Abstract
Microbial metabolites in rhizosphere soil are important to plant growth. In this study, microbial diversity in blueberry plant rhizosphere soil was characterized using high-throughput amplicon sequencing technology. There were 11 bacterial phyla and three fungal phyla dominating in the soil. In addition, inorganic-phosphate-solubilizing bacteria (iPSB) in the rhizosphere soil were isolated and evaluated by molybdenum-antimony anti-coloration method. Their silicate solubilizing, auxin production, and nitrogen fixation capabilities were also determined. Eighteen iPSB in the rhizosphere soil strains were isolated and identified as Buttiauxella, Paraburkholderia and Pseudomonas. The higher phosphorus-solubilizing capacity and auxin production in blueberry rhizosphere belonged to genus Buttiauxella sp. The strains belong to genus Paraburkholderia had the same function of dissolving both phosphorus and producing auxin, as well as silicate and nitrogen fixation. The blueberry seeds incubated with the strains had higher germination rates. The results of this study could be helpful in developing the plant growth-promoting rhizobacteria (PGPR) method for enhancing soil nutrients to blueberry plant.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Blueberry (Vaccinium Spp.) is one of the most popular fruits in the world and possesses many highly nutritive compounds including minerals, vitamins, fatty acids, antioxidant phenolic compounds which have antitumor, and anti-inflammatory properties [1, 2]. As most plants, blueberry plant rhizosphere is inhabited by millions of parasitic, commensal, and mutualistic microorganisms. They coexist in the complex ecological communities and profoundly influence the plant growth [3]. For example, maize rhizosphere soil microorganisms have been recognized as the main driver of soil nutrients cycling and play a critical role in regulating soil fertility and plant health [4]. Therefore, a better understanding of rhizosphere microbiome is very useful to rationally exploit the plant–microbe interactions and establish a new perspective way to promote plant growth and improve fruit yield through its beneficial rhizosphere microorganisms recently named as plant growth-promoting rhizobacteria (PGPR) [5].
Bacillus, Pseudomonas, Enterobacter, Acinetobacter, Burkholderia, and Arthrobacter are the most common microorganisms present in rhizosphere and referred as the PGPR to improve soil nutritional quality for better plant growth [6, 7]. As an important member of PGPR, iPSB (inorganic-phosphate-solubilizing bacteria) is particularly effective in releasing phosphorus from pools of inorganic phosphorus and plays an important role in plant growth [8, 9]. For chemical fertilizer, the majority of its soluble inorganic phosphorus is rapidly immobilized by soil fixation and becomes unavailable for plant uptake, which results in low phosphorus-use efficiency and potentially excess phosphorus in rhizosphere [10]. Thus, soil phosphorus must be managed to minimize its loss and immobilization and increase its using efficiency for plant growth. iPSB strains can solve the issue and improve the available phosphorus content in soil for plants intake at any time. Some iPSB strains have also been reported to have other pro-growth abilities, such as production of auxin and dissolution of silicate and nitrogen fixation [11, 12].
In this study, microbial diversity in the rhizospheres of three most common blueberry varieties (Vaccinium corymbosum, Vaccinium austral and Vaccinium ashei) was investigated through high-throughput sequencing technology. As the nutrients requirement of soil for developing root system of blueberry plant is very demanded, the enrichment of nutrient contents in blueberry plant soil usually is the major cost for blueberry cultivation. In this study, the iPSBs in blueberry rhizosphere were identified and isolated. The growth-promoting capabilities in production of auxin and dissolution of silicate and nitrogen fixation of these iPSB strains were evaluated as well. Furthermore, effects of the iPSB strains on the germination rate of blueberry seeds were studied. In general, the results of this study could be very useful to develop efficient and environmentally friendly PGPR fertilizer and increase the essential soil nutrients for blueberry plant growth.
