Abstract
Each symbiotic Chlorella variabilis associated with the ciliate Paramecium bursaria is enclosed in a symbiosome called the perialgal vacuole. Various potential symbionts, such as bacteria, yeasts, other algae, and free-living Chlorella spp., can infect P. bursaria. However, the detailed infection process of each of them in algae-free P. bursaria is unknown. Here, we aimed to elucidate the difference of the infection process between the free-living C. sorokiniana strain NIES-2169 and native symbiotic C. variabilis strain 1N. We investigated the fate of ingested algae using algae-free P. bursaria exposed separately to three types of algal inocula: NIES-2169 only, 1N only, or a mixture of NIES-2169 and 1N. We found that (1) only one algal species, preferably the native one, was retained in host cells, indicating a type of host compatibility and (2) the algal localization style beneath the host cell cortex varied between different Chlorella spp. showing various levels of host compatibilities, which was prospectively attributable to the difference in the formation of the perialgal vacuole membrane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Symbiotic interactions are spread ubiquitously in nature, whether they are parasitic or mutualistic. While the term “symbiosis” has frequently been used to describe mutually beneficial associations, symbiosis means “living together” regardless of whether the outcome is beneficial, neutral, or detrimental [1]. In marine environments, symbioses between invertebrates and photosynthetic dinoflagellate algae belonging to the genus Symbiodinium are frequent; mutualistic association between Symbiodinium spp. and Cnidarians, such as hard and soft corals, sea anemones, jellyfish, and hydrocorals, is common [2]. In freshwater environments, endosymbiotic Chlorella or Chlorella-like algae are present in many protists, including Paramecium bursaria, Climacostomum virens, Coleps hirtus, Euplotes daidaleos, Frontonia sp., Ophrydium sp., Stentor polymorphus, Difflugia sp., and Mayorella viridis, and hydras [3, 4].
Previously, symbiotic associations between P. bursaria and coccoid green algae belonging to the genera Chlorella and Micractinium were reported [5]. Each symbiotic alga is enveloped with a perialgal vacuole (PV) membrane derived from the host digestive vacuole (DV) membrane; it corresponds to the symbiosome membrane and protects the alga from digestion by host lysosomal fusion [6]. Paramecium bursaria and the symbiotic algae retain the ability to divide in the absence of the other. Reestablishment of endosymbiosis between algae-free P. bursaria and symbiotic Chlorella cells isolated from algae-bearing P. bursaria can be synchronously induced by pulse labeling of symbiotic Chlorella cells, chasing them for different periods [7], and infection can be observed using light and transmission electron microscopy [8]. Four cytological events were identified to be crucial for establishing endosymbiosis [8,9,10,11,12]: (1) Approximately 3 min after inoculation of algae-free P. bursaria with isolated symbiotic algae, some algae in DVs acquire resistance to the host lysosomal enzymes. (2) Approximately 30 min after inoculation, the algae escape from the DVs through the budding of DV membranes. This event is strongly inhibited by a dynamin inhibitor, Dynasore, suggesting that dynamin may be involved in DV budding. (3) Approximately 45 min after inoculation, the DV membrane enclosing a single undigested green algal cell differentiates into a PV membrane, which protects the algal cell from host lysosomal fusion. (4) Subsequently, the alga localizes beneath the host cell cortex via adhesion of the PV membrane to the host mitochondrial outer membrane. Approximately 24 h after inoculation, the algal cells localized beneath the host cell cortex start dividing and establish endosymbiosis. P. bursaria is currently used as a model organism for studying the induction of endosymbiosis. Recently, the nuclear genomes of the symbiotic C. variabilis [13] and host P. bursaria [14] have been revealed.
Previously, we reported that free-living algal strains, C. sorokiniana NIES-2169 (formerly IAM C-212) and Parachlorella kessleri (formerly Chlorella kessleri) C-531, were maintained in the algae-free P. bursaria strain OS1w (syngen R3 or B1, mating type IV) for more than 2 years after mixing [15]. Cells of C. variabilis strain NC64A, isolated from P. bursaria nearly 50 years ago and cultivated outside the host, showed low infectivity to algae-free P. bursaria in the algal logarithmic phase of growth, whereas those in the algal early stationary phase showed high infectivity [16]. Although the permanent establishment of a stable symbiosis seems to be restricted to Chlorella spp., various potential symbionts/parasites, such as bacteria (Pseudomonas sp.), yeasts (Rhodotorula rubra and Yarrowia lipolytica), and other algae (Scenedesmus sp.), are known to infect P. bursaria [17,18,19]. However, the detailed infection process of each of them in algae-free P. bursaria is unknown, and some are considered contaminants. To examine the infection process of the free-living C. sorokiniana strain NIES-2169 and native symbiotic C. variabilis strain 1N, the fate of algae, ingested in algae-free P. bursaria cells, was investigated in three experimental sets involving three different algal inocula: NIES-2169 only, 1N only, and a mixture of NIES-2169 and 1N. Based on the results obtained, we discussed the differential roles of Chlorella sp., having either high or low host compatibility, in establishing endosymbiosis with algae-free P. bursaria and the mechanism regulating this phenomenon.
Materials and Methods
Cultivation of Algae-Free and Algae-Bearing P. bursaria
Two P. bursaria strains were used in this study: algae-free Yad1w and algae-bearing Yad1g1N (syngen R3 or B1, mating type III). The Yad1g1N strain was produced by infecting cloned symbiotic Chlorella variabilis strain 1 N cells with the Yad1w cells [9]. Paramecium cell culture, prepared using red pea (Pisum sativum) extract culture medium [20] in modified Dryl’s solution (MDS: NaH2PO4·2H2O replaced with KH2PO4) [21], was inoculated with non-pathogenic Klebsiella aerogenes (ATCC35028) as food bacteria 1 day before use [22]. For ordinary cultures, several hundred P. bursaria cells were used to inoculate 2-mL aliquots of the culture medium in test tubes. Subsequently, 2 mL of fresh culture medium was added daily for 12 days. Cultures in the early stationary phase were used in experiments 1 day after the last feeding and cultured at 24 ± 1 °C. Algae-bearing P. bursaria cells were cultured under fluorescent light maintained at 20–30-μmol photons m−2s−1 using an incandescent lamp.
