Abstract
A Gram-stain-negative, light yellow, aerobic, non-motile, short rod-shaped bacterium named strain Y-23T with iprodione-degrading capability was isolated from a soil under a greenhouse in Tibet, PR China. Strain Y-23T grew at 4–37 ℃ and pH 5.0–9.0 (optimum, 25 ℃ and pH 7.0) with 0–3% (w/v) NaCl (optimum, 0%). Phylogenetic analysis based on 16S rRNA gene and chromosome genome indicated that strain Y-23T formed a stable evolutionary branch with Acinetobacter tandoii DSM 14970T. The 16S rRNA gene similarity, digital DNA-DNA hybridization and average nucleotide identity values between strain Y-23T and Acinetobacter tandoii DSM 14970T were 98.31%, 43.2% and 91.2%, respectively. The genome size was 3.39 Mbp with a genomic DNA G+C content of 40.59 mol%. The predominant fatty acids were C18:1 ω9c, Summed feature 3 (C16:1 ω7c/C16:1 ω6c), C12:0, C12:0 3-OH and C16:0. The polar lipids were diphosphatidyl glycerol, phosphatidyl glycerol, phosphatidyl ethanolamine, phosphatidyl choline, unidentified phospholipid, four unidentified aminophospholipids and two unidentified lipids. The isoprenoid quinone was Q-8 (19.43%) and Q-9 (80.57%). Based on phenotypic, phylogenetic, and genotypic data, strain Y-23T is considered to represent a novel species of the genus Acinetobacter, for which the name Acinetobacter tibetensis sp. nov. is proposed. The type strain is Y-23T (= CICC 25150T = JCM 35630T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The highly diverse genus of Acinetobacter was first described by Brisou and Prevot [1]. Acinetobacter spp. are Gram-negative, non-motile, strictly aerobic, non-fermenting and non-fastidious [2, 3]. The genus is heterogeneous and highly diverse metabolically as well as physiologically [4]. As of November 2022, the classification of the genus Acinetobacter included 75 validly published species and 29 not validly published species (https://lpsn.dsmz.de/genus/Acinetobacter). Iprodione (C13H13Cl2N3O3), is a dicarboxamide fungicide that inhibits DNA and RNA synthesis, cell division, and cellular metabolism in fungi [5], which is commonly used to control fungal infestations by Botrytis cinerea, Alternaria sp., Monilinia fructigena, Rhizoctonia solani, Sclerotinia sclerotiorum, Penicillium sp., Sclerotinia sp., and other fungal pathogens in crops [6,7,8,9]. During the investigation of iprodione-degrading bacteria using enrichment culture technique from the soil under a greenhouse, a light yellow bacterium, named Y-23, was isolated. The taxonomic characterization of strain Y-23 suggested that it represented a novel species of genus Acinetobacter, for which the name Acinetobacter tibetensis sp. nov. is proposed.
Materials and Methods
Isolation and Cultivation of Strain Y-23
In April 2021, a study of iprodione-degrading bacteria using enrichment culture technique led to the isolation of strain Y-23. The samples were collected from the soil for vegetable growing under a greenhouse at Lhasa, Tibet (29°66′84.4″N, 90°94′27.6″E, Altitude: 3667 m). A 5.0 g amount of soil sample was added into a 250 mL flask with 100 mL of sterile MSM containing 100 mg/L iprodione and was incubated on a rotary shaker (180 rpm) at 25 ℃ for 5 d [10]. The suspension (5 mL) was successively transferred to fresh MSM containing 200 mg/L, 300 mg/L, 400 mg/L iprodione and incubated for another 5 d, respectively. After 4 rounds of enrichment, the culture was diluted and spread onto TSA agar at 25 ℃. After 7 d of incubation, a light yellow colony was collected and named Y-23. Strain Y-23 was routinely cultured on TSA agar medium at 25 ℃ after repeated purifying. The purified strain was preserved at − 80 ℃ with 25% (v/v) glycerol. Strain Y-23 has been deposited at CICC (China Center of Industrial Culture Collection) and JCM (Japan Collection of Microorganisms).