Materials and Methods
Collection of Soil Samples
Rhizosphere soil samples were collected from the roots of three blueberry species, Vaccinium corymbosum (Northern highbush blueberry, NHB), Vaccinium austral (Southern highbush blueberry, SHB) and Vaccinium ashei (Rabbiteye blueberry, RB) in April 2020 according to the method described in previous study [13]. The geographical locations of sampling sites are shown in Table 1. The depth of collected soil was 5 to 20 cm under the ground surface surrounding blueberry plant trunks. Thirty samples from each blueberry species were collected and mixed homogeneously. The control check (CK) soil samples were also collected at 5 to 20 cm under the ground surface without growing vegetation beside the blueberry plants area. They were also mixed homogeneously. Soil pH value was determined using a pH meter (PHS-3C, INESA Scientific Instrument Co, Ltd, Shanghai, China). The soil solution was prepared by dissolving soil sample in distilled water at a ratio of 1:5 [14]. The pH measurement was repeated in triplicates, and pH value was expressed by mean with standard derivation.
Analysis of DNA Sequences of Microbes in Soil Sample Using High-Throughput Amplicon Sequencing Technology
The genome of microbes were extracted using a DNA extraction kit (Fast DNA Spin Kit for Soil, MP Biomedicals, Santa Ana, CA, USA). 16S rRNA genes of the V4 regions were amplified by specific primers (515F/806R) (Invitrogen, Carlsbad, CA, USA), and ITS genes of the ITS4 regions were amplified by specific primers (ITS3-F/ITS4R) (Invitrogen, Carlsbad, CA, USA). PCR reaction solution consisted of 25 μL of 2 × Premix Taq (Takara Biotech, Dalian, China), 1 μL of each primer (10 mM), 3 μL of DNA (20 ng/μL) template and 21 μL of double distilled (dd) H2O. The reaction took place in BioRad S1000 instrument (Bio-Rad Laboratory, CA, USA) with a thermos-cycling program of 5 min at 94 °C for initialization, 30 cycles of 30 s denaturation at 94 °C, 30 s annealing at 52 °C, 30 s extension at 72 °C and 10 min final elongation at 72 °C.
Gel (1% Agarose) electrophoresis was applied to detect the lengths and concentrations of the PCR products. The products with significant concentrations at 290–310 bp length for 16S or 370–450 bp length for ITS, were mixed at the same density ratio. Then, the mixture of PCR products was purified by the EZNA Gel Extraction Kit (D2500-02, Omega, USA). The DNA libraries were set up using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, MA, USA). After the evaluation of library quality, the final sequences were output by an Illumina HiSeq 2500 platform (Illumina, CA, USA).
The sequences sharing high similarity (> 97%) were assigned to the same operational taxonomic unit (OTU) to represent a species. The most occurring sequence was taken out as a representative for each OTU. The chimera sequence and singleton OTU were removed at the same time, and then statistics of Paired-end raw reads, Paired-end clean reads, Raw Tags, Clean Tags, Length of Clean Tags, GC percent of Clean Tags, OTU number were carried out. For each representative sequence, the Silva database (https://www.arb-silva.de/) was used to annotate taxonomic information (set the confidence threshold to default to ≥ 0.5). After that, the pollution OTU and its Tags, which are annotated as chloroplasts or mitochondria were removed. Then the OTU taxonomy synthesis information table was obtained for the final analysis. The total number of OTU sequences and OTU types were counted. A normalized OTU table was obtained based on the sample with the least sequences. These Hiseq sequencing results in double-ended sequence data (pairwise. Fastq files) were submitted to the Sequence Read Archive (https://submit.ncbi.nlm.nih.gov/subs/sra/), and the accession number was obtained. According the OTU table, the annotation ratio on each classification level was calculated to obtain the sequence composition of each sample at each classification level. The relative abundance of each OTU to the lowest sequence was applied for analyzing the alpha and beta diversity of microbes.