Cultivation of Free-Living C. sorokiniana
Chlorella sorokiniana strain NIES-2169 provided by the NIES through the NBRP of the MEXT, Japan. It was cultured using a modified Bold’s basal medium (MBBM) [23]. Two-hundred µL of 7–14-day-old NIES-2169 suspension was transferred to 10 mL of MBBM and incubated at 24 ± 1 °C for 5 days under a forementioned constant light conditions.
Isolation of Native Symbiotic C. variabilis from Algae‐Bearing P. bursaria
For the isolation of native symbiotic C. variabilis strain 1N cells, cell culture of algae-bearing P. bursaria Yad1g1N (300 mL) was strained through two layers of KimWipes to remove gross debris and subsequently, filtered using a centrifuge tube equip with a 15-μm pore nylon mesh. Yad1g1N cells were washed using 50 mL of MDS, harvested using a hand-operated centrifuge (UKG-2; Uchida Rikakiki, Tokyo, Japan), and resuspended in 1 mL of MDS. The sedimented cells were suspended in 0.1-mM phenylmethylsulphonyl fluoride (Sigma-Aldrich, St. Louis, MO, USA) containing 1-mL MDS and manually homogenized in a Teflon homogenizer placed on ice. The homogenates were then filtered using a centrifuge tube equipped with a 15-μm nylon mesh. The filtrate was transferred to 1.5-mL tubes, washed three times using 1.5-mL MDS via centrifugation at 4350×g for 1 min, and then reduced to a final volume of 500 μL. The algal cell density was calculated using a Thoma blood-counting chamber.
Labeling C. variabilis Strain 1N with Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE)
The isolated C. variabilis 1N cells were stained using CFSE (Dojindo, Japan) [24]. Suspension of the 1N cells was incubated with twice the volume of 2 × 10–3 mg/mL CFSE for 10 min in the dark at 24 ± 1 °C. Subsequently, the cells were washed twice with MDS, centrifuged at 4350×g for 1 min, and observed using differential interference contrast (DIC) and fluorescence microscope (BX53; EVIDENT, Tokyo, Japan) equipped with fluorescence mirror units U-FBNA (excitation 470–495 nm, emission 510–550 nm) for CFSE fluorescence and U-FGW (excitation 530–550 nm, emission 575 nm) for algal autofluorescence. Images were captured using a DP74 microscope (EVIDENT, Tokyo, Japan).
Analyzing the Infection Process of Free-Living C. sorokiniana and Native Symbiotic C. variabilis to the Algae-Free P. bursaria Cells
We conducted the three kinds of algal infection experiments as follows. In the infection experiments using only one Chlorella strain, the isolated native symbiotic C. variabilis, 1N, or free-living C. sorokiniana, NIES-2169, was mixed with algae-free P. bursaria (5 × 107 algal cells mixed with 5 × 103 paramecia/mL) in a centrifuge tube (10 mL) for 1 h. After mixing, the paramecia were separated from uningested algae using a 15-μm pore size nylon mesh. The paramecia retained in the mesh were transferred to a centrifuge tube (10 mL), resuspended in 1-mL MDS, and investigated at 1 and 48 h after the algal mixing. A 100-μL aliquot of the cell suspension was fixed with 100 μL of 8% (w/v) paraformaldehyde (PFA) 1 h and 48 h after the mixing, and the fixed P. bursaria cells were observed using the DIC and fluorescence microscopy and determined the algal infection rate. The infection rate refers to the percentage of P. bursaria that retained multiple Chlorella sp. in the cell. In the infection experiments using two strains of Chlorella mixture, equal volumes of CFSE-labeled 1N and NIES-2169 (2.5 × 107 algal cells/mL of each strain) were mixed with algae-free P. bursaria (5 × 103 cells/mL) as described above. Throughout this experiment, all setups were maintained at 24 ± 1 °C under a forementioned constant light conditions.
Indirect Immunofluorescence Microscopy
Indirect immunofluorescence microscopy was performed as described previously [25, 26]. Approximately one hundred P. bursaria cells were dried on coverslips (4.5 × 24 mm) using a Slide Glass Dryer Fan (AS ONE Corporation, Japan) 48 h after mixing with NIES-2169. The cells were then fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) (137-mM NaCl, 2.68-mM KCl, 8.1-mM NaHPO4·12H2O, 1.47-mM KH2PO4, pH 7.2) for 10 min at 4 °C. The fixed cells were then washed with PBST (PBS containing 0.05% Tween 20) and PBS for 10 min at 4 °C. The cells were then treated with anti-Paramecium mitochondrial monoclonal antibody [26] overnight at 4 °C. Cells were washed twice with PBS. The cells were then treated with Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes) diluted 1000-fold in PBS for 2 h at 24 ± 1 °C and washed twice with PBS for 10 min. The samples were examined by DIC and fluorescence microscopy. Professor Emeritus Masahiro Fujishima (Yamaguchi University, Japan) provided the monoclonal antibody.
Quantification and Statistical Analysis
The Welch’s t test and two-sided Fisher’s exact test was used for the statistical evaluation of the results. Values are shown for data that reached a significance of P ≤ 0.01 (∗∗). All data are shown as the mean ± SD. All statistical analyses were performed using R (R Ver 4.1.3) [27].
