Abstract
The rapid spread of the SARS-CoV-2 virus has emphasized the urgent need for effective therapies to combat COVID-19. Investigating the potential targets, inhibitors, and in silico approaches pertinent to COVID-19 are of utmost need to develop novel therapeutic agents and reprofiling of existing FDA-approved drugs. This article reviews the viral enzymes and their counter receptors involved in the entry of SARS-CoV-2 into host cells, replication of genomic RNA, and controlling the host cell physiology. In addition, the study provides an overview of the computational techniques such as docking simulations, molecular dynamics, QSAR modeling, and homology modeling that have been used to find the FDA-approved drugs and other inhibitors against SARS-CoV-2. Furthermore, a comprehensive overview of virus-based and host-based druggable targets from a structural point of view, together with the reported therapeutic compounds against SARS-CoV-2 have also been presented. The current study offers future perspectives for research in the field of network pharmacology investigating the large unexplored molecular libraries. Overall, the present in-depth review aims to expedite the process of identifying and repurposing drugs for researchers involved in the field of COVID-19 drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The worldwide pandemic that began with the emergence of the novel coronavirus SARS-CoV-2 in late 2019 has deep and far-reaching effects on public health, society, and the economy [1, 2]. The virus has affected virtually every country, resulting in millions of deaths, hospitalizations, and long-term health complications [3]. The pandemic has also caused significant disruptions to daily life, including lockdowns, travel restrictions, and economic downturns [4]. The impact of the pandemic has underscored the urgent need to develop effective strategies to control the spread of the virus and mitigate its impact on human health [5]. SARS-CoV-2 is a highly infectious virus that primarily spreads through respiratory droplets and close contact with infected individuals [6]. The virus’s ability to infect asymptomatic individuals has made it challenging to control its spread. Moreover, the emergence of new variants of the virus has complicated efforts to develop effective vaccines and treatments [7].
The pandemic has highlighted the importance of scientific research in combating infectious diseases and has created a pressing need for disease-fighting medications and vaccines [8]. Scientists worldwide have worked tirelessly to gain a better understanding of the virus's biology, transmission, and pathogenesis [9]. Such research has led to the development of effective vaccines and treatments that have saved countless lives [10]. However, there is still much to learn about SARS-CoV-2, and ongoing research is critical to developing better prevention and treatment strategies. One area of research that has gained increased attention in recent years is computational studies [11]. These studies utilize computer-based simulations to model the behavior of molecules and target proteins, allowing scientists to identify potential drug candidates and optimize their properties [12, 13]. Computational studies have played an essential role in developing effective treatments for various diseases, including cancer, HIV/AIDS, and hepatitis C [14]. These virtual approaches have accelerated the drug discovery process, as they allow researchers to rapidly screen large numbers of potential hit candidates and identify promising lead compounds for further testing [15]. In the context of COVID-19 research, in silico studies have played a critical role in the search for potential inhibitors and targets for SARS-CoV-2 [16]. Through these studies, researchers have been able to model and simulate the interactions between the promising targets and potential drug candidates [17].
Identification of a potential approach or therapy against COVID-19 is still under consideration. Therefore, gaining insights into SARS-CoV-2, its interacting partners, and functional domains is crucial for developing effective treatments. The article provides a comprehensive review on virus enzymes and their interacting host molecules that play the major role to enter, replicate its genomic material within the host and regulate host cell biology. Further, this review also summarizes the potential inhibitors that are being studied and identified to block these target molecules for the treatment of COVID-19. The deep insight of various computational approaches is also emphasized for repurposing existing drugs or designing new therapeutic options with special reference to SARS-CoV-2. By emphasizing the critical role of computational studies in combating the ongoing pandemic, this article aims to provide a valuable resource for scientists and pharmacologists worldwide.
Mechanisms of SARS-CoV-2 Entry into Host Cell
SARS-CoV-2 also known as β-coronavirus is an enveloped positive-strand RNA virus with an unusually large genome length of 29,881 bp of coronaviridae family [18].
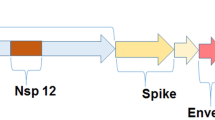
The viral genome is structurally attached to Nucleocapsid (N) protein, and the double-layered lipid viral envelope is formed by the combination of Spike (S), Envelope (E), and Membrane (M) proteins (Fig. 1). The genome has 14 open reading frames (ORFs), which encode different structural proteins such as the S, E, M, and N proteins along with 16 non-structural proteins (NSP 1–16) as shown in Table 1 (Fig. 1).
Illustration of the structural composition and genomic organization within SARS-CoV-2. A An overview of structural features of SARS-CoV-2 and primary key proteins. B The genomic organization of SARS-CoV-2. (The Figure was created with https://www.biorender.com)
The viral life cycle of SARS-CoV-2 is complex and involves several distinct steps, including viral entry, replication, assembly, and release [19]. To make effective drugs against COVID-19, it is necessary to have an in-depth knowledge of the targets of the virus and the receptors of the host cell. For this, the detailed mechanism of virus entry into the host cell, its growth and multiplication is given here (Fig. 2).
SARS-CoV-2 infiltrates host cells through endocytosis or binding to the transmembrane angiotensin converting enzyme 2 (ACE2) receptors on cell surface. This triggers membrane fusion via host proteases, enabling viral entry, replication, maturation, and relentlessly efficient release to nearby cells. Note, the inhibitor arrows are pointing the activity of FDA-approved drugs against promising targets. (The Figure was created with https://www.biorender.com)
The entry of SARS-CoV-2 into host cells is a crucial step in the viral life cycle and is mediated by the virus's spike protein, which binds to the ACE2 receptor on the surface of the host cell [20]. The affinity of the spike protein for ACE2 is an important factor in the viral entry process [21]. The receptor-binding domain (RBD) in the S1 subunit of the spike protein recognizes and binds to the host cell’s ACE2 receptor [22]. The S1 RBD of the spike protein binds to the ACE2 receptor with high affinity, allowing the virus to enter host cells even at low viral loads. This high affinity is thought to be a key factor in the high infectivity of SARS-CoV-2 [20, 23]. The fusion peptide and other sequences required for fusion of the viral membrane with the host cell membrane are found in the S2 subunit [24]. Once the virus is attached to the ACE2 receptor, the S2 subunit undergoes a conformational change that allows the virus to fuse its membrane with the host cell membrane [25]. This fusion step is critical for the release of the large viral genome into the host cell and the initiation of viral replication. The ACE2-mediated entry of SARS-CoV-2 into host cells is also affected by the distribution and differential expression levels of ACE2 receptors in different tissues and cells. The ACE2, and the protease enzymes of the host cell play important role for the cleavage and fusion of the viral spike protein [26]. The location and transcript amount of ACE2 receptors, crucial in the viral entry process [27], vary across different tissues and cell types, including the lungs, heart, kidneys, and intestines [28]. The expression levels of ACE2 can also vary greatly depending on the individual [29]. These variations in expression levels of ACE2 may be correlated with the observed differences in COVID-19 severity among patients [30].
One of the important factor in the viral entry process is the proteases enzymes that cleave the viral spike protein and activate its fusion activity [31]. The spike protein of SARS-CoV-2 is cleaved by host cell proteases, including TMPRSS2 and cathepsin L (Fig. 2) [32]. The cleavage of the spike protein by these proteases is required for the fusion of the virus membrane with the host cell membrane [33].
In addition to ACE2-mediated entry, there is evidence to suggest that SARS-CoV-2 can enter cells through alternative pathways [20, 34]. One of these pathways is TMPRSS2-mediated entry, where the spike protein is cleaved by the host cell protease TMPRSS2 inducing the fusion activity of the spike protein and facilitating the fusion of the virus membrane with the host cell membrane [35] Another possible pathway is endocytosis, where the virus is taken up by the host cell in a vesicle [31]. The virus can then fuse with the endosomal membrane, releasing its genetic material into the cytoplasm (Fig. 2). There is also evidence to suggest that SARS-CoV-2 may be able to enter cells through a mechanism called membrane fusion without endocytosis, where the virus fuses directly with the host cell membrane.
Once the virus has entered the host cell, it begins to replicate and produce new viral particles. The viral genome is translated into viral proteins, which assemble into new viral particles. The new viral particles then leave the host cell through a process called budding, which involves the incorporation of viral proteins into the host cell membrane [36]. The virus can then spread to other nearby cells thereby infecting additional host cells, perpetuating the cycle of infection [37]. Given our understanding of these various entry pathways for SARS-CoV-2, targeted drug therapy plays the crucial role for developing effective treatments for COVID-19 [38]. By targeting specific steps in these pathways, researchers can develop interventions that limit the viral entry and replication to spread the virus, and mitigate the impact of the COVID-19 pandemic [6, 39].