Screen and Isolation of Inorganic-Phosphate-Solubilizing Bacteria (iPSB)
The soil sample (10) g was added in a sterilized flask and mixed with 100 mL of sterilized distilled water. The flask was incubated at 28 °C for 30 min with a shaking rate at 150 rpm. Then the soil suspension (5 mL) was sampled to carry out multiple dilutions using sterilized distilled water until the soil suspension was diluted to a final dilution level of 10–4. The final diluted solution was used as a soil stock solution for the following experiment. The prepared soil stock solution (100 µL) was spread on 20 mL of sterilized inorganic phosphorus bacteria medium (HB8670; Hope BioTech, Shandong, China). After incubation at 28 °C for 3 days, the monoclonal microbes with soluble phosphorus circle on the medium were selected and inoculated into inorganic phosphorus bacteria medium. The selected bacteria were named as P1, P2, and so on. For confirming the iPSB strains, three inoculation points from each iPSB strain were used to cultivate on inorganic phosphorus medium plate at incubation temperature 28 °C for 7 days. The experiment for each strain was repeated in triplicates. Those strains, which were with soluble phosphorus circle, were selected for the follow-up experiments.
The 16S rRNA Sequencing of iPSB Strain
All the iPSB strains were incubated in liquid beef extract peptone medium at 28 °C for 24 h, then the supernatant of the incubated medium was removed. The cell pellets were collected for DNA extraction. The DNA in each iPSB was extracted according to the manufacturer's instruction (TIANamp Bacteria DNA Kit, DP302; Tiangen BioTech, Beijing, China). The 16S rRNA genes were amplified by primers of 27F (5′ -AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′ -GGT TAC CTT GTT ACG ACT T-3′) (Invitrogen, Carlsbad, CA, USA). The PCR reaction solution consisted of 25 µL of 2xPremix Taq (Takara Biotech, Dalian, China), 2 µL of each primer (10 mM), 1 µL of DNA (20 ng/mL) template and 22 µL of double distilled H2O. The solution was amplified by BioRad S1000 (Bio-Rad Laboratory, Hercules, CA, USA) with a thermos-cycling program of 5 min at 94 °C for initialization, 40 cycles of 30 s denaturation at 94 °C, 30 s annealing at 56.2 °C, and 90 s extension at 72 °C and 10 min final elongation at 72 °C. The PCR products were detected by using 0.8% agarose of gel electrophoresis method. The data of sequence analysis was searched in the BLAST (www.blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with the corresponding sequence in the GenBank database. The type strains of these strains were obtained in EZ BioCloud (https://www.ezbiocloud.net/), and compared with the sequences.The representative sequence of each species was selected, submitted to the NCBI (www.submit.ncbi.nlm.nih.gov/subs/) website to obtain the GenBank accession numbers [15].
Determinations of Phosphorus and Silicate Solubilizing, Auxin Production and Nitrogen Fixation Capabilities of iPSB Strains
The isolated iPSB strains were inoculated into liquid inorganic phosphorus bacteria medium (HB8670-1; Hope BioTceh, Shandong, China) at 28 °C for 3 days. The inoculated medium was centrifuged at 4 °C at 6000 g for 15 min. The supernatant was collected to determine the soluble phosphorus content using Molybdenum-antimony anti-coloration method [16].
For evaluation of silicate dissolution of the iPSB strains, three inoculation points from each iPSB strain were inoculated into silicate bacteria agar medium (HB8548; Solarbio, Beijing, China). After inoculation at 28 °C for 4 days, the colony diameter was recorded to determine silicate-solubilizing capability.
The iPSB strains were inoculated into liquid beef extract peptone medium with 100 mg/L L-Tryptophan (T0011; Solarbio, Beijing, China) and incubated at 28 °C for 3 days for evaluation of its capability of auxin production. After incubation, the inoculated medium was centrifuged at 4 °C at 6000 g for 15 min. The amount of auxin in supernatant was determined according to the colorimetric method by Salkowski colorimetric method [17].
For evaluation of the capability of nitrogen fixation, three inoculation points from each iPSB strain were also used into A Sugai’s medium (HB8540; Solarbio, Beijing, China). After the inoculated medium was incubated at 28 °C for 4 days, the colony diameter was recorded to express nitrogen fixation capability. The determinations for each strain were repeated in triplicates.