Results
Growth Curve of C. sorokiniana Strain NIES-2169 in MBBM
As indicated previously, the rate of C. variabilis strain NC64A infection in algae-free P. bursaria depends on the algal growth phase [16]. Therefore, we first examined the growth curve of C. sorokiniana strain NIES-2169 in MBBM; the culture entered the early stationary and stationary phases at 5 and 8 days after inoculation, respectively. The culture entered the decay phase 19 days after inoculation, and the cell density gradually decreased. According to Welch’s t test at a 5% significance level, the cell density did not vary significantly between the 5 and 19 days after inoculation. These results indicated that the early stationary phase of NIES-2169 started on day 5 (Fig. 1).
Growth curves of Chlorella sorokiniana strain NIES-2169. The culture entered early stationary, stationary, and decay phases at 5, 8, and 19 days after cultivation, respectively; at the decay phase, the cell density began to decrease slightly. The bars in the graph indicate the standard deviation (SD) for three samples. The reproducibility of this result was confirmed three times
The Process of Free-Living C. sorokiniana and Native Symbiotic C. variabilis Infecting the Algae-Free P. bursaria
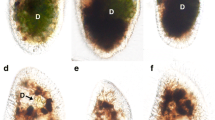
NIES-2169 cells in the early stationary phase were mixed with algae-free P. bursaria for 1 h (Fig. 2a, b). Non-ingested NIES-2169 by the algae-free P. bursaria cells were removed, and then P. bursaria cells were fixed using PFA and analyzed using DIC and fluorescence microscopy. DIC microscopy revealed many DVs enclosing green NIES-2169 cells in the P. bursaria 1 h after mixing algae (Fig. 2a). These DV membranes were moving in a constant direction in the cytoplasm due to the host cytoplasmic streaming (Fig. S1, upper). The fluorescence image demonstrated algal autofluorescence (Fig. 2b). All P. bursaria (n = 60, three independent experiments) cells contained several hundreds of ingested algae. Many crystals (Fig. 2a, r; polarized rainbow-colored granules) were observed in P. bursaria. P. bursaria that ingested NIES-2169 were incubated under light conditions without feeding and investigated 48 h after mixing with algae; DIC microscopic image revealed multiple green NIES-2169 in P. bursaria cells (Fig. 2c), and multiple green NIES-2169 cells enclosed by a single DV membrane localized near the host cell cortex (Fig. 2c'). This phenomenon is evident in the magnified image showing algal autofluorescence (Fig. 2d, d′). The DV membrane enclosing multiple green NIES-2169 cells did not flow by the host cytoplasmic streaming and attached beneath the host cell cortex as like as a PV membrane (Fig. S1, lower). Figure 2e shows a DIC image of some vacuoles of algae-free P. bursaria 48 h after mixing with NIES-2169, and Fig. 2f shows a merged image of the autofluorescence of chlorophyll in algal chloroplasts of Fig. 2e and immunofluorescence microscopic image of Fig. 2e. Host mitochondria are indicated by green immunofluorescence in Fig. 2f. In the case of the PV membrane, the host mitochondria are localized around the PV membrane enclosing a symbiotic alga as shown in [26]. Immunofluorescence showed that the DV containing multiple NIES-2169 localizes closely to the mitochondria of the host cell (Fig. 2f). This mitochondrial adjacency is not found in the DV membrane but only in the PV membrane [26]. Crystals, observed 1 h after inoculation, were retained in the host cells at 48 h after inoculation (Fig. 2c). The NIES-2169 were maintained in the P. bursaria cell for several weeks (data not shown). However, when P. bursaria cells divided, non-algal P. bursaria cells frequently appeared. This observation suggests that host and NIES-2169 cell divisions may not be synchronized.
Differential interference contrast (a, c, c′, g, i, i′) and chlorophyll autofluorescence (b, d, d′, h, j, j′) and micrographs of algae-free P. bursaria mixed with free-living C. sorokiniana strain NIES-2169 and symbiotic C. variabilis strain 1N. e Shows a DIC image of some DVs of algae-free P. bursaria 48 h after mixing with NIES-2169 and f show a merged image of the autofluorescence of chlorophyll in algal chloroplasts of (e) and the immunofluorescence microscopic image of (e). Immunofluorescence (IF) of goat anti-mouse IgG indicates the host mitochondria in (f). One hour after mixing with algae, many cells of NIES-2169 (a, b) and 1N (g, h) were ingested by algae-free P. bursaria. Digestive vacuoles (a, D) and crystals (a, r) were observed in cells that ingested NIES-2169. Both NIES-2169 (c, d) and 1N (i, j) were retained in the P. bursaria after 48 h of mixing, but the patterns of their localization beneath the host cell cortex differed. The magnified image c′, d′, i′, j′ show dotted square area of c, d, i, j revealed that multiple NIES-2169 cells wrapped in a single DV membrane localized beneath the host cell cortex (d′, D). Note that the host mitochondria are localized around the DV membrane which containing NIES-2169 (f). On the other hand, 1N localized as individual cells (i′ black arrowheads and j′ white arrowheads). Ma macronucleus (Color figure online)
Figure 2g and h shows native symbiotic C. variabilis strain 1N freshly isolated from algae-bearing P. bursaria, mixed with algae-free P. bursaria for 1 h. Non-ingested 1N by the algae-free P. bursaria cells were removed and then P. bursaria cells were fixed using PFA and analyzed using DIC and fluorescence microscopy. DIC microscopy revealed a lot of 1N cells in the cytoplasm of P. bursaria (Fig. 2g). However, in contrast to the images showing NIES-2169, the shape of each DV was barely observable. The digested 1N cells could be distinguished based on their brown color (Fig. 2g). Whether the 1N was digested or not could not be distinguished based on autofluorescence (Fig. 2h). As shown in [7], each DV containing multiple 1N cells flowed by the host cytoplasmic streaming. These P. bursaria cells lacked the crystals observed in their counterparts that ingested NIES-2169. Forty-eight h after mixing with the algae-free P. bursaria, each 1N cell was individually attached beneath the host cell cortex (Fig. 2i–j′). The membrane enveloping each 1N cell did not flow, so this type of algal localization indicates the establishment of endosymbiosis with algae-free P. bursaria and the 1N cells were considered enveloped by the PV membranes [7].