Potential Targets for COVID-19
SARS-CoV-2 interacts with various host cell components to enter, replicate, and control the host cellular mechanism. Hence, all protein enzymes of the coronavirus and the involved host cell molecules are potentially druggable targets for the development of promising molecules against COVID-19. In principle, understanding the viral proteins and their interacting partners within host cell are essential to develop effective therapeutic strategies against the SARS-CoV-2 virus [40]. Herein, we have summarized the virus-based and host-based potential therapeutic targets in the search of therapeutic options to tackle with the SARS-CoV-2.
The virus's genome encodes for a range of structural and non-structural proteins that are essential for viral entry into host cell and replication, thus, these proteins represent promising targets in the search for drug-like molecules [41]. The viral genomic RNA begins with a 5′ un-translated region (UTR), then the ORF1a and ORF1b replication complexes followed by the four structural protein regions and 3′ un-translated region (UTR), ending with non-structural ORFs and a poly(A) tail. ORF1a contains 10 non-structural proteins and ORF1b contains16 non-structural proteins (Table 1). The combination of ORF1a and ORF1b codes pp1a (440–500 kDa) and pp1b (740–810 kDa) polyproteins that form the viral replication complex.
The present section is divided into two main categories, including SARS-CoV-2 virus-based proteins (Table 1) and host-based prominent receptors (Supporting Table 1) as targets for COVID-19. This section recapitulates the structural and non-structural proteins from virus and interacting receptors from host which are being promisingly targeted to design the novel drug candidates or for the repurposing of existing drugs against SARS-CoV-2 [42].
SARS-CoV-2 Components as Targets for COVID-19
Structural Proteins
Structural proteins are a category of molecular proteins that help organs and cells to maintain their molecular integrity, shape, and ability to withstand external stress [43].These proteins are involved in forming the physical structure of cells, tissues, and organs in living organisms. Overall, structural proteins are typically considered by their high tensile strength and rigidity, which is due to their unique molecular structure [44, 45]. Most structural proteins are composed of long, linear chains of amino acids that are cross-linked by strong covalent bonds called peptide bonds resisting mechanical stress [46].These amino acids are typically arranged in repeating patterns, forming a secondary structure such as an alpha-helix or beta-sheet [47]. These secondary structures then fold into a three-dimensional structure, which is stabilized by additional non-covalent interactions such as hydrogen bonds and van der Waals forces [48]. The most common structural proteins in living organisms, such as collagen, elastin, and keratin, not only serve structural roles but also have specific functional roles. For example, collagen provides strength, elasticity and involved in wound healing to tissues such as skin, bones, lungs, and cartilage [49].
In the context of SARS-CoV-2, the four key structural proteins are the Nucleocapsid protein (N), Spike (S), Envelope (E), and Membrane (M) proteins, which are essential for the virus's structural integrity and functional aspects [42]. The spike protein (S) is responsible for binding the virus to the host cell and initiating the infection process [50]. The S protein (spike protein) is the primary target for neutralizing antibodies against SARS-CoV-2. The envelope protein (E) and membrane protein (M) are involved in the assembly of the virus and are important for maintaining its structural integrity (Fig. 1). The nucleocapsid protein (N) is responsible for packaging the virus's genetic material (Fig. 1) [41]. Hence, with the extensive role of structural proteins to enter and multiplication of SARS-CoV-2 into the host cell, these molecules are the potential targets for the development of promising molecules against COVID-19. In principle, understanding the structural proteins is essential to develop effective therapeutic strategies to tackle SARS-CoV-2 infection.
Non-structural Proteins
Non-structural proteins are a group of proteins that are encoded by the single-stranded coronavirus (CoV) genome. These viral proteins are involved in various aspects of the viral life cycle and play the crucial role in viral replication and pathogenesis [41, 51]. In general, the non-structural proteins are essential for proliferation of SARS-CoV-2 and COVID-19. Open reading frames (ORFs) of viral genome encode the 16 different types of non-structural proteins (NSP 1–16) proteins. ORF1a contains 10 non-structural proteins and ORF1b contains 16 non-structural proteins (Table 1). The RNA-dependent RNA polymerase (RdRp), helicase (Hel), exonuclease (ExoN), MPro papain-like protease (PLpro), 3C-like protease (3CLpro), and ORF-8 protein (NS8 protein) are some of the key non-structural proteins encoded by the CoV genome [19, 52]. Scientists have paid their attention to design lead molecules against SARS-CoV-2 by targeting these NSPs.
Mpro, also known as the main protease or 3CLpro, is a key enzyme in the replication of coronaviruses, including the SARS-CoV-2 virus responsible for the COVID-19 pandemic [53]. Mpro plays a crucial role in the cleavage of viral polyproteins at specific sites, which is required for the formation of new viral particles. RdRp (NSP12) or RNA-dependent RNA polymerase enzyme catalyzes the synthesis of the genomic RNA from a template of viral RNA and plays the key role in producing multiple copies of the viral RNA [54,55,56]. CoV2 contains a non-structural large viral polyprotein PLpro (Mpro papain-like protease/NSP3) that forms the replicase complex. This protein is essential for processing viral polyproteins, which are translated from the viral RNA. Mpro papain-like protease cleaves these polyproteins into individual functional viral proteins. Spike gly2/Receptor binding domain (RBD) is a clove-shaped kind of transmembrane protein (type I-TM) on the surface of the SARS-CoV-2 virus that allows it to enter human cells. It is the target of many vaccines and therapeutic antibodies. Helicase (Hel/NSP13)/NTPase is essential for the formation of the replicase complex, which is responsible for viral RNA replication [57]. NTPase/helicase plays a key role in the central viral dogma unwinding of double-stranded DNA to replicate viral RNA. The helicase enzyme of SARS-CoV-2 is part of superfamily I (SF1) that has a common core with two RecA-like motifs involving in ATP binding, nucleic acid binding, and unwinding. Exoribonuclease (ExoN) is involved in the subgenomic micro RNA (sgmRNA) recombination which is required for normal multiplication of coronavirus. This protein plays an important role in the fidelity of viral RNA replication and transcription, eliminating non-compatible nucleotides.
With this in-depth knowledge about important role of different structural and non-structural proteins in viral entry and replication in host cell, researchers would be able to design medicines and vaccines against SARS-CoV-2 more efficiently.
Host Cell Receptors as Targets for COVID-19
As discussed above, SARS-CoV-2 viral proteins and enzymes interact with various host cell receptors to enter, replicate, and control the cellular mechanism in COVID-19 patient. Several human corona viruses (HCoVs) functional receptors play a critical role in viral replication in an infected host cell. Hence, along with the CoV protein enzymes, interacting host cell partners are also promising drug targets for the development of effective medications against COVID-19. Herein, an effort has been made to review host-based potential therapeutic targets collected from several studies and listed in Supporting Table 1. These host cell receptors are studied and targeted to develop novel drug candidates or repurposing the existing drug to combat COVID-19. Biomedical researchers are working extensively using computational methods to find effective antiviral agents against various human corona viruses (HCoVs). Scientists continuously simulate small and large molecules with different PDB IDs of various virus-based and host-based targets to analyze hotspot residues and their interactions to discover novel and potent inhibitors against SARS-CoV-2 as well as to study the binding of interacting amino acid site residues of protein with any ligand molecule (Supporting Table 2) [58].
Inhibitors of SARS-CoV-2 Targets
Following the detailed discussion about druggable targets, potential therapeutic agents targeting either the viral proteins or interacting partners in host cell could be effective inhibiting the viral replication [20]. The rapid emergence of the pandemic left the scientists with limited time to design new drug molecules. As of now, there is no specific approved therapy for treating COVID-19. Concerning the emergence of pandemic and time-consuming traditional drug designing process, reprofiling existing drugs have become the most practiced and effective strategy. Therefore, different categories of drugs are being repurposed in different in vitro and in vivo studies to combat SARS-CoV-2 [59]. In this regard, the World Health Organisation (WHO) has preapproved the repositioning of Remdesivir, Hydroxy-chloroquine, and Ribavirin as a temporary therapy to cure COVID-19 patients. Nevertheless, additional medications are also being re-profiled via if they pass clinical trials and demonstrate efficacy in in silico, in vitro and in vivo research.
Ongoing clinical trials offer a range of therapeutic alternatives including various Food and Drug Administration (FDA)-approved drugs, convalescent plasma therapy, cell therapy, monoclonal antibodies, and immunoglobulin therapy. The current section summarized a comprehensive description about the available potential therapeutics against SARS-CoV-2 to assist researchers in their current and future endeavours to treat COVID‑19 patients. Table 2 lists the many kind of drugs that have been reiterated for the treatment of COVID-19 including anti-inflammatory, anti-malarial drugs as well as numerous other prescribed drugs (Table 2).
FDA-Approved Drugs
Several FDA-approved drugs have shown significant potential to inhibit the SARS-CoV-2 activity. These drugs have been repurposed from their original indication and are being used in clinical trials to treat COVID-19 [60]. In this regard, few examples such as Remdesivir, Lopinavir/ritonavir, and chloroquine/hydroxychloroquine are being discussed here as a time being therapy that has preapproved by World Health Organization.