Evaluation of iPSB Strains on Promoting Seed Germination
The isolated iPSB strains were inoculated into liquid inorganic phosphorus bacteria medium individually and incubated at 28 °C for 3 days. Mature blueberry seeds of three blueberry species were selected and soaked in sterilized water for 24 h at 25 °C. Forty seeds of each of three blueberry species were mixed and soaked in the prepared liquid inorganic phosphorus bacteria medium at 25 °C for 24 h. Another forty seeds were soaked in sterilized liquid inorganic phosphorus bacteria medium at 25 °C for 24 h and used as a control group (CK). Then, the seeds were washed and planted in sterilized petri dishes and incubated at 25 °C under continuous illumination (~ 1500 lx). Seed germination was evaluated every day for a period of 45 days and recorded when a sprout was appeared. Germination rate was expressed by percentage of the number of germinated seeds on the total number of inoculated seeds.
Data Analysis
All experiments were repeated in triplicates. Alpha diversity was applied in analyzing the complexity of the diversity through three indices: Chao1, Shannon and Simpson [18,19,20]. All the indices were calculated with QIIME (Quantitative Insights Into Microbial Ecology) (V1.9.1) and displayed with R software (V2.15.3). They were expressed by mean with standard derivation, tested for statistical distribution and compared by ANOVA analysis with a significant difference between two data at p < 0.05 or p < 0.01. The significant differences between different species were determined by linear discriminant analysis (LDA) effect size (LEfSe) (https://github.com/biobakery/lefse) with two as the default setting filter value for LDA score. The dissolution of phosphorus and auxin production, the colony diameters, and the colony diameters of iPSB strains were expressed by mean with standard derivation. The seed germination rates were expressed by mean with standard derivation, tested for statistical distribution and compared by ANOVA analysis with a significant difference between two data at p < 0.05 or p < 0.01.
Results
Microbial Diversity in Blueberry Plant Rhizosphere Soil
In this study, microbial diversity in the rhizosphere of blueberry was investigated by high-throughput sequencing technology. The results were submitted to the Sequence Read Archive first, and the accession number was PRJNA864869. There were 11 bacterial phyla (Thaumarchaeota, Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, Verrucomicrobia, and WPS-2) with an average relative abundance of above 0.1%. These 11 phyla collectively accounted for more than 96.4% of all sequencing reads in the rhizosphere samples form the three blueberry species (Fig. 1A). Meanwhile, there were ten bacterial phyla with an average relative abundance of above 0.1% which collectively accounted for more than 89.8% of all sequencing reads in the CK samples (Fig. 1A). Proteobacteria was the most prominent phyla with more than 30% relative abundance in each sample (Fig. 1A). The relative abundances of Actinobacteria and Acidobacteria were higher than other phyla in the soil samples form the three blueberry species, while the relative abundances of Acidobacteria and Bacteroidetes were the highest among the phyla in CK soil samples (Fig. 1A).
A total of 889 bacterial genera were identified in the blueberry rhizospheres. Among them, 20 genera (Nitrosotalea, Acidipila, Granulicella, Occallatibacter, Koribacter, Solibacter, Actinospica, Acidothermus, Streptacidiphilus, Mucilaginibacter, Ruminococcaceae_UCG-005, Ruminococcaceae_UCG-010, Burkholderia, Acidibacter, Chujaibacter, Sulfuriferula, Conexibacter, Bryobacter, Rhodoplanes, and Bradyrhizobium) had relative abundance greater than 1%, while Nitrosotalea, Occallatibacter, Acidothermus, Burkholderia and Chujaibacter were genera with relative abundance greater than 3% in the soil samples (Fig. 1C). Bradyrhizobium, Arthrobacter, Flavobacterium, Klebsiella, Lysobacter, Mesorhizobium, MND1, Pseudomonas, Massilia, Nitrospira, Clostridium, Ellin 6067, and Flavitalea were the genera with relative abundance greater than 1% in the CK soil samples (Fig. 1C).
For fungal communities, most of the OTUs were classified as Ascomycota, Zygomycota and Basidiomycota at phylum level (Fig. 1B). The communities were dominated by Cladophialophora, Fusarium, Oidiodendron, Penicillium, Pseudogymnoascus, Solicoccozyma, Trichoderma, Geomyces, Guehomyces, Humicola, Lecythophora, Lentinus and Exophiala in blueberry rhizosphere at genus level (Fig. 1D).