The Infection Process Analyzed Using the Algae-Free P. bursaria and the Mixture of Equal Volumes of NIES-2169 and 1N
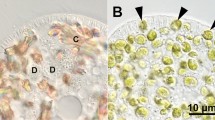
As cells of both NIES-2169 and 1N strains are green and coccoid, only 1N cells were labeled with CFSE for identification. All 1N cells (Fig. 3a) exhibited CFSE-green fluorescence (Fig. 3b). It has been confirmed that the CFSE does not leak out from the stained 1N cells and is not taken up by the NIES-2169 cells when mixed together (Tsuda and Kodama unpublished data). After mixing NIES-2169 culture and equal volumes of 1N culture with the algae-free P. bursaria for 1 and 48 h and removing non-ingested algae, P. bursaria cells were fixed with PFA for DIC (Fig. 3c, f) and fluorescence (Fig. 3d, e, g, h) microscopic analyses. One hour after mixing, several algal cells were ingested by the P. bursaria and appeared green (undigested) or brown (digested) in the DIC microscopic image (Fig. 3c). Although CFSE-green fluorescence was detected, the algal autofluorescence image aided differentiation between the two strains; only 1N cells showed yellow fluorescence due to the overlap of strong CFSE-green and red autofluorescence (Fig. 3e). Green algae localized beneath the host cell cortex were observed 48 h after algal mixing (Fig. 3f). Notably, all localized algae showed CFSE-green fluorescence (Fig. 3g), indicating that they belonged to the strain 1N strain. When algae-free P. bursaria was fed the NIES-2169 strain alone, these algal cells were observed in the P. bursaria at 48 h (Fig. 2c–d′). However, Fig. 3g shows when P. bursaria cells were fed with both NIES-2169 and 1N cells, only 1N cells are present at 48 h. This result suggests that if a native symbiont is contained within the P. bursaria’s DVs, the other organisms may be expelled or digested as soon as possible. CFSE can also be used to stain other green algae, such as Chlamydomonas reinhardtii [24] and Chlorogonium sp. (Kodama unpublished data). Furthermore, we have confirmed that CFSE stains only live Chlorella spp. rather than boiled or digested ones (Kodama unpublished data). Therefore, during the algal infection process, live and dead Chlorella cells can be distinguished using CFSE.
Differential interference contrast (a) and fluorescence (b) micrographs of symbiotic 1N cells treated with CFSE. All the cells displayed green fluorescence indicating the presence of CFSE. The reproducibility of this result was confirmed five times, and more than 500 cells were observed at each experiment. Differential interference contrast (c, f) and fluorescence (d, e, g, h) micrographs of algae-free P. bursaria mixed with a 1:1 mixture of NIES-2169 and 1N cells treated with CFSE for 1 h. Paramecia were observed at 1 h (c–e) and 48 h (f–h) after algal mixing. Only 1N cells, labeled with CFSE to distinguish them from NIES-2169, showed yellow fluorescence due to the overlap of strong green fluorescence of CFSE (d) and red autofluorescence (e). The algae, retained 48 h after mixing, localized beneath the host cell cortex (f black arrowheads) and showed CFSE fluorescence (g white arrowheads); this result indicates that these were 1N cells (Color figure online)
Infection Rates of NIES-2169, 1N, and Their Combination in the Algae-Free P. bursaria
Figure 4 shows the average infection rates detected in three independent experiments performed using algae-free P. bursaria and either of NIES-2169, 1N, and a mixture of NIES-2169 and 1N. The infection rate refers to the percentage of P. bursaria infected with green algae. Multiple green algae observed in P. bursaria indicates that the P. bursaria was being infected with Chlorella [7]. Forty-eight hours after mixing with Chlorella, the infection rates were 42%, 75%, and 43% for NIES-2169, 1N, and NIES-2169 combined with 1N, respectively. The infection rate of 1N differed significantly (Fisher’s exact test; P < 0.01) from that of NIES-2169 and the mixture of both strains (NIES-2169 and 1N). These results suggest two aspects. (i) The native symbiotic Chlorella (1N) infected algae-free P. bursaria at a higher level than the free-living Chlorella (NIES-2169). (ii) Fig. 3 shows when the addition of a mixture of NIES-2169 and 1N, only 1N cells successfully infected algae-free P. bursaria. Notably, the infection rate of 1N alone was significantly higher than that of 1N combined with NIES-2169.