Remdesivir
Remdesivir is an antiviral drug that has shown promise in inhibiting the replication of several RNA viruses, including SARS-CoV-2 [61]. The researchers have found potent activity of remdesivir against SARS-CoV-2 in silico, in vitro, as well as in animal models of MERS and SARS [62, 63]. In May 2020, the US Food and Drug Administration (FDA) granted emergency use authorization (EUA) for remdesivir for the treatment of hospitalized COVID-19 patients [64]. This authorization was based on data from a clinical trial conducted by the National Institute of Allergy and Infectious Diseases (NIAID), which showed that remdesivir reduced the median time to recovery in hospitalized COVID-19 patients from 11 to 15 days [64]. Overall, remdesivir remains an important therapeutic option for the treatment of COVID-19, particularly in hospitalized patients.
Hydroxychloroquine
Hydroxychloroquine is an antimalarial drug that has been repurposed as a potential treatment for COVID-19, caused by SARS-CoV-2 [65]. The drug works by altering the pH of intracellular compartments, leading to inhibition of viral entry into cells and reduced replication of the virus [66]. Several in vitro investigations have shown that hydroxychloroquine has antiviral activity against SARS-CoV-2, which has led to interest in using the drug as a potential treatment for COVID-19 [65]. In addition, the drug and its combination with azithromycin has been associated with several adverse effects, including cardiac arrhythmias and gastrointestinal symptoms [65].
Ribavirin
Another antiviral drug is ribavirin that has been used to treat a range of viral infections, including respiratory syncytial virus (RSV), hepatitis C, and viral hemorrhagic fevers [67]. It is a broad-spectrum antiviral drug that works by inhibiting viral RNA synthesis, and has shown in vitro activity against SARS-CoV-2 [68]. In the context of COVID-19, ribavirin has been evaluated in combination with other drugs such as interferon and lopinavir/ritonavir [69]. However, due to its potential side effects and lack of efficacy in COVID-19 patients, ribavirin is not a recommended treatment option for COVID-19 [69].
Other Potential Inhibitors
Several other potential therapeutic agents have also shown promising effects in inhibiting SARS-CoV-2 replication in vitro and in preclinical investigations. These agents are currently being evaluated in clinical trials for their safety and efficacy in treating COVID-19 [70].
Tocilizumab
Tocilizumab is a monoclonal antibody that targets the interleukin-6 (IL-6) receptor, which is involved in the body's immune response to infection and inflammation [71]. It is a FDA-approved medication for the treatment of rheumatoid arthritis and other autoimmune diseases. Tocilizumab has also been investigated as a potential therapeutic agent for COVID-19 due to its ability to reduce inflammation and cytokine release syndrome (CRS), a severe immune overreaction that can occur in some patients with COVID-19 [72]. Several clinical trials have evaluated the safety and efficacy of tocilizumab in the treatment of COVID-19. Some studies have reported that tocilizumab can reduce the risk of mechanical ventilation or death in COVID-19 patients with severe symptoms [71]. For example, a randomized controlled trial demonstrated that tocilizumab significantly reduced the risk of death and the need for mechanical ventilation in hospitalized COVID-19 patients with severe respiratory disease [71]. However, there are also concerns about the potential side effects of tocilizumab, including an increased risk of serious infections, liver toxicity, and a reduction in white blood cell count [71]. Therefore, the use of tocilizumab in COVID-19 patients needs to be carefully evaluated on a case-by-case basis. The drug is currently not approved for the treatment of COVID-19, but it has received emergency use authorization from the FDA in some circumstances.
Galidesivir
Galidesivir is an antiviral drug that belongs to a class of drugs called nucleoside analogs and acts by inhibiting the replication of RNA viruses [73]. Galidesivir was originally developed to treat the Ebola virus and has shown promising results in preclinical studies against a broad range of RNA viruses, including SARS-CoV-2 [73]. In vitro studies have shown that galidesivir has potent activity against SARS-CoV-2, and this drug has also demonstrated efficacy in animal models of COVID-19. Galidesivir has been shown to reduce viral load and lung damage in hamsters infected with SARS-CoV-2. Clinical trials of galidesivir for the treatment of COVID-19 are ongoing, with results from phase 1 and 2 studies indicating that the drug is safe and well-tolerated. A phase 3 trial of galidesivir in hospitalized COVID-19 patients is currently underway [73]. Galidesivir's mechanism of action and broad-spectrum antiviral activity make it a promising candidate for the treatment of COVID-19. However, further studies are needed to evaluate its efficacy and safety in larger clinical trials [60, 74].
Sofosbuvir
Sofosbuvir is a direct-acting antiviral drug that has been used to treat chronic hepatitis C infection. It is a nucleotide analog inhibitor that acts by blocking the replication of the hepatitis C virus (HCV) by targeting the NS5B RNA polymerase enzyme, which is essential for HCV replication [75]. More recently, sofosbuvir has shown potential in inhibiting SARS-CoV-2 replication in vitro, and some clinical trials have suggested its potential benefits in treating COVID-19 [76]. In vitro studies have shown that sofosbuvir can inhibit the replication of SARS-CoV-2 by interfering with the virus's RNA-dependent RNA polymerase (RdRp) [76]. Several clinical trials have evaluated the safety and efficacy of sofosbuvir in combination with other drugs such as daclatasvir, ribavirin, and interferon in treating COVID-19 patients [75]. A randomized controlled trial conducted and reaveled that the combination of sofosbuvir and daclatasvir, along with standard care, was associated with a significant reduction in mortality and hospital stay in COVID-19 patients compared to standard care alone [76]. Another study evaluated the efficacy of sofosbuvir and ribavirin in treating COVID-19 patients with moderate to severe disease [75]. The study revealed that the combination of sofosbuvir and ribavirin was associated with a significant reduction in the time to clinical recovery and viral clearance. Sofosbuvir is generally well-tolerated, but some of its side effects include headache, fatigue, and gastrointestinal disturbances. It is also expensive and may not be widely available in some regions, which could limit its use as a potential therapeutic agent for COVID-19 [75]. Therefore, further clinical trials are needed to establish the safety and efficacy of sofosbuvir in treating COVID-19.
Tenofovir
Tenofovir is a nucleotide analog reverse transcriptase inhibitor that is commonly used as an antiviral drug to treat human immunodeficiency virus (HIV) infection. It works by inhibiting the reverse transcriptase enzyme, which is essential for viral replication [77]. Tenofovir has also been investigated for its potential to inhibit the replication of other viruses, including SARS-CoV-2. In vitro studies have shown that tenofovir can effectively inhibit SARS-CoV-2 replication [78]. Clinical trials have been conducted to evaluate the safety and efficacy of tenofovir in the treatment of COVID-19. In one randomized, controlled trial, patients with mild to moderate COVID-19 were treated with a combination of tenofovir and interferon alfa-2b. The study revealed that the combination therapy resulted in a shorter time to clinical recovery, a shorter duration of hospitalization, and a higher rate of viral clearance compared to standard of care [79]. Another study investigated the use of tenofovir disoproxil fumarate (TDF) as pre-exposure prophylaxis (PrEP) for SARS-CoV-2 in healthcare workers [78]. The study found that TDF treatment was associated with a lower risk of SARS-CoV-2 infection in healthcare workers compared to those who did not receive TDF, while Tenofovir shows good agreement as a potential therapeutic agent for COVID-19, however, further research is needed to fully evaluate its safety and efficacy. Additionally, it is important to note that tenofovir is not currently FDA-approved for the treatment of COVID-19, and it should only be used under the supervision of a healthcare provider [78, 80].
In Silico Approaches to Combat SARS-CoV-2
In silico approaches to drug discovery involve the application of computational methods of screening and optimize the potential therapeutic agents [81, 82]. These methods encompass a range of techniques that leverage our understanding of the biological systems involved in the virus–host interaction, as well as our knowledge of the physicochemical properties of small molecules [17]. In recent years, researchers have used a combination of in vitro and in silico studies to investigate efficacy and the pharmacokinetic properties of lead molecules. In vitro studies involve experiments conducted in a laboratory setting, such as cell culture experiments or biochemical assays, whereas in silico studies involve computational methods, such as hierarchical virtual screening of small and large molecules or inhibitors, homology modeling, molecular docking, pharmacokinetic (ADMET) prediction, QSAR study and molecular dynamics simulations [83, 84]. Virtual screening involves the use of computer algorithms to search large databases of compounds to identify potential drug candidates that are likely to bind to specific viral proteins. There are two main types of virtual screening: ligand-based and structure-based. Ligand-based virtual screening uses the known structure of a ligand that binds to the target protein as a template to search for compounds that have similar structures and are therefore likely to bind to the target protein. On the other hand, structure-based virtual screening uses the 3-D structure of the target protein to search for compounds that can bind to specific regions of the protein.