Phylogenetic Analysis Based on 16S rRNA Gene
Genomic DNA of strain Y-23T was extracted using MiniBEST Bacterial Genomic DNA Extraction Kit Version 2.0 (TaKaRa Biotechnology Co., Tokyo, Japan). The amplification of 16S rRNA gene was done according to Lane [11]. The 16S rRNA gene was aligned in EzBioCloud [12]. Maximum-likelihood, neighbor-joining and maximum evolution trees were constructed using MEGA7.0 software with bootstrap values of 1000 replicates [13].
Genome Sequencing and Comparative Genomic Analysis
Strain Y-23T was sequenced with PacBio Sequel II platform in Shanghai Lingen Biological Technology Co., Ltd. The genomic sequence information of Y-23T has been submitted to the National Center for Biotechnology Information (NCBI) database. Genome assemblies were prepared from the ONT reads using Apades v3.11.0, gene prediction using Glimmer 3.02 software.
Based upon the close relationship with the test strain in phylogenetic analyses, the genome sequence of related Acinetobacter spp. were obtained from NCBI database. The digital DNA-DNA hybridization (dDDH) values and confidence intervals were calculated using the recommended settings of Genome-to-Genome Distance Calculator [14]. The average nucleotide identity (ANI) was determined between strain Y-23T and closely related strains of the genus Acinetobacter using OrthANIu [15]. The whole-genome evolution tree were constructed using Type (Strain) Genome Server [16].
Phenotypic Characterization
The phenotypic characteristics of strain Y-23T were tested on TSA agar after incubation for 24 h at 30 ℃. Cell morphology was observed by both light microscopy (CX31, Olympus) and scanning electron microscopy (Hitachi FE-SEM SU8010). The temperature for optimal growth was tested at 4–55 ℃ (4, 10, 15, 20, 25, 30,37, 40, 45, 50 and 55 ℃). The pH range for growth was determined by measuring the OD600 of the culture grown in TSA broth, which was adjusted prior to sterilization to various pH values (pH 3.0–12.0 with an interval of 1.0 units) using appropriate biological buffers [17]. The salt tolerance was determined with various NaCl concentrations (0, 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10%, w/v). Gram-straining reaction was carried out according to Claus [18]. Anaerobic growth was checked using the Oxoid AnaeroGen system. Other tests to determine the biochemical characteristics were carried out using API 50CH, API 20E, API 20NE, and API ZYM strips according to the manufacturer’s instructions (BioMérieux).
Determination of Fatty Acids, Polar Lipids and Isoprenoid Quinones
After incubation on TSA at 28 ℃ for 3 d, cells were collected for fatty acids test. Fatty acids were saponified, methylated and extracted according to the standard protocol of the Sherlock Microbial Identification System (MIDI), analyzed via gas chromatography and identified using the Sherlock Aerobic Bacterial Database (RTSBA 6.2B) [19]. Polar lipids of cell wall were detected using thin layer chromatography method. Isoprenoid quinones were identified using high-performance liquid chromatography method [20].
Degradation of Iprodione by Strain Y-23 T
Cells of strain Y-23 T were cultured in liquid LB medium for 24 h at 25 °C and then collected by centrifugation at 7000 rpm for 4 min. The cell pellets were washed twice with sterilized MSM, adjusted to an optical density at 600 nm (OD600) of approximately 1.5, and used as the inoculant. An aliquot of the cells (5%, vol/vol) was inoculated into a 100 mL Erlenmeyer flask containing 30 mL of MSM supplemented with 50 mg/L iprodione as the sole source of carbon. The flasks were then incubated at 25 °C with shaking (180 rpm). At each sampling point, three flasks were used to measure the iprodione concentration by GC-ECD and other three flasks were used to determine the values of OD600 of strain Y-23T. Control experiments (medium without inoculum) were carried out under the same conditions.