Infection rates of NIES-2169, 1N, and the combination of both algal strains in the algae-free P. bursaria. Infection rates were estimated as the percentage of P. bursaria cells containing Chlorella sp. 48 h after mixing with them. The infection rate of the original symbiotic 1N was high, whereas that of the free-living NIES-2169 was low; the infection rate detected using the mixture of NIES-2169 and 1N was as low as that of NIES-2169 alone; as also shown in Fig. 3, in a mixture of NIES-2169 and 1N, only 1N can infect P. bursaria cells. Error bars indicate SD. Asterisks indicate significant differences (two-tailed Fisher’s exact test, **P < 0.01). The reproducibility of this result was confirmed three times and more than 20 cells were observed at each experiment
Discussion
In the previous studies, the sequence of the internal transcribed spacer region 1 of the 18S rRNA gene [28] and a monoclonal antibody specific for symbiotic Chlorella sp. [29] are used to identify native symbiotic algae, and the both papers have reported that only the native symbionts could establish endosymbiosis with algae-free P. bursaria. What are the triggers for the establishment of the endosymbiosis with the algae-free P. bursaria cells? First, it is known that both symbiont density and contact time are important factors for successful infection to the algae-free P. bursaria [30]. In fact, Summerer et al. [28] have successfully infected algal strains that had reportedly failed to infect to algae-free P. bursaria cells, when the algal mixing time was much longer (7 days). Secondly, maltose released by Chlorella spp. appears to act as a recognition signal and as a signal that induces the recognition ability of algae-free P. bursaria [31]. Muscatine et al. [32] reported that all symbiotic strains of the host P. bursaria, the sponge Spongilla, and the hydra Chlorohydra viridissima liberated soluble maltose or glucose to the extent of 5.4–86.7% of their total photosynthate. On the other hand, all the free-living Chlorella spp. and Selenastrum sp. released 0.4–7.6% of their total photosynthate mainly in the form of glycolic acid. Therefore, the release of soluble carbohydrates by symbiotic algae is considered to be an adaptation to the symbiosis. However, free-living Chlorella spp. do not release maltose, but they have been shown to have infectivity [17, 33]. Thus, the ability to release maltose may be advantageous but not really necessary for the infection process [18]. Third, Takeda et al. [34] reported that “infection-capable” Chlorella species to the algae-free P. bursaria cells, including the original symbiotic ones, could be distinguished by the presence of glucosamine as a chemical component in their rigid walls (alkali-insoluble part of the Chlorella cell wall), whereas the rigid walls of “infection-incapable” species to the algae-free P. bursaria cell contained glucose and mannose. They suggested that the presence of glucosamine in the rigid walls of algae is a prerequisite for determining the symbiotic association between P. bursaria and Chlorella species. Subsequently, it was reported that the infectivity of Chlorella species toward P. bursaria depends on their ability to localize beneath the host cell cortex after budding from the host DVs [15].
The following mechanisms have been reported in symbioses between other algae and other animals: Pattern recognition receptors on the host cell membrane and microbe-associated molecular patterns on the symbiont cell surface have been proposed to be critical for initiating symbiotic relationships [2], such as the symbiosis between green Hydra and Chlorella [35] or that between marine cnidarians and Symbiodinium [36, 37]. Molecular patterns, such as glycans, differ among Symbiodinium strains [37, 38], thus suggesting that the combination of host and symbiont cell surface molecules may mediate species specificity. In addition to cell surface molecules, the Symbiodinium cell size has been reported to influence its infectivity in three cnidarian species (Aiptasia sp., Acropora tenuis, and Cyphastrea serailia) [39].
Previously, we demonstrated that the rate of algal re-endosymbiosis with the algae-free P. bursaria significantly decreased when microspheres (diameter: 0.20 μm) were mixed with isolated symbiotic algae [40]. Karakashian and Karakashian [41] reported that symbiotic algae can delay the digestion of heat-killed algae when coexisting in a DV and proposed that symbiotic Chlorella actively interferes with an early digestive event in the host. We noticed a similar phenomenon; it suggests that algae release an unknown factor in response to light exposure, which may delay the host digestive process [42]. In this study, 1N that was mixed with NIES-2169 showed a significantly lower infection rate than 1N alone. This result can be discussed as follows. Lysosomes were reported to be fused to DV membranes enclosing latex beads that are not food for Paramecium; moreover, acid phosphatase activity was observed within the vacuole [43]. Microspheres, of course, cannot establish endosymbiosis. NIES-2169 also cannot establish a permanent endosymbiosis; however, their energy, molecules, or some other factor, including a digestive enzyme, such as acid phosphatase, might have been consumed, which resulted in a lower infection rate of 1N in the mixture than that of 1N alone. However, the detailed mechanism of this phenomenon remains unknown.
In cells of P. bursaria, the symbiotic alga was enclosed in a PV membrane. Previously, the following characteristics of the PV membrane have been identified: (1) The PV membrane encloses a single algal cell [44, 45]. (2) The PV membrane is directly associated with the algal cell wall and connects to the host mitochondrial membrane [46]. (3) The diameter of the PV does not vary considerably, except that observed during the division of the enclosed symbiotic alga [47]. (4) The PV is not involved in cyclosis but localized immediately beneath the host cell cortex [7, 47]. (5) The particle density and its distribution on the PV membrane show little evidence of any endocytotic or exocytotic activity, which can be observed in the DV membrane [48]. (6) PV membrane contains an active proton pump and establishes a gradient that drives maltose transport from the symbiotic algae to the host cell [49]. At 48 h after mixing NIES-2169 and algae-free P. bursaria cells, Figs. 2f and S1 lower show that the membrane surrounding NIES-2169 has the characteristics of the PV membrane, not the DV membrane. Figure 5 shows a schematic representation of the infection process of NIES-2169, 1N, and Francisella novicida strain U112 in algae-free P. bursaria; the infection process of U112 has been depicted according to the results reported in Watanabe et al. [50]. F. novicida is an intracellular pathogen and the causative agent of tularemia, and Watanabe et al. [50] had found a stable intracellular relationship established between U112 and algae-free P. bursaria under experimental conditions. Moreover, the intracellular localization of U112 in P. bursaria was observed using transmission electron microscopy; U112 localized beneath the host cell cortex 48 h after mixing with algae-free P. bursaria. Small vesicles, each enclosing multiple U112 cells, were frequent, whereas isolated U112 cells were rare. However, these vesicles were smaller than DVs observed during the early infection process and contained fewer bacteria than those in the DVs. Considering these results, the authors concluded that the vesicles were equivalent to the PV membranes. After 48 h, the number of U112 per a host cell gradually decreased with cell division of P. bursaria, possibly because U112 cannot replicate efficiently inside P. bursaria or because exocytosis of bacterial cells occurred [50]. The diagram (Fig. 5) shows that the original Chlorella sp., free-living Chlorella sp., and F. novicida were similarly ingested by the host. However, the original Chlorella spp. and other species differ in their patterns of adherence to the host cell cortex. Furthermore, free-living Chlorella sp., which can individually infect algae-free P. bursaria, cannot infect when mixed with the original ones. The establishment of the stable symbiotic association depends on the type of attachment to the host cell cortex. The failure of differentiation from the DV membrane to the permanent PV membrane, which is a crucial cytological event for establishing endosymbiosis, prospectively affects the foundation of the association. In this study, it was not possible to determine whether the NIES-2169-containing vacuolar membrane localized beneath the host cell cortex was a DV or PV membrane. Future development of a monoclonal antibody specific for the PV membrane of P. bursaria would be helpful in elucidating the PV membrane.