A recent survey of COVID-19 drug discovery studies published in the Journal of Medicinal Chemistry found that the majority of studies (54%) used a combination of in vitro and in silico methods, whereas 28% of studies used only in silico methods and 18% of studies used only in vitro methods [85, 86]. Computational tools have been used to extract and analyze the structural information about pharmacophores and drug–receptor complexes. Some of the commonly used computational tools, program codes, and web servers are listed in Supporting Table 3.
In the context of SARS-CoV-2 drug discovery, virtual screening has been employed greatly to identify potential drug candidates that can bind to the druggable targets (discussed in section “Potential Targets for COVID-19”). Once potential lead compounds are identified, they can be further tested and optimized for their effectiveness and safety in treating COVID-19. One advantage of virtual screening is that it can rapidly identify potential drug candidates, reducing the time and cost required for drug discovery. However, it is important to note that virtual screening is a computational prediction and further experimental studies are needed to validate the effectiveness and safety of the identified compounds [14]. Looking an important role of in silico approaches using computational tools in the drug discovery, herein, we discussed the commonly used computational methods by researchers for curing from Covid-19 to reduce the failures in clinical trials and speed-up the drug design process.
Molecular Docking Simulations
Molecular docking simulation is a computational technique used to predict the binding affinity of small molecules to protein targets. These molecular simulations have played a crucial role in the drug discovery process for SARS-CoV-2, the virus responsible for COVID-19 [87,88,89,90]. By using docking simulations, researchers can identify potential drug candidates that can bind to viral proteins and inhibit their activity, ultimately preventing the virus from replicating and spreading. Docking simulations have been used to identify small molecules that can bind to specific regions of the spike protein and prevent it from binding to the human ACE2 receptor [91]. These studies have also identified small molecules that prevent proteases from functioning properly [92, 93]. Overall, docking studies can help to identify potential drug candidates for SARS-CoV-2 by predicting their binding affinity to viral proteins. However, it is important to note that these predictions need to be validated through further experimental studies to confirm their effectiveness and safety [94].
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are a computational technique used to study the behavior of molecules over small fraction of time (nano seconds). By simulating the movements and vibrations of the atoms in the protein and drug, researchers can determine the strength and stability of their interactions, as well as identify any potential sites for optimization [95]. These simulations can be used to investigate the interactions between a drug and its target protein, providing valuable insights for drug discovery. Using MD simulations, researchers can study how drug molecules interact with the SARS-CoV-2 determining the potential targets at an atomic level. Keretsu & co-workers have used MD simulations to study the interaction of the 19 antiviral drugs including remdesivir with the MProtease (3CL Pro protein) [62]. They observed the good binding affinity of saquinavir and three other investigational drugs aclarubicin, TMC‑310911, and faldaprevir. MD simulations can also be used to screen large libraries of potential drug compounds for their ability to bind to the spike protein. By simulating the binding affinity and stability of thousands of compounds, researchers became able to identify promising candidates for further testing and development. Overall, MD simulations offer a powerful tool for drug discovery in the fight against SARS-CoV-2. By providing insights into the atomic-level interactions between drugs and their target proteins, these simulations has accelerate the discovery of effective therapeutics and helped to combat the COVID-19 pandemic [96].
Quantitative Structure–Activity Relationship (QSAR) Modeling
Quantitative structure–activity relationship (QSAR) modeling is a computational approach used to reveal the associated biological activity of a compound with its chemical structure [97, 98]. This technique has been widely used in drug discovery and development to identify compounds with specific biological activities and optimize their structures. In the case of SARS-CoV-2, researchers have applied QSAR modeling to identify potential drug candidates that can target the virus [99]. QSAR modeling involves building a mathematical relationship between the chemical structure of a compound and its biological activity against the virus. This relationship can then be used to predict the biological activity of new compounds that have not yet been synthesized or tested. To build a QSAR model for SARS-CoV-2 drug discovery, researchers first collect a dataset of compounds with known biological activities against the virus. This dataset is typically a combination of experimental data and in silico calculations. The dataset should also include information on the chemical structures of the compounds, such as their molecular weight, size, shape, and functional groups. Once the QSAR model is built, it can be used to screen large databases of compounds to identify potential lead candidates. These candidates can then be synthesized and tested in vitro and in vivo to confirm their biological activity and optimize their chemical structure. By predicting the biological activity of compounds based on their chemical structure, QSAR modeling can accelerate the discovery of effective therapeutics and help to combat the COVID-19 pandemic [99, 100]. Overall, QSAR modeling offers a powerful tool for drug discovery in the fight against SARS-CoV-2.
Homology Modeling
Homology modeling is a comparative computational modeling to predict the three-dimensional structure of a protein that has not been experimentally determined [99]. This technique relies on the assumption that the structure of a protein is more conserved than its sequence across evolution. Therefore, if the sequence of a target protein is known, and a homologous protein with a similar sequence and known structure exists, the structure of the target protein can be predicted by aligning the target sequence with the template sequence and transferring the template structure to the target sequence [101].
Several research groups have used homology modeling, comparative modeling (CM), template-based modeling (TBM), and template-free modeling (TFM) (ab initio or de novo modeling methods), to predict the structure of Mpro and screen a library of compounds for potential inhibitors against SARS-CoV-2 [102,103,104]. Homology modeling has been used extensively to study the structure of the virus's proteins, including the spike protein, main protease (Mpro), RNA-dependent RNA polymerase (RdRp), and papain-like protease (PLpro). As we have discussed in section “Potential Targets for COVID-19”, these proteins are essential for the virus's replication and are considered important targets for drug development against COVID-19. Modeling studies of RdRp protein showed functional consequences for RdRp targeting potential therapeutics by investigating the alteration in the stability of RdRp protein by mutations [105, 106]. Another study by Dai & coworkers using homology modeling revealed the structure of RdRp and designed drugs that could potentially bind to the active site of the enzyme [107]. In some other studies the researchers used homology modeling to predict the structure of the spike protein receptor-binding domain (RBD) and design neutralizing antibodies that could bind to the RBD and block the virus's entry into cell [108,109,110]. Overall, homology modeling is a valuable tool for predicting the structure of COVID-19 targets and designing drugs that could potentially block the virus's replication. Combined with other in vitro and in silico methods, homology modeling has played a critical role in the development of potential COVID-19 therapeutics.
Using the above theoretical approaches, scientists have paid their attention for new lead molecules as well as repurposing the existing drugs for the medication of Covid-19. Remdesivir, an antiviral medication created initially to treat infections caused by the Ebola virus, is a possible inhibitor discovered through in silico research. In silico studies have shown that it has potent activity against SARS-CoV-2 by inhibiting the virus's ability to replicate. Clinical studies have demonstrated its effectiveness in treating COVID-19, and governing organizations in many nations have authorized its usage in emergency situations [64]. In the search for potential inhibitors of SARS-CoV-2, another illustration was hydroxychloroquine, an antimalarial drug that gained widespread attention. In silico studies suggested that hydroxychloroquine could potentially interfere with the virus's ability to enter human cells. In addition to hydroxychloroquine and remdesivir, many other approved drugs such as antiviral drugs, natural products, and small molecules have been investigated as potential inhibitors of SARS-CoV-2 through in silico studies [63]. Number of flavonoids and alkaloids from natural sources have also been identified as potential hit molecules to inhibit Mpro activity [111, 112]. In silico studies have identified several compounds that can bind to RdRp and inhibit its activity, including remdesivir, which has been shown to be effective in clinical trials [113, 114]. In silico studies have identified several potential targets within the non-structural proteins (NSPs) that could be exploited for the development of antiviral drugs against SARS-CoV-2. Several studies have identified potential monoclonal antibodies, inhibitors of S protein, Nsp12 an RNA polymerase that are essential for viral replication [115, 116]. Overall, in silico studies have been helpful in the hunt for possible SARS-CoV-2 inhibitors and targets, offering insightful information about the biology of the virus and facilitating the speeding up of the drug discovery process.
Conclusion and Future Prospects
The current review emphasizes on SARS-CoV-2, potential druggable targets, inhibitors, and deep insights into in silico studies that are being used to drug design and repurposing of existing FDA-approved drugs. The comprehensive collection of viral enzymes and their counter receptors that are involved in SARS-CoV-2 entry in human cells, replication of genomic RNA, and controlling the host cell physiology provide the future direction to medicinal chemists toward drug design. The detailed insights about FDA-approved and other potential inhibitors that have been identified through various studies and are under the clinical trial stage have also been complied and are of interest for the researchers and budding scientific brains. The application of computational approaches such as docking simulations, molecular dynamics, QSAR modeling, and homology modeling that have been used to find plausible inhibitors against SARS-CoV-2 will also provide the future directions for research to investigate the large mountains of untouched molecules. Overall, this review fulfills its objective providing vital resources of promising drug targets, potential inhibitors, and advanced computational approaches to strengthen the researchers working on drug discovery for COVID-19 and other lethal diseases. The application of in silico investigation prior to the laboratory experiments will reduce the time, cost, and excessive use of living organisms to identify the potential drug candidates and their effectiveness with lower toxicity.