Sample preparation of fermentation broth: 20.0 g sample were placed in 150 mL beaker, then 40 mL acetonitrile and 5–6 g NaCl were added, vibration at 180 rpm for 10 min, after 30 min of stratification, 10 mL of supernatant were rotatably evaporated to nearly dry, 5.0 mL acetone with n-hexane (1:9) was used as constant volume for GC-ECD analysis. The test conditions by Gas chromatography are as follows: HP-5 capillary column (30 m × 0.25 mm × 0.45 μm), carrier gas (N2, 99.999% purity), flow rate (3.0 mL/min), flow mode (10:1), sample volume (1 μL), inlet temperature (280 ℃), heating process: 150 ℃ for 0 min, 15 ℃/min to 210 ℃ and 10 ℃/min to 260℃, 20 ℃/min to 300 ℃ for 6 min, electron capture detector temperature (230 ℃) [21].
Results and Discussion
Phylogenetic Analysis
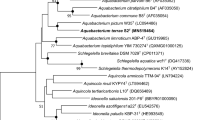
Compared to the sequences deposited in EzBioCloud, the 16S rRNA gene sequence of strain Y-23T shared highest similarity with Acinetobacter tandoii DSM 14970T (98.31%), followed by Acinetobacter piscicola LW15T (98.31%) and Acinetobacter beijerinckii CIP 110307T (98.24%). Phylogenetic analysis of Y-23T based on 16S rRNA genes confirmed its placement within the Acinetobacter genus, to form a separate branch of evolution with Acinetobacter tandoii DSM 14970T. A neighbor-joining tree derived from full 16S rRNA alignments is shown in Fig. 1, similar results were obtained using maximum-likelihood and maximum evolution methods (Fig. S1 and S2).
Genomic Characteristics and Comparative Genomics Analysis
The whole genome of strain Y-23T contained one chromosome and one plasmid with an N50 value of 3381792 bp. The genome size of strain Y-23T is 3.39 Mb. A total of 3175 genes were predicted in the genome of strain Y-23T. The genomic DNA G+C content of Y-23T is 40.59 mol%, which is similarity to Acinetobacter tandoii DSM 14970T (40.2%). The whole-genome evolution tree of Y-23T and 34 related bacteria shown that strain Y-23T and Acinetobacter tandoii DSM 14970T formed a stable evolutionary branch (Fig. 2). Furthermore, the dDDH and ANI values between Y-23T and other related strains were 20.1–43.2% and 74.7–91.2%, which were lower than the threshold values of 70% and 95–96% for species discrimination. Both the 16S rRNA gene and whole-genome in the phylogenetic trees demonstrated that strain Y-23T had the closest phylogenetic relationship with Acinetobacter tandoii DSM 14970T.
Morphological, Cultural, Physiological and Biochemical Characteristics
Colonies of strain Y-23T were light yellow, round, moist, opacity, neat edges on TSA solid medium (Fig. S3A). Strain Y-23T was Gram-stain-negative, aerobic, non-motile, short rod-shaped, single or paired, 0.7–1.0 μm × 0.9–2.0 μm (Fig. S3B). Strain Y-23T grew at 4–37 ℃ and pH 5.0–9.0 (optimum, 25 ℃ and pH 7.0) with 0–3% (w/v) NaCl (optimum, 0%). Strain Y-23T and Acinetobacter tandoii DSM 14970T had a few differential phenotypic characteristics (Table 1), they could not be identified with biolog system which was consistent with previous research [22, 23]. The negative properties of strain Y-23T to API 50CH, API 20E, API ZYM and API 20NE are listed in Table S1.