A schematic diagram of the infection process of free-living C. sorokiniana strain NIES-2169 (a), original symbiotic C. variabilis strain 1N (b), NIES-2169, and 1N (c) and Francisella novicida strain U112 (d) in the algae-free P. bursaria. The schematic representation of U112 infection is based on the results reported by Watanabe et al. [50]. a The NIES-2169 cells were taken up through the host cytopharynx and enclosed in a DV membrane. The initial DV membrane flowed by the host cytoplasmic streaming and some algae were digested during it. Subsequently, some NIES-2169 cells escaped the host digestion and localized beneath the host cell cortex by wrapping the membrane which enclosing multiple NIES-2169. The membrane surrounding multiple NIES-2169 had characteristics of both DV and PV membranes. As a characteristic of the DV membrane is that it contains multiple algae and as a characteristic of the PV membrane is that it adheres directly to the host cell cortex as shown in Fig. S1 and that the proximity of the host mitochondria as shown in Fig. 2f. b The 1N cells were taken up and enclosed in a DV membrane. As in NIES-2169, the initial DV membrane enclosing multiple 1N cells flowed by the host cytoplasmic streaming and some algae were digested during this process. Some of the 1N cells were also able to escape host digestion and were localized beneath the host cell cortex. In contrast to NIES-2169, each alga was individually localized beneath the host cell cortex; therefore, they were considered to be wrapped with PV membranes (blue membrane in the figure). c Both NIES-2169 and 1N cells, added as a mixture, were ingested and enclosed in the same DV membrane. Although some algal cells belonging to both the strains escaped host digestion, only 1N cells, wrapped with the PV membrane, successfully infected the P. bursaria and established endosymbiosis. d U112 cells were ingested and enclosed in a DV membrane. U112 cells were able to infect the P. bursaria and localized beneath the host cell cortex in a manner similar to that observed in case of NIES-2169 (a) (Color figure online)
Conclusion
This study revealed the following: (1) Only one species, preferably the native one, was retained in the algae-free P. bursaria involved in symbiosis, which indicates a type of host compatibility. (2) A difference in the style of localization under the host cell cortex was noticed between Chlorella species with different levels of host compatibility, which was possibly attributable to the difference in the perialgal vacuolar membrane formation. P. bursaria, which serves as a host for a variety of microorganisms, can be easily cultured. Future research to elucidate the mechanism of microbial infection of P. bursaria would aid in the effective control of microbial infection and facilitate the prevention of the spread of pathogenic bacteria using the paramecium as a host in the environment. Conversely, it may also be possible to create P. bursaria infected with beneficial microorganisms in any combination.
Data Availability
Upon reasonable request, the datasets used in the current study are available from the corresponding author.
References
Leung TLF, Poulin R (2008) Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Vie et Milieu. pp 107–115. https://hal.sorbonne-universite.fr/hal-03246057
Davy SK, Allemand D, Weis VM (2012) Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–261. https://doi.org/10.1128/MMBR.05014-11
Goetsch W (1924) Die Symbiose der Süsswasser-Hydroiden und ihre künstliche Beeinflussung. Z Morphol Ökol Tiere 1:660
Lee JJ, Soldo AT, Reisser W, Lee M, Jeon KW, Görtz H-D (1985) The extent of algal and bacterial endosymbioses in protozoa. J Protozool 32:391–403. https://doi.org/10.1111/j.1550-7408.1985.tb04034.x
Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L (2011) The systematics of “Zoochlorella” revisited employing an integrative approach. Environ Microbiol 13:350–364. https://doi.org/10.1111/j.1462-2920.2010.02333.x
Kodama Y, Fujishima M (2009) Localization of perialgal vacuoles beneath the host cell surface is not a prerequisite phenomenon for protection from the host’s lysosomal fusion in the ciliate Parameium bursaria. Protist 160:319–329. https://doi.org/10.1016/j.protis.2008.06.001
Kodama Y, Fujishima M (2005) Symbiotic Chlorella sp. of the ciliate Paramecium bursaria do not prevent acidification and lysosomal fusion of host digestive vacuole during infection. Protoplasma 225:191–203. https://doi.org/10.1007/s00709-005-0087-5
Kodama Y, Fujishima M (2010) Secondary symbiosis between Paramecium and Chlorella cells. Int Rev Cell Mol Biol 279:33–77. https://doi.org/10.1016/S1937-6448(10)79002-X
Kodama Y, Fujishima M (2009) Timing of perialgal vacuole membrane differentiation from digestive vacuole membrane in infection of symbiotic algae Chlorella vulgaris of the ciliate Paramecium bursaria. Protist 160:65–74. https://doi.org/10.1016/j.protis.2008.06.001
Kodama Y, Fujishima M (2011) Four important cytological events needed to establish endosymbiosis of symbiotic Chlorella sp. to the algae-free Paramecium bursaria. Jpn J Protozool 44:1–20
Kodama Y, Fujishima M (2012) Characteristics of the digestive vacuole membrane of the alga-bearing ciliate Paramecium bursaria. Protist 163:658–670. https://doi.org/10.1016/j.protis.2011.10.004