Although, in silico studies offer significant advantages for drug discovery, they also have certain limitations. These limitations include limited accuracy, which depends on the quality of the data and models used, and can be impacted by various factors such as protein flexibility and solvation effects. Additionally, the findings of in silico studies need to be validated experimentally to confirm their accuracy and to ensure the efficacy of potential drug candidates. Inability to account for unknown factors is another limitation, as computational studies rely on existing data and knowledge, and may not be able to account for unknown factors that may impact the effectiveness of potential drug candidates. Finally, there are ethical considerations associated with in silico studies, as they can identify potential drug candidates with ethical implications, such as drugs derived from endangered species or that are not economically viable for widespread use.
Amidst the continuing evolution of the SARS-CoV-2, the development of effective treatments and the discovery of possible drug targets and inhibitors is of paramount significance. In silico research can play a significant role in the process of finding possible targets and inhibitors. Several future directions have been identified to advance the effectiveness of computational studies in drug discovery for SARS-CoV-2 including the integration of multiple computational techniques, the utilisation of machine learning and artificial intelligence, exploring the repurposing of existing drugs and integration of experimental validation. These directions highlight the potential for advanced computational techniques to identify novel therapeutic targets and candidates for COVID-19 treatment.
References
Roychoudhury S, Das A, Sengupta P, Dutta S, Roychoudhury S, Choudhury AP et al (2020) Viral pandemics of the last four decades: pathophysiology, health impacts and perspectives. Int J Environ Res Public Health 17:9411. https://doi.org/10.3390/ijerph17249411
Kumar S, Singh R, Kumari N, Karmakar S, Behera M, Siddiqui AJ et al (2021) Current understanding of the influence of environmental factors on SARS-CoV-2 transmission, persistence, and infectivity. Environ Sci Pollut Res 28:6267–88. https://doi.org/10.1007/s11356-020-12165-1
Kaye AD, Okeagu CN, Pham AD, Silva RA, Hurley JJ, Arron BL et al (2021) Economic impact of COVID-19 pandemic on healthcare facilities and systems: International perspectives. Best Pract Res Clin Anaesthesiol 35:293–306. https://doi.org/10.1016/j.bpa.2020.11.009
Sohrabi C, Mathew G, Franchi T, Kerwan A, Griffin M, Del Mundo SC et al (2021) Impact of the coronavirus (COVID-19) pandemic on scientific research and implications for clinical academic training—a review. Int J Surg 86:57–63. https://doi.org/10.1016/j.ijsu.2020.12.008
Wilder-Smith A, Gubler DJ, Weaver SC, Monath TP, Heymann DL, Scott TW (2017) Epidemic arboviral diseases: priorities for research and public health. Lancet Infect Dis 17:e101-6. https://doi.org/10.1016/S1473-3099(16)30518-7
Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C et al (2020) The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg 78:185–93. https://doi.org/10.1016/j.ijsu.2020.04.018
Cascella M, Rajnik M, Aleem A, Dulebohn SC, Di Napoli R (2023) Features, evaluation, and treatment of coronavirus (COVID-19). StatPearls, Treasure Island
Moradian N, Ochs HD, Sedikies C, Hamblin MR, Camargo CA, Martinez JA et al (2020) The urgent need for integrated science to fight COVID-19 pandemic and beyond. J Transl Med 18:1–7. https://doi.org/10.1186/s12967-020-02364-2
Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109:102433. https://doi.org/10.1016/j.jaut.2020.102433
Kangabam R, Sahoo S, Ghosh A, Roy R, Silla Y, Misra N et al (2021) Next-generation computational tools and resources for coronavirus research: from detection to vaccine discovery. Comput Biol Med 128:104158. https://doi.org/10.1016/j.compbiomed.2020.104158
Chen X, Zhang R, Xing Y, Jiang B, Li B, Xu X et al (2021) The efficacy of different seed priming agents for promoting sorghum germination under salt stress. PLoS ONE 16:1–14. https://doi.org/10.1371/journal.pone.0245505
Honarparvar B, Govender T, Maguire GEM, Soliman MES, Kruger HG (2014) Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem Rev 114:493–537. https://doi.org/10.1021/cr300314q
Kazan MM, Asmare MM, Mahapatra RK (2023) Identification of potential drug targets in erythrocyte invasion pathway of Plasmodium falciparum. Curr Microbiol 80:1–10. https://doi.org/10.1007/s00284-023-03282-4
Lin X, Li X, Lin X (2020) A review on applications of computational methods in drug screening and design. Molecules 25:1–17. https://doi.org/10.3390/molecules25061375
Parvathaneni V, Gupta V (2020) Utilizing drug repurposing against COVID-19—efficacy, limitations, and challenges. Life Sci 259:118275. https://doi.org/10.1016/j.lfs.2020.118275
Sekaran K, Karthik A, Varghese RP, Sathiyarajeswaran P, Shree Devi MS, Siva R et al (2024) In silico network pharmacology study on Glycyrrhiza glabra: analyzing the immune-boosting phytochemical properties of Siddha medicinal plant against COVID-19. Elsevier, Amsterdam, pp 233–55
Cobre AF, Maia Neto M, de Melo EB, Fachi MM, Ferreira LM, Tonin FS et al (2023) Naringenin-4’-glucuronide as a new drug candidate against the COVID-19 Omicron variant: a study based on molecular docking, molecular dynamics, MM/PBSA and MM/GBSA. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2229446
Franchini M, Farrugia A, Velati C, Zanetti A, Romanò L, Grazzini G et al (2020) The impact of the SARS-CoV-2 outbreak on the safety and availability of blood transfusions in Italy. Vox Sang 115:603–5. https://doi.org/10.1111/vox.12928
Vkovski P, Kratzel A, Steiner S, Stalder H, Thiel V (2021) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–70. https://doi.org/10.1038/s41579-020-00468-6
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. https://doi.org/10.1038/s41580-021-00418-x
Rahman MM, Hasan M, Ahmed A (2021) Potential detrimental role of soluble ACE2 in severe COVID-19 comorbid patients. Rev Med Virol 31:1–12. https://doi.org/10.1002/rmv.2213
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S et al (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–20. https://doi.org/10.1038/s41586-020-2180-5
Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y et al (2020) Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci 117:28046–55. https://doi.org/10.1073/pnas.2016093117
Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S (2020) Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res 178:104792. https://doi.org/10.1016/j.antiviral.2020.104792
Zhang Q, Xiang R, Huo S, Zhou Y, Jiang S, Wang Q, et al (2021) Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. https://doi.org/10.1038/s41392-021-00653-w
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Saiz ML, DeDiego ML, López-García D, Corte-Iglesias V, Baragaño Raneros A, Astola I et al (2021) Epigenetic targeting of the ACE2 and NRP1 viral receptors limits SARS-CoV-2 infectivity. Clin Epigenet 13:187. https://doi.org/10.1186/s13148-021-01168-5
Qi F, Qian S, Zhang S, Zhang Z (2020) Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 526:135–40. https://doi.org/10.1016/j.bbrc.2020.03.044
Li M, Li L, Zhang Y, Wang X (2020) An investigation of the expression of 2019 novel coronavirus cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9(1):1–7. https://doi.org/10.1186/s40249-020-00662-x
Beyerstedt S, Casaro EB, Rangel ÉB (2021) COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis 40:905–19. https://doi.org/10.1007/s10096-020-04138-6
Takeda M (2022) Proteolytic activation of SARS-CoV-2 spike protein. Microbiol Immunol 66:15–23. https://doi.org/10.1016/j.jbc.2022.101710
Hoffmann M, Pöhlmann S (2021) How SARS-CoV-2 makes the cut. Nat Microbiol 6:828–9. https://doi.org/10.1038/s41564-021-00931-x
Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC et al (2021) The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6:899–909. https://doi.org/10.1038/s41564-021-00908-w
Liu J, Lu F, Chen Y, Plow E, Qin J (2022) Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J Biol Chem 298:101710. https://doi.org/10.1016/j.jbc.2022.101710
Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85:873–82. https://doi.org/10.1128/JVI.02062-10
Welsch S, Müller B, Kräusslich H-G (2007) More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett 581:2089–97. https://doi.org/10.1016/j.febslet.2007.03.060
Machhi J, Herskovitz J, Senan AM, Dutta D, Nath B, Oleynikov MD et al (2020) The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J Neuroimmune Pharmacol 15:359–86. https://doi.org/10.1007/s11481-020-09944-5
Khan S, Liu J, Xue M (2020) Transmission of SARS-CoV-2, required developments in research and associated public health concerns. Front Med. https://doi.org/10.3389/fmed.2020.00310
Speiser DE, Bachmann MF (2020) Covid-19: mechanisms of vaccination and immunity. Vaccines 8:1–22. https://doi.org/10.3390/vaccines8030404
Farahani M, Niknam Z, Mohammadi Amirabad L, Amiri-Dashatan N, Koushki M, Nemati M et al (2022) Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets. Biomed Pharmacother 145:112420. https://doi.org/10.1016/j.biopha.2021.112420
Yadav R, Chaudhary JK, Jain N, Chaudhary PK, Khanra S, Dhamija P et al (2021) Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 10:1–16. https://doi.org/10.3390/cells10040821
Satarker S, Nampoothiri M (2020) Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch Med Res 51:482–91. https://doi.org/10.1016/j.arcmed.2020.05.012
Pluskota-Karwatka D, Hoffmann M, Barciszewski J (2021) Reducing SARS-CoV-2 pathological protein activity with small molecules. J Pharm Anal 11:383–97. https://doi.org/10.1016/j.jpha.2021.03.012
Karygianni L, Ren Z, Koo H, Thurnheer T (2020) Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol 28:668–81. https://doi.org/10.1016/j.tim.2020.03.016
Dutour-Provenzano G, Etienne-Manneville S (2021) Intermediate filaments. Curr Biol 31:R522-9. https://doi.org/10.1016/j.cub.2021.04.011
Mouw JK, Ou G, Weaver VM (2014) Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol 15:771–85. https://doi.org/10.1038/nrm3902
Newberry RW, Raines RT (2019) Secondary forces in protein folding. ACS Chem Biol 14:1677–86. https://doi.org/10.1021/acschembio.9b00339
Harrington MJ, Fratzl P (2021) Natural load-bearing protein materials. Prog Mater Sci 120:100767. https://doi.org/10.1016/j.pmatsci.2020.100767
Xu Q, Torres JE, Hakim M, Babiak PM, Pal P, Battistoni CM et al (2021) Collagen- and hyaluronic acid-based hydrogels and their biomedical applications. Mater Sci Eng R Rep 146:100641. https://doi.org/10.1016/j.mser.2021.100641
Gómez SA, Rojas-Valencia N, Gómez S, Egidi F, Cappelli C, Restrepo A (2021) Binding of SARS-CoV-2 to cell receptors: a tale of molecular evolution. ChemBioChem 22:724–32. https://doi.org/10.1002/cbic.202000618
Mousavizadeh L, Ghasemi S (2021) Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect 54:159–63. https://doi.org/10.1016/j.jmii.2020.03.022
Jamir E, Sarma H, Priyadarsinee L, Kiewhuo K, Nagamani S, Sastry GN (2022) A structure-based drug repurposing approach by considering the twenty four SARS-CoV2 targets: a consensus scoring approach. 382:727. https://doi.org/10.21203/rs.3.rs-2083023/v1
Mody V, Ho J, Wills S, Mawri A, Lawson L, Ebert MCCJC et al (2021) Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun Biol 4:93. https://doi.org/10.1038/s42003-020-01577-x
Venkataraman S, Prasad BVLS, Selvarajan R (2018) RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses 10:1–23. https://doi.org/10.3390/v10020076
Zhu W, Chen CZ, Gorshkov K, Xu M, Lo DC, Zheng W (2020) RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov 25:1141–51. https://doi.org/10.1177/2472555220942123
Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P et al (2020) Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med 18:179. https://doi.org/10.1186/s12967-020-02344-6
Kumar P, Parveen, Raj N, Kumar M, Fakhri KU, Kumar S et al (2024) Natural products from Streptomyces spp. as potential inhibitors of the major factors (holoRdRp and nsp13) for SARS-CoV-2 replication: an in silico approach. Arch Microbiol 206:1–23. https://doi.org/10.1007/s00203-023-03820-5
Polak S, Wisniowska B, Fijorek K, Glinka A, Polak M, Mendyk A (2011) The open-access dataset for insilico cardiotoxicity prediction system. Bioinformation 6:244. https://doi.org/10.6026/97320630006244
Sinha A, Vaggu RG, Swain R, Patnaik S (2023) Repurposing of RAS-pathway mediated drugs for intestinal inflammation related diseases for treating SARS-CoV-2 infection. Curr Microbiol 80:1–14. https://doi.org/10.1007/s00284-023-03304-1
Ng YL, Salim CK, Chu JJH (2021) Drug repurposing for COVID-19: approaches, challenges and promising candidates. Pharmacol Ther 228:107930. https://doi.org/10.1016/j.pharmthera.2021.107930
Malin JJ, Suárez I, Priesner V, Fätkenheuer G, Rybniker J (2021) Remdesivir against COVID-19 and other viral diseases. Clin Microbiol Rev 34:1–21. https://doi.org/10.1128/CMR.00162-20
Keretsu S, Bhujbal SP, Cho SJ (2020) Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci Rep. https://doi.org/10.1038/s41598-020-74468-0
Frediansyah A, Nainu F, Dhama K, Mudatsir M, Harapan H (2021) Remdesivir and its antiviral activity against COVID-19: a systematic review. Clin Epidemiol Glob Health 9:123–7. https://doi.org/10.1016/j.cegh.2020.07.011
Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S et al (2020) Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci 6:672–83. https://doi.org/10.1021/acscentsci.0c00489
Tripathy S, Dassarma B, Roy S, Chabalala H, Matsabisa MG (2020) A review on possible modes of action of chloroquine/hydroxychloroquine: repurposing against SAR-CoV-2 (COVID-19) pandemic. Int J Antimicrob Agents 56:106028. https://doi.org/10.1016/j.ijantimicag.2020.106028
Hashem AM, Alghamdi BS, Algaissi AA, Alshehri FS, Bukhari A, Alfaleh MA et al (2020) Therapeutic use of chloroquine and hydroxychloroquine in COVID-19 and other viral infections: a narrative review. Travel Med Infect Dis 35:101735. https://doi.org/10.1016/j.tmaid.2020.101735
Zhu Y, Li J, Pang Z (2021) Recent insights for the emerging COVID-19: drug discovery, therapeutic options and vaccine. Asian J Pharm Sci 16:4–23. https://doi.org/10.1016/j.ajps.2020.06.001
Gorems A, Fentahun E, Demelash Z (2021) Roles of existing drug and drug targets for COVID-19 management. Metab Open 11:100103. https://doi.org/10.1016/j.metop.2021.100103
Malhani AA, Enani MA, Sharif-Askari FS, Alghareeb MR, Bin-Brikan RT, AlShahrani SA et al (2021) Combination of (interferon beta-1b, lopinavir/ritonavir and ribavirin) versus favipiravir in hospitalized patients with non-critical COVID-19: a cohort study. PLoS ONE 16:1–10. https://doi.org/10.1371/journal.pone.0252984
Niknam Z, Jafari A, Golchin A, Danesh Pouya F, Nemati M, Rezaei-Tavirani M et al (2022) Potential therapeutic options for COVID-19: an update on current evidence. Eur J Med Res 27:1–15. https://doi.org/10.1186/s40001-021-00626-3
Malgie J, Schoones JW, Pijls BG (2021) Decreased mortality in coronavirus disease 2019 patients treated with tocilizumab: a rapid systematic review and meta-analysis of observational studies. Clin Infect Dis 72:e742-9. https://doi.org/10.1093/cid/ciaa1445
Magro G (2020) SARS-CoV-2 and COVID-19: Is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides tocilizumab? SGP130Fc. Cytokine X 2:100029. https://doi.org/10.1016/j.cytox.2020.100029
Ghanbari R, Teimoori A, Sadeghi A, Mohamadkhani A, Rezasoltani S, Asadi E et al (2020) Existing antiviral options against SARS-CoV-2 replication in COVID-19 patients. Future Microbiol 15:1747–58. https://doi.org/10.2217/fmb-2020-0120
Roy V, Agrofoglio LA (2022) Nucleosides and emerging viruses: a new story. Drug Discov Today 27:1945–53. https://doi.org/10.1016/j.drudis.2022.02.013
Zein AFMZ, Sulistiyana CS, Raffaello WM, Wibowo A, Pranata R (2022) Sofosbuvir with daclatasvir and the outcomes of patients with COVID-19: a systematic review and meta-analysis with GRADE assessment. Postgrad Med J 98:509–14. https://doi.org/10.1136/postgradmedj-2021-140287
Jockusch S, Tao C, Li X, Chien M, Kumar S, Morozova I et al (2020) Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci Rep 10:16577. https://doi.org/10.1038/s41598-020-73641-9
Fung HB, Stone EA, Piacenti FJ (2002) Tenofovir disoproxil fumarate: a nucleotide reverse transcriptase inhibitor for the treatment of HIV infection. Clin Ther 24:1515–48. https://doi.org/10.1016/S0149-2918(02)80058-3