Fatty Acids, Polar Lipids and Isoprenoid Quinone
The predominant fatty acids of strain Y-23T (> 5.0% of the total amounts) were C18:1 ω9c (35.5%), Summed feature 3 (C16:1 ω7c/C16:1 ω6c) (20.1%), C16:0 (8.9%), C12:0 3-OH (8.5%) and C12:0 (7.2%). The fatty acids composition of Y-23T is consistent with previous results for recognized species of the genus Acinetobacter, justifying its placement in this genus [3]. The detailed fatty acid profiles of strain Y-23T is summarized in Table S2. The polar lipids were diphosphatidyl glycerol, phosphatidyl glycerol, phosphatidyl ethanolamine, phosphatidyl choline, unidentified phospholipid, four unidentified aminophospholipids and two unidentified lipids (Fig. S4), the lipid profile of Y-23T was similar to Acinetobacter tandoii DSM 14970T [3]. The isoprenoid quinone was Q-8 (19.43%) and Q-9 (80.57%). The chemotaxonomic characteristics of Y-23T compared with the closely related type strain Acinetobacter tandoii DSM 14970T further support the hypothesis that strain Y-23T represents a member of the genus Acinetobacter.
Degradation of Iprodione by Strain Y-23 T
The degradation kinetics of iprodione and growth of strain Y-23T were simultaneously investigated (Fig. 3). During the first 12 h, strain Y-23T grew faster, and then the growth rate was reduced. After 96 h of incubation, 25.03 mg/L iprodione was degraded by strain Y-23T and the degradation rate was about 50.07%. The results had shown that strain Y-23T could utilize iprodione to support its growth. But no previously reported iprodione-degrading genes found in other strains are present in the genome of strain Y-23T.
Description of Acinetobacter tibetensis sp. nov.
Acinetobacter tibetensis (ti.bet.en’sis. N.L. masc./fem. adj. tibetensis, pertaining to Tibet, an autonomous region in southwest of China).
Cells are Gram-stain-negative, light yellow, aerobic, non-motile, short rod-shaped, 0.7–1.0 μm in width, and 0.9–2.0 μm in length. Optimal growth was observed at 4–37 ℃ and pH 5.0–9.0 (optimum, 25 ℃ and pH 7.0) with 0–3% (w/v) NaCl (optimum, 0%). In API 20NE kit, a positive reaction for esculin hydrolysis and utilization of malic acid, phenylacetic acid and decanoic acid. In the API ZYM kit, esterase (C4), lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase are positive. All the other tests are negative in API 20E and API 50CH kit. After 96 h of incubation, 25.03 mg/L (50.07%) iprodione was degraded at 25 ℃.
The major cellular fatty acids are C18:1 ω9c, Summed feature 3 (C16:1 ω7c/C16:1 ω6c), C16:0, C12:0 3-OH and C12:0. The polar lipids are diphosphatidyl glycerol, phosphatidyl glycerol, phosphatidyl ethanolamine, phosphatidyl choline, unidentified phospholipid, four unidentified aminophospholipids and two unidentified lipids. The isoprenoid quinone are Q-8 (19.43%) and Q-9 (80.57%). The genome size is 3.39 Mbp with a genomic DNA G+C content of 40.59 mol%.
The type strain is Y-23T (= CICC 25150T = JCM 35630T), which was isolated from a soil under a greenhouse in Tibet, PR China.
The GenBank accession numbers for the genome and 16S rRNA gene sequences of Acinetobacter tibetensis strain Y-23T are CP098732-CP098733 and ON138910, respectively.
Data Availability
The 16S rRNA gene sequence and genome of Acinetobacter tibetensis strain Y-23T have been deposited in GenBank.