Fujishima M, Kodama Y (2022) Mechanisms for establishing primary and secondary endosymbiosis in Paramecium. J Euk Microbiol. https://doi.org/10.1111/jeu.12901
Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon G, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosynthesis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955. https://doi.org/10.1105/tpc.110.076406
Cheng YH, Liu CFJ, Yu YH, Jhou YT, Fujishima M, Tsai I, Leu JY (2020) Genome plasticity in Paramecium bursaria revealed by population genomics. BMC Biol 18:180. https://doi.org/10.1186/s12915-020-00912-2
Kodama Y, Fujishima M (2007) Infectivity of Chlorella species for the ciliate Paramecium bursaria is not based on sugar residues of their cell wall components, but based on their ability to localize beneath the host cell membrane after escaping from the host digestive vacuole in the early infection process. Protoplasma 231:55–63. https://doi.org/10.1007/s00709-006-0241-8
Kodama Y, Fujishima M (2016) Differences in infectivity between endosymbiotic Chlorella variabilis cultivated outside host Paramecium bursaria for 50 years and those immediately isolated from host cells after one year of reendosymbiosis. Biol Open 5(1):55–61. https://doi.org/10.1242/bio.013946
Bomford R (1965) Infection of algae-free Paramecium bursaria with strains of Chlorella, Scenedesmus, and a yeast. J Protozool 12:221–224. https://doi.org/10.1111/j.1550-7408.1965.tb01840.x
Görtz H-D (1982) Infection of Paramecium bursaria with bacteria and yeasts. J Cell Sci 58:445–453. https://doi.org/10.1242/jcs.58.1.445
Suzaki T, Omura G, Görtz H-D (2003) Infection of symbiont-free Paramecium bursaria with yeasts. Jpn J Protozool 36:17–18 (in Japanese)
Tsukii Y, Harumoto T, Yazaki K (1995) Evidence for viral macronuclear endosymbiont in Paramecium caudatum. J Euk Microbiol 42:109–115. https://doi.org/10.1111/j.1550-7408.1995.tb01550.x
Dryl S (1959) Antigensic transformation in Paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J Protozool 6:25
Fujishima M, Nagahara K, Kojima Y (1990) Changes in morphology, buoyant density and protein composition in differentiation from the reproductive short form to the infectious long form of Holospora obtusa, a macronucleus-specific symbiont of the ciliate Paramecium caudatum. Zool Sci 7(5):849–860. https://doi.org/10.34425/zs000785
Agarkova I, Dunigan D, Gurnon J, Greiner T, Barres J, Thiel G, Van Etten JL (2008) Chlorovirus-mediated membrane depolarization of Chlorella alters secondary active transport of solutes. J Virol 82:12181–12190. https://doi.org/10.1128/JVI.01687-08
Iwai S, Fujita K, Takanishi Y, Fukushi K (2019) Photosynthetic endosymbionts benefit from host’s phagotrophy, including predation on potential competitors. Curr Biol 29:3114.e3–3119.e3. https://doi.org/10.1016/j.cub.2019.07.074
Kodama Y, Fujishima M (2011) Endosymbiosis of Chlorella species to the ciliate Paramecium bursaria alters the distribution of the host’s trichocysts beneath the host cell cortex. Protoplasma 248:325–337. https://doi.org/10.1007/s00709-010-0175-z
Kodama Y, Fujishima M (2022) Endosymbiotic Chlorella variabilis reduces mitochondrial number in the ciliate Paramecium bursaria. Sci Rep 12:8216. https://doi.org/10.1038/s41598-022-12496-8
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comp Graph Stat 5:299–314
Summerer M, Sonntag B, Sommaruga R (2007) An experimental test of the symbiosis specificity between the ciliate Paramecium bursaria and strains of the unicellular green alga Chlorella. Environ Microbiol 9(8):2117–2122. https://doi.org/10.1111/j.1462-2920.2007.01322.x
Nishijima A, Fujishima M (2007) Chlorella vulgaris and C. sorokiniana cannot be maintained in Paramecium bursaria cell. Jpn J Protozool 40(1):28–29 (in Japanese)
Weis DS, Ayala A (1979) Effect of exposure period and algal concentrations on the frequency of infection of aposymbiotic ciliates by symbiotic algae from Paramecium bursaria. J Protozool 26:245–248. https://doi.org/10.1111/j.1550-7408.1979.tb02772.x
Weis DS (1980) Hypothesis: free maltose and algal cell surface sugars are signals in the infection of Paramecium bursaria by algae. In: Schwemmler W, Schenk HEA (eds) Endocytobiology, vol 1. Walter de Gruyter & Co, Berlin/New York, pp 105–112
Muscatine L, Karakashian SJ, Karakashian MW (1967) Soluble extracellular products of algae symbiotic with a ciliate, a sponge and a mutant hydra. Comp Biochem Physiol 20(1):1–12. https://doi.org/10.1016/0010-406X(67)90720-7
Karakashian SJ (1963) Growth of Paramecium bursaria as influenced by the presence of algal symbionts. Physiol Zool 36:52–68
Takeda H, Sekiguchi T, Nunokawa S, Usuki I (1998) Species-specificity of Chlorella for establishment of symbiotic association with Paramecium bursaria. Does infectivity depend upon sugar components of the cell wall? Eur J Protistol 34:133–137. https://doi.org/10.1016/S0932-4739(98)80023-0
Meints RH, Pardy RL (1980) Quantitative demonstration of cell surface involvement in a plant-animal symbiosis: lectin inhibition of reassociation. J Cell Sci 43:239–251. https://doi.org/10.1242/jcs.43.1.239
Lin KL, Wang JT, Fang LS (2000) Participation of glycoproteins on zooxanthellal cell walls in the establishment of a symbiotic relationship with the sea anemone, Aiptasia pulchella. Zool Stud 39:172–178
Wood-Charlson EM, Hollingsworth LL, Krupp DA, Weis VM (2006) Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell Microbiol 8:1985–1993. https://doi.org/10.1111/j.1462-5822.2006.00765.x
