Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME (2007) Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 176:2139–2150
Acknowledgements
This project was funded by NIH-AREA research Grant 2R15GM085794-02 to E.G. and NSF-MRI grant DBI0722757 for confocal microscopy. D.E.E. was supported and K.M.L. was partially supported by the above NIH grant; F.J.R. was supported by NIH-RISE grant 5R25-GM071638-07; D.K.O was supported by Beckman Scholars Program. We thank Dr. Greg Payne (UCLA) for the yeast strain library, Dr. Claudio De Virgilio (University of Geneva) for TOR1-3XGFP strain and for plasmid p1462 encoding Sch9-HA, Dr. Christian Ungermann (University of Osnabruck) for PEP12-RFP and VAM3-RFP expressing plasmids.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
D. E. Ejzykowicz and K. M. Locken contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ejzykowicz, D.E., Locken, K.M., Ruiz, F.J. et al. Hygromycin B hypersensitive (hhy) mutants implicate an intact trans-Golgi and late endosome interface in efficient Tor1 vacuolar localization and TORC1 function. Curr Genet 63, 531–551 (2017). https://doi.org/10.1007/s00294-016-0660-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0660-9