Parienti JJ, Prazuck T, Peyro-Saint-Paul L, Fournier A, Valentin C, Brucato S et al (2021) Effect of tenofovir disoproxil fumarate and emtricitabine on nasopharyngeal SARS-CoV-2 viral load burden amongst outpatients with COVID-19: a pilot, randomized, open-label phase 2 trial. eClinicalMedicine 38:100993. https://doi.org/10.1016/j.eclinm.2021.100993
de la Torre BG, Albericio AF (2021) The pharmaceutical industry in 2021. An analysis of FDA drug approvals from the perspective of molecules. Molecules 2022(27):1075. https://doi.org/10.3390/molecules27031075
Ayerdi O, Puerta T, Clavo P, Vera M, Ballesteros J, Fuentes ME et al (2020) Preventive efficacy of tenofovir/emtricitabine against severe acute respiratory syndrome coronavirus 2 among pre-exposure prophylaxis users. Open Forum Infect Dis. https://doi.org/10.1093/ofid/ofaa455
Sivakumar K, Kannappan S, Vijayakumar B (2023) Docking studies on biomolecules from marine microalga skeletonema costatum against hemolysin protein of bioluminescence disease-causing Vibrio harveyi. Curr Microbiol. 80:1–16. https://doi.org/10.1007/s00284-023-03372-3
Arokia Rajan MS, Thirunavukkarasu R, Joseph J, Palliyath GK, Somarathinam K, Kothandan G et al (2023) Identification of the seaweed metabolites as potential anti-tubercular agents against human pantothenate synthetase: an in silico approach. Curr Microbiol 80:1–9. https://doi.org/10.1007/s00284-023-03422-w
Sadybekov AV, Katritch V (2023) Computational approaches streamlining drug discovery. Nature 616:673–85. https://doi.org/10.1038/s41586-023-05905-z
Mohamed EAR, Abdel-Rahman IM, Zaki MEA, Al-Khdhairawi A, Abdelhamid MM, Alqaisi AM et al (2023) In silico prediction of potential inhibitors for SARS-CoV-2 omicron variant using molecular docking and dynamics simulation-based drug repurposing. J Mol Model. https://doi.org/10.1007/s00894-023-05457-z
Rolta R, Yadav R, Salaria D, Trivedi S, Imran M, Sourirajan A et al (2020) In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J Biomol Struct Dyn 0:1–18. https://doi.org/10.1080/07391102.2020.1804457
Muratov EN, Amaro R, Andrade CH, Brown N, Ekins S, Fourches D et al (2021) A critical overview of computational approaches employed for COVID-19 drug discovery. Chem Soc Rev 50:9121–51. https://doi.org/10.1039/d0cs01065k
Harwansh RK (2022) In-silico bioprospecting of taraxerol as a main protease inhibitor of SARS-CoV-2 to develop therapy against COVID-19. Struct Chem. https://doi.org/10.1007/s11224-022-01943-x
Borkotoky S, Banerjee M, Modi GP, Dubey VK (2021) Identification of high affinity and low molecular alternatives of boceprevir against SARS-CoV-2 main protease: a virtual screening approach. Chem Phys Lett. 770:138446. https://doi.org/10.1016/j.cplett.2021.138446
Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MÁ et al (2020) COVID-19: drug targets and potential treatments. J Med Chem 63:12359–86. https://doi.org/10.1021/acs.jmedchem.0c00606
Teli DM, Shah MB, Chhabria MT (2021) In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: targets for COVID-19. Front Mol Biosci. https://doi.org/10.3389/fmolb.2020.599079
Freidel MR, Armen RS (2021) Mapping major SARS-CoV-2 drug targets and assessment of druggability using computational fragment screening: Identification of an allosteric small-molecule binding site on the Nsp13 helicase. PLoS ONE 16:e0246181. https://doi.org/10.1371/journal.pone.0246181
Deniz S, Uysal TK, Capasso C, Supuran CT, Ozensoy Guler O (2021) Is carbonic anhydrase inhibition useful as a complementary therapy of Covid-19 infection? J Enzyme Inhib Med Chem 36:1230–5. https://doi.org/10.1080/14756366.2021.1924165
Rogers DM, Agarwal R, Vermaas JV, Smith MD, Rajeshwar RT, Cooper C et al (2023) SARS-CoV2 billion-compound docking. Sci Data 10:1–12. https://doi.org/10.1038/s41597-023-01984-9
Amin SA, Banerjee S, Ghosh K, Gayen S, Jha T (2021) Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg Med Chem 29:115860. https://doi.org/10.1016/j.bmc.2020.115860
Romeo A, Iacovelli F, Falconi M (2020) Targeting the SARS-CoV-2 spike glycoprotein prefusion conformation: virtual screening and molecular dynamics simulations applied to the identification of potential fusion inhibitors. Virus Res 286:198068. https://doi.org/10.1016/j.virusres.2020.198068
Saurabh S, Sivakumar PM, Perumal V, Khosravi A, Sugumaran A, Prabhawathi V (2020) Molecular dynamics simulations in drug discovery and drug delivery. Springer, Cham, pp 275–301
Chang Y, Hawkins BA, Du JJ, Groundwater PW, Hibbs DE, Lai F (2022) A guide to in silico drug design. Pharmaceutics 15:49. https://doi.org/10.3390/pharmaceutics15010049
Bai Q, Li L, Liu S, Xiao S, Guo Y (2018) Drug design progress of in silico, in vitro and in vivo researches. In-vitro In-vivo In-silico J pp 16–37
Edache EI, Uzairu A, Mamza PA, Shallangwa GA (2022) QSAR, homology modeling, and docking simulation on SARS-CoV-2 and Pseudomonas aeruginosa inhibitors, ADMET, and molecular dynamic simulations to find a possible oral lead candidate. J Genet Eng Biotechnol. https://doi.org/10.1186/s43141-022-00362-z
Kanan T, Kanan D, Al Shardoub EJ, Durdagi S (2021) Transcription factor NF-κB as target for SARS-CoV-2 drug discovery efforts using inflammation-based QSAR screening model. J Mol Graph Model 108:107968. https://doi.org/10.1016/j.jmgm.2021.107968
Haddad Y, Adam V, Heger Z (2020) Ten quick tips for homology modeling of high-resolution protein 3D structures. PLoS Comput Biol 16:1–13. https://doi.org/10.1371/journal.pcbi.1007449
Xiang R, Yu Z, Wang Y, Wang L, Huo S, Li Y, et al (2022) Recent advances in developing small-molecule inhibitors against SARS-CoV-2. Acta Pharm Sin B. https://doi.org/10.1016/j.apsb.2021.06.016
Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y et al (2020) Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582:289–93. https://doi.org/10.1038/s41586-020-2223-y
Pearce R, Zhang Y (2021) Deep learning techniques have significantly impacted protein structure prediction and protein design. Curr Opin Struct Biol 68:194–207. https://doi.org/10.1016/j.sbi.2021.01.007
Chand GB, Banerjee A, Azad GK (2020) Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ 8:e9492. https://doi.org/10.7717/peerj.9492
Zeng L, Li D, Tong W, Shi T, Ning B (2021) Biochemical features and mutations of key proteins in SARS-CoV-2 and their impacts on RNA therapeutics. Biochem Pharmacol 189:114424. https://doi.org/10.1016/j.bcp.2021.114424
Shen W, Shi Y, Dai Z, Wang A (2020) The RNA-dependent RNA polymerase NiB of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses 12:77. https://doi.org/10.3390/v12010077
Sun C, Chen L, Yang J, Luo C, Zhang Y, Li J et al (2020) SARS-CoV-2 and SARS-CoV Spike-RBD structure and receptor binding comparison and potential implications on neutralizing antibody and vaccine development. bioRxiv 25:2000044. https://doi.org/10.1101/2020.02.16.951723
Luan J, Jin X, Lu Y, Zhang L (2020) SARS-CoV-2 spike protein favors ACE2 from bovidae and cricetidae. J Med Virol 92:1649–56. https://doi.org/10.1002/jmv.25817
Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M et al (2020) Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183:1024-1042.e21. https://doi.org/10.1016/j.cell.2020.09.037
Sayed AM, Khattab AR, AboulMagd AM, Hassan HM, Rateb ME, Zaid H et al (2020) Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale. RSC Adv 10:19790–802. https://doi.org/10.1039/D0RA04199H
Chaturvedi M, Nagre K, Yadav JP (2021) In silico approach for identification of natural compounds as potential COVID 19 main protease (Mpro) inhibitors. VirusDisease 32:325–9. https://doi.org/10.1007/s13337-021-00701-7
Buonaguro L, Tagliamonte M, Tornesello ML, Buonaguro FM (2020) SARS-CoV-2 RNA polymerase as target for antiviral therapy. J Transl Med. BioMed Central 18:1–8. https://doi.org/10.1186/s12967-020-02355-3
Vicenti I, Zazzi M, Saladini F (2021) SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin Ther Pat 31:325–37. https://doi.org/10.1080/13543776.2021.1880568
Low ZY, Zabidi NZ, Yip AJW, Puniyamurti A, Chow VTK, Lal SK (2022) SARS-CoV-2 non-structural proteins and their roles in host immune evasion. Viruses 14:1–27. https://doi.org/10.3390/v14091991