Logan DDK, LaFlamme AC, Weis VM, Davy SK (2010) Flow-cytometric characterisation of the cell-surface glycans of symbiotic dinoflagellates (Symbiodinium spp.). J Phycol 46:525–533. https://doi.org/10.1111/j.1529-8817.2010.00819.x
Biquand E, Okubo N, Aihara Y, Rolland V, Hayward DC, Hatta M, Minagawa J, Maruyama T, Takahashi S (2017) Acceptable symbiont cell size differs among cnidarian species and may limit symbiont diversity. ISME J 11:1702–1712. https://doi.org/10.1038/ismej.2017.17
Kodama Y, Sumita H (2022) The ciliate Paramecium bursaria allows budding of symbiotic Chlorella variabilis cells singly from digestive vacuole membrane into the cytoplasm during algal reinfection. Protoplasma 259:117–125. https://doi.org/10.1007/s00709-021-01645-x
Karakashian MW, Karakashian SJ (1973) Intracellular digestion and symbiosis in Paramecium bursaria. Exp Cell Res 81:111–119. https://doi.org/10.1016/0014-4827(73)90117-1
Kodama Y, Nagase M, Takahama A (2017) Symbiotic Chlorella variabilis strain, 1N, can influence the digestive process in the host Paramecium bursaria during early infection. Symbiosis 71:47–55. https://doi.org/10.1007/s13199-016-0411-1
Fok AK, Lee Y, Allen RD (1982) The correlation of DV pH and size with the digestive cycle in Paramecium caudatum. J Euk Microbiol 29:409–414. https://doi.org/10.1111/j.1550-7408.1982.tb05423.x
Gu FK, Chen L, Ni B, Zhang X (2002) A comparative study on the electron microscopic enzymo-cytochemistry of Paramecium bursaria from light and dark cultures. Eur J Protistol 38:267–278. https://doi.org/10.1078/0932-4739-00875
Karakashian SJ, Rudzinska MA (1981) Inhibition of lysosomal fusion with symbiont containing vacuoles in Paramecium bursaria. Exp Cell Res 131:387–393. https://doi.org/10.1016/0014-4827(81)90242-1
Song C, Murata K, Suzaki T (2017) Intracellular symbiosis of algae with possible involvement of mitochondrial dynamics. Sci Rep 7:1221. https://doi.org/10.1038/s41598-017-01331-0
Reisser W (1992) Endosymbiotic associations of algae with freshwater protozoa and invertebrates. In: Reisser W (ed) Algae and symbioses: plants, animals, fungi, viruses, interactions explored, vol 1.1. Biopress, Bristol, pp 1–19
Meier R, Lefort-Tran M, Pouphile M, Reisser W, Wiessner W (1984) Comparative freeze-fracture study of perialgal and digestive vacuoles in Paramecium bursaria. J Cell Sci 71:121–140. https://doi.org/10.1242/jcs.71.1.121
Schüßler A, Schnepf E (1992) Photosynthesis dependent acidification of perialgal vacuoles in the Paramecium bursaria/Chlorella symbiosis: visualization by monensin. Protoplasma 166:218–222. https://doi.org/10.1007/BF01322784
Watanabe K, Motonaga A, Tachibana M, Shimizu T, Watarai M (2022) Francisella novicida can utilize Paramecium bursaria as its potential host. Environ Microbiol Rep 14(1):50–59. https://doi.org/10.1111/1758-2229.13029
Acknowledgements
The authors thank Professor Emeritus Masahiro Fujishima (Yamaguchi University, Japan) for giving us the valuable monoclonal antibody against mitochondria. The authors would like to thank Editage (www.editage.com) for the English-language editing of the manuscript. The authors thank the faculty of Life and Environmental Sciences in Shimane University for financial supports in publishing this report.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (C) (Grant Number 20K06768) and (B) (Grant Number 23H02529) from the Japan Society for the Promotion of Science (JSPS) and the Oshimo Foundation (Hiroshima, Japan) to YK.
Author information
Authors and Affiliations
Contributions
YK conceived and designed the experiments. YK and YE performed the experiments. YK wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2023_3590_MOESM1_ESM.tif
Supplementary file1 (TIF 4874 KB)
Figure S1. Cytoplasmic streaming in P. bursaria after mixing with NIES-2169 1 h (upper) and 48 h (lower). Micrographs were taken at 10-s intervals at each time point. One hour after mixing with NIES-2169, numerous DV membranes containing NIES-2169 had formed. These DV membranes were rapidly flowing in the cytoplasm due to host cytoplasmic streaming. The black arrow indicates a representative DV. Forty-eight hours after mixing, the vacuoles enclosing multiple NIES-2169 were not flowing by the cytoplasmic streaming, because the vacuole was localized beneath the host cell cortex. Black arrowhead and double black arrowhead show representative vacuoles. This attachment of the vacuole beneath the host cell cortex is a typical feature of PV membrane. Because the host cytoplasmic streaming was suppressed compared to 1 h after the algal mixing, not only the vacuoles enclosing NIES-2169, but also other vacuoles or crystals were barely moving (Data not shown). On the other hand, when 1N cells were ingested, the initial DV membrane flowed by the host cytoplasmic streaming, and the membrane that later encloses a single 1N cell localized beneath the host cell cortex and did not flow as shown in [7]. Ma macronucleus.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kodama, Y., Endoh, Y. Comparative Analyses of the Symbiotic Associations of the Host Paramecium bursaria with Free-Living and Native Symbiotic Species of Chlorella. Curr Microbiol 81, 66 (2024). https://doi.org/10.1007/s00284-023-03590-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03590-9