There were significant differences in microbial diversity between blueberry rhizosphere and CK soil. Ascomycota was the most prominent phyla in the soil samples collected from rhizospheres of blueberry plants with more than 40% relative abundance in each sample (Fig. 1B). Mortierellomycota (with 45.15% relative abundance) was the most prominent phyla in the CK soil samples. However, the relative abundance of Mortierellomycota in the soil samples collected from rhizospheres of blueberry plants were lower than 1% (Fig. 1B). Furthermore, the relative abundance of Zygomycota were more than 37.13% in the soil samples from the three blueberry plant rhizospheres, but lower than 1% in the CK soil samples (Fig. 1B). There were 11 genera (Mortierella, Tetracladium, Trichocladium, Cladosporium, Cystofilobasidium, Paraphoma, Sphaerulina, Spizellomyces, Vishniacozyma, Diversispora, and Fusarium) with relative abundance greater than 1% (Fig. 1D) in the CK soil sample. Fusarium was the only genus with relative abundance greater than 1% in both of the CK soil samples and the soil samples collected from rhizospheres of blueberry plants.
Alpha Diversity Index was calculated to compare the microbial diversity in the blueberry plant rhizosphere soil and CK soil. The highest number of microbe was found in the soil samples from SHB by the highest Chao one index, whereas the lowest was found in the CK soil samples (Fig. 2A). It was significantly different in Chao 1 index between the soil samples from the three blueberry plant rhizospheres and the CK soil samples (Fig. 2A). However, there was no significant difference of microbial diversity in the blueberry plant rhizosphere and CK soil samples according to the results of Shannon and Simpson (Appendix Fig. 6A and B). The highest number of fungus was found in the soil samples from RB by the highest Chao 1 index. It was significantly different between the soil samples from the three blueberry plant rhizospheres and CK samples (Fig. 2B), while the Shannon and Simpson indices of fungal communities were found no significant difference (Appendix Fig. 6C and D).
A total of 52 distinct bacterial biomarkers were identified by using the LDA threshold score of ≥ 2.0. Most of them were Proteobacteria (especially Alphaproteobacteria), Chloroflexi and Actinobacteria (Fig. 3A). The NHB-enriched phylotypes belonged to the Proteobacteria (Alphaproteobacteria), Chloroflexi, Firmicutes (Bacilli), Cyanobacteria (Oxyphotobacteria), and Acidobacteria (Acidobacteriia). The SHB rhizosphere bacteria were abundant by several Proteobacteria (Alphaproteobacteria and Gammaproteobacteria), Chloroflexi, Actinobacteria (Microbacteriaceae), Firmicutes (Lachnospiraceae), Spirochaetes (Spirochaeta_2), Latescibacteria, Bacteroidetes (Ohtaekwangia), and Planctomycetes (Pir4_lineage). The RB specific phylotypes were taxonomically diverse and included members of Actinobacteria, Proteobacteria (Nitrincolaceae) and Firmicutes (Clostridiales).
The analysis of fungal communities revealed 53 distinct biomarkers that unevenly distributed among the microorganisms in rhizospheres of the blueberry species (Fig. 3B). The rhizosphere fungi of NHB was rich in diverse Ascomycota (Sordariomycetes, Eurotiomycetes, and Leotiomycetes) and Basidiomycota (Piskurozymaceae). In contrast, the SHB specific fungi included an abundance of Ascomycota (Sordariomycetes, Pezizomycotina, and Dothideomycetes) and Basidiomycota (Sebacinales). There were 27 distinct biomarkers which differentially distributed in the rhizosphere samples from RB compared with the rhizospheres of the other two blueberry species (Fig. 2D). These rhizosphere fungi were Ascomycota (Saccharomycetes, Lecanoromycetes, Dothideomycetes, Sordariomycetes, Orbiliomycetes, Leotiomycetes, and Pezizomycotina), Basidiomycota (Ustilaginomycetes) and Zygomycota (Mucorales).