Xiaojie S, Yu L, Lei Y, Guang Y, Min Q (2021) Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res 50:102125. https://doi.org/10.1016/j.scr.2020.102125
Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M et al (2020) Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun 527:618–23. https://doi.org/10.1016/j.bbrc.2020.04.136
Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CTK et al (2008) Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–9. https://doi.org/10.1128/JVI.02472-07
Hagemeijer MC, Verheije MH, Ulasli M, Shaltiël IA, de Vries LA, Reggiori F et al (2010) Dynamics of coronavirus replication-transcription complexes. J Virol 84:2134–49. https://doi.org/10.1128/JVI.01716-09
Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ (2013) Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. https://doi.org/10.1128/mBio.00524-13
Liu Y, Qin C, Rao Y, Ngo C, Feng JJ, Zhao J et al (2021) SARS-CoV-2 Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. MBio. https://doi.org/10.1128/mBio.02335-21
Raj R (2021) Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochem Biophys Rep 25:100847. https://doi.org/10.1016/j.bbrep.2020.100847
Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E (2012) RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci 109:9372–7. https://doi.org/10.1073/pnas.1201130109
Wang D, Chen J, Yu C, Zhu X, Xu S, Fang L et al (2019) Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J Virol. https://doi.org/10.1128/JVI.00623-19
Tahir M (2021) Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol 93:4258–64. https://doi.org/10.1002/jmv.27009
Chang L-J, Chen T-H (2021) NSP16 2′-O-MTase in coronavirus pathogenesis: possible prevention and treatments strategies. Viruses 13:538. https://doi.org/10.3390/v13040538
Yue Y, Nabar NR, Shi C-S, Kamenyeva O, Xiao X, Hwang I-Y et al (2018) SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis 9:904https://doi.org/10.1038/s41419-018-0917-y
Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–57https://doi.org/10.1128/JVI.01782-06
Shiraki K, Sato N, Sakai K, Matsumoto S, Kaszynski RH, Takemoto M (2022) Antiviral therapy for COVID-19: derivation of optimal strategy based on past antiviral and favipiravir experiences. Pharmacol Ther 235:108121https://doi.org/10.1016/j.pharmthera.2022.108121
Chen J, Lin S, Niu C, Xiao Q (2021) Clinical evaluation of Shufeng jiedu capsules combined with umifenovir (Arbidol) in the treatment of common-type COVID-19: a retrospective study. Expert Rev Respir Med 15:257–65https://doi.org/10.1080/17476348.2020.1822741
Shoemaker RH, Panettieri RA, Libutti SK, Hochster HS, Watts NR, Wingfield PT et al (2022) Development of an aerosol intervention for COVID-19 disease: tolerability of soluble ACE2 (APN01) administered via nebulizer. PLoS ONE 17:e0271066https://doi.org/10.1371/journal.pone.0271066
Rogosnitzky M, Okediji P, Koman I (2020) Cepharanthine: a review of the antiviral potential of a Japanese-approved alopecia drug in COVID-19. Pharmacol Rep 72:1509–16https://doi.org/10.1007/s43440-020-00132-z
Vangeel L, Chiu W, De Jonghe S, Maes P, Slechten B, Raymenants J et al (2022) Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 omicron and other variants of concern. Antiviral Res 198:105252https://doi.org/10.1016/j.antiviral.2022.105252
Zhang Z, Wang S, Tu X, Peng X, Huang Y, Wang L et al (2020) A comparative study on the time to achieve negative nucleic acid testing and hospital stays between danoprevir and lopinavir/ritonavir in the treatment of patients with COVID-19. J Med Virol 92:2631–6https://doi.org/10.1002/jmv.26141
Doi K, Ikeda M, Hayase N, Moriya K, Morimura N, Maehara H et al (2020) Nafamostat mesylate treatment in combination with favipiravir for patients critically ill with Covid-19: a case series. Crit Care 24:392https://doi.org/10.1186/s13054-020-03078-z
Sarthi P, Gupta S, Biswal S, Panda SK, Ray K, Rana MK (2022) Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. J Biomol Struct Dyn 40:2217–26. https://doi.org/10.1080/07391102.2020.1839564
Cao Y, Wei J, Zou L, Jiang T, Wang G, Chen L et al (2020) Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J Allergy Clin Immunol 146:137-146.e3. https://doi.org/10.1016/j.jaci.2020.05.019
Guimarães PO, Quirk D, Furtado RH, Maia LN, Saraiva JF, Antunes MO et al (2021) Tofacitinib in patients hospitalized with covid-19 pneumonia. N Engl J Med 385:406–15. https://doi.org/10.1056/NEJMoa2101643
Aman J, Duijvelaar E, Botros L, Kianzad A, Schippers JR, Smeele PJ et al (2021) Imatinib in patients with severe COVID-19: a randomised, double-blind, placebo-controlled, clinical trial. Lancet Respir Med 9:957–68. https://doi.org/10.1016/S2213-2600(21)00237-X
Lenze EJ, Mattar C, Zorumski CF, Stevens A, Schweiger J, Nicol GE et al (2020) Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID-19. JAMA 324:2292. https://doi.org/10.1001/jama.2020.22760
Wang Y, Jiang W, He Q, Wang C, Wang B, Zhou P et al (2020) A retrospective cohort study of methylprednisolone therapy in severe patients with COVID-19 pneumonia. Signal Transduct Target Ther 5:57. https://doi.org/10.1038/s41392-020-0158-2
Ramakrishnan S, Nicolau DV, Langford B, Mahdi M, Jeffers H, Mwasuku C et al (2021) Inhaled budesonide in the treatment of early COVID-19 (STOIC): a phase 2, open-label, randomised controlled trial. Lancet Respir Med 9:763–72. https://doi.org/10.1016/S2213-2600(21)00160-0
Lin Y, Wu F, Xie Z, Song X, Zhu Q, Wei J et al (2020) Clinical study of artesunate in the treatment of coronavirus disease 2019. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 32:417–20. https://doi.org/10.3760/cma.j.cn121430-20200312-00412
Nakhlband A, Fakhari A, Azizi H (2021) Interferon-beta offers promising avenues to COVID-19 treatment: a systematic review and meta-analysis of clinical trial studies. Naunyn Schmiedebergs Arch Pharmacol 394:829–38. https://doi.org/10.1007/s00210-021-02061
Al-kuraishy HM, Al-Gareeb AI, Elekhnawy E, Batiha GE-S (2022) Nitazoxanide and COVID-19: a review. Mol Biol Rep. 49:11169–76. https://doi.org/10.1007/s11033-022-07822-2
Singh S, Weiss A, Goodman J, Fisk M, Kulkarni S, Lu I et al (2022) Niclosamide—a promising treatment for COVID-19. Br J Pharmacol 179:3250–67. https://doi.org/10.1111/bph.15843
Ansarin K, Tolouian R, Ardalan M, Taghizadieh A, Varshochi M, Teimouri S et al (2020) Effect of bromhexine on clinical outcomes and mortality in COVID-19 patients: a randomized clinical trial. BioImpacts 10:209–15. https://doi.org/10.34172/bi.2020.27
Earhart AP, Holliday ZM, Hofmann HV, Schrum AG (2020) Consideration of dornase alfa for the treatment of severe COVID-19 acute respiratory distress syndrome. New Microbes New Infect 35:100689. https://doi.org/10.1016/j.nmni.2020.100689
Zhao H, Davies R, Ma D (2021) Potential therapeutic value of dexmedetomidine in COVID-19 patients admitted to ICU. Br J Anaesth 126:e33-5. https://doi.org/10.1016/j.bja.2020.09.031
Mahdi M, Hermán L, Réthelyi JM, Bálint BL (2022) Potential role of the antidepressants fluoxetine and fluvoxamine in the treatment of COVID-19. Int J Mol Sci 23:3812. https://doi.org/10.3390/ijms23073812
Acknowledgements
Authors LKA & KS sincerely acknowledges the Ministry of Education, Government of India, and the Ministry of Higher Education, Government of Rajasthan, for providing grant under RUSA 2.0, Research and Innovation project.
Funding
The Authors Lokesh Kumar Agarwal and Kuldeep Sharma are grateful to RUSA 2.0 of Ministry of Education (formerly known as MHRD), New Delhi, INDIA (F30(16)SPD/RUSA/2016/178 Dated on 31 March, 2020) for financial support.
Author information
Authors and Affiliations
Contributions
P.K. wrote the original draft, Data curation; K.S. reviewed the manuscript L.K.A. conceptualized, supervised, data analysis, and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no known competing financial and/or non-financial interests that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumawat, P., Agarwal, L.K. & Sharma, K. An Overview of SARS-CoV-2 Potential Targets, Inhibitors, and Computational Insights to Enrich the Promising Treatment Strategies. Curr Microbiol 81, 169 (2024). https://doi.org/10.1007/s00284-024-03671-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-024-03671-3