References
Brisou J, Prevot AR (1954) Studies on bacterial taxonomy.X. The revision of species under Acromobacter group. Ann Inst Pasteur (Paris) 86(6):722–728
Khatun A, Mehta S, Singh VA, Pottathil S (2018) Characterisation of clinical isolates of Acinetobacter species with emphasis on multidrug resistant, extensively drug resistant and pan-drug resistant strains. JK Sci 20(2):100–105
Das L, Deb S, Das SK (2021) Description of Acinetobacter kanungonis sp. nov., based on phylogenomic analysis. Int J Syst Evol Microbiol 71:004833
Marie T, Jean C, Eun-jeong Y, Lenka K, Cerqueira GC, Cheryl M, Michael F, Jennifer W, Dominique C, Thierry L (2014) The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882
Davidse LC (1986) Benzimidazole fungicides: mechanism of action and biological impact. Annu Rev Phytopathol 24(1):43–65
Mukherjee I, Gopal M, Chatterjee SC (2003) Persistence and effectiveness of iprodione against Alternaria blight in mustard. Bull Environ Contam Toxicol 70(3):586–591
Morales J, Manso JA, Cid A, Mejuto JC (2013) Stability study of iprodione in alkaline media in the presence of humic acids. Chemosphere 92(11):1536–1541
Grabke A, Fernández-Ortuño D, Amiri A, Li X, Peres NA, Smith P, Schnabel G (2014) Characterization of iprodione resistance in Botrytis cinerea from strawberry and blackberry. Phytopathology 104(4):396–402
Campos M, Perruchon C, Vasilieiadis S, Menkissoglu-Spiroudi U, Karpouzas DG, Diez MC (2015) Isolation and characterization of bacteria from acidic pristine soil environment able to transform iprodione and 3,5-dichloraniline. Int Biodeterior Biodegrad 104:201–211
Yang Z, Jiang W, Wang X, Cheng T, Zhang D, Wang H, Qiu J, Cao L, Wang X, Hong Q (2018) An amidase gene, ipaH, is responsible for the initial step in the iprodione degradation pathway of Paenarthrobacter sp. strain YJN-5. Appl Environ Microbiol 84(19):e01150-18
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons Ltd, New York, pp 115–148
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613–1617
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Meier-Kolthof JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confdence intervals and improved distance functions. BMC Bioinf 14:60–73
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton Leeuw 110:1281–1286
Meier-Kolthoff JP, Sardà CJ, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res 50(D1):D801–D807
Chung YC, Kobayashi T, Kanai H, Akiba T, Kudo T (1995) Purification and properties of extracellular amylase from the hyperthermophilic archaeon Thermococcus profundus DT5432. Appl Environ Microbiol 61:1502–1506
Claus D (1992) A standardized gram staining procedure. World J Microb Biot 8:451–452
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Komagata K, Suzuki K (1987) Lipids and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–203
Kim NH, Lee JS, Park KA, Kim YH, Lee SR, Lee JM, Yu IS, Jung K, Lee YK (2016) Determination of matrix effects occurred during the analysis of organochlorine pesticides in agricultural products using GC-ECD. Food Sci Biotechnol 25(1):33–40
Knight GC, McDonnell SA, Seviour RJ, Soddell JA (1993) Identification of Acinetobacter isolates using the biolog identification system. Lett Appl Microbiol 16:261–264
Carr EL, Kämpfer P, Patel BKC, Gürtler V, Seviour RJ (2003) Seven novel species of Acinetobacter isolated from activated sludge. Int J Syst Evol Microbiol 53:953–963
Funding
This research was financially supported by the National Natural Science Foundation of China (No. 32060025), the National Natural Science Foundation of Tibet (No. XZ202101ZR0090G), the Training Program for Excellent Young Innovators of Changsha (No. kq2106049), the Provincial College Students Innovative Entrepreneurial Training Plan Program of Hunan, China (No. S202110537006X), the College students’ Innovative Entreneurial Training Plan Program of Hunan Agricultural University, China (No. S202010537078) and the Double First-Class Construction Project of Hunan Agricultural University (No. SYL201802002).
Author information
Authors and Affiliations
Contributions
YT, YZ and HP conceived the project. HP, XL and HL performed the experiments. YZ and HP analyzed the data, and HP and YT drafted and revised the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that there is no conflict of interest.
Ethical Approval
No animals or human participants were included in the present study.
Consent for Publication
All the authors agree to submit for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, H., Li, J., Liu, HH. et al. Acinetobacter tibetensis sp. nov., Isolated from a Soil Under a Greenhouse in Tibet. Curr Microbiol 80, 51 (2023). https://doi.org/10.1007/s00284-022-03158-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03158-z