Phylogenetic Identification of iPSB Strains and Their Effects on Phosphorus- and Potassium- Solubilizing, Auxin Production, Nitrogen Fixation and Seed Germination
A total of 18 iPSB strains were isolated from the blueberry plant rhizosphere samples. The typical phosphorus decomposition halos for iPSB strains are showed in Appendix Fig. 7A–C. Based on their sequences, they were phyla Proteobacteria and classified into Buttiauxella, Paraburkholderia, and Pseudomonas (Table 2). The representative sequence of each inorganic-phosphate-solubilizing microbes was submitted (Table 2).
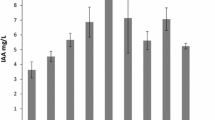
Based on the concentration of soluble phosphorus in the culture mediums, different strains had different inorganic phosphate-solubilizing activity. The concentrations of phosphorus in the supernatant collected from liquid medium were 0.03–4.99 mg/L, (Fig. 4A). The concentrations of phosphorus extracted from isolates P6, P7, P8, P37, P40 and P42 strain culture mediums were higher than 3 mg/L. Other strains were less than 1 mg/L (Fig. 4A). The strains P6, P7, P37, P40 and P42 were all Buttiauxella sp., while P8 belonged to the genus Pseudomonas sp.
Capabilities of phosphorus- and silicate- solubilizing, auxin production and nitrogen fixation analysis of isolated iPSB strains. A The dissolution of phosphorus and auxin production of iPSB strains; B The colony diameters of iPSB strains in silicate bacteria mediums; C The colony diameters of iPSB strains in A Sugai’s medium
The levels of auxin produced by the strains are shown in Fig. 4A. Isolate P7 (Buttiauxella brennerae) had the highest level at 58.07 mg/L, whilst the isolate P17 (Paraburkholderia insulsa) produced the lowest level at 5.53 mg/L amongst the selected isolates (Fig. 3A). The other strains that produced auxin more than 50 mg/L were P40 (Buttiauxella sp.) and P42 (Buttiauxella gaviniae), while auxin produced by other strains were lower than 25 mg/L (Fig. 4A).
After inoculation and incubation at 28 °C for 4 days, the visible colonies were observed in the silicate bacteria mediums inoculated with P10, P11, P17, P21 and P26. P17 strain had the largest diameter (4.25 mm) with a silicate-decomposition halo around colonies appeared on the medium (Fig. 4B). Visible colonies were also observed on the A Sugai’s medium inoculated with P12, P17 and P22 strains. P17 had the largest colony diameters (3.67 mm) in all the three strains (Fig. 4C).
The seed incubated with the isolate strains had better germination, compared with the control treatment (CK), although their germination rates varied with different strains (Fig. 5). Among all the iPSB strains, the best germination was found in P42 strain inoculation with germination rate at 44.37% and followed by P40 and P7 strain inoculations (Fig. 5). The seed germination with inoculation of the isolated iPSB strains were significantly improved. The best germination rate was found in the seed with P42 inoculation.
Discussion
Microbial Diversity in Blueberry Plant Rhizosphere Soil
Blueberries encompass several wild species of shrubs of the genus Vaccinium L. that are native to eastern North America and now are widely cultivated all over the world [21]. Blueberry rhizosphere microenvironment is very important for its growing, especially in germinating, because of high nutrient requirements and underdeveloped root system of blueberry plant. Bacterial and fungal communities play important roles in rhizosphere for plant nutrient acquisition [22]. In this study, microbial diversity in the rhizosphere of blueberry was investigated by high-throughput sequencing technology. There were 11 prominent bacterial phyla found in the collected rhizosphere samples. It was in agreement with previous studies on the rhizosphere microbial diversity of different blueberry species [23,24,25]. In addition, these were common phyla in other plant rhizosphere microorganisms [14].
Thaumarchaeota was found as one of prominent phyla in rhizosphere of SHB (6.45%) and RB (2.25%) blueberry species. Thaumarchaeota is mainly present in acidic soils, such as ocean and plankton sediment [26]. In addition, the relative abundance of bacteroidetes in the rhizosphere was significantly higher than that in the CK soil. It may be due to many bacteroidetes play a key role in carbohydrate turnover in plant rhizosphere, for example, identified Chitinophaga pinensis could participate in the stability of plant rhizosphere microecological environment and interactions with plants [26]. Although the relative abundance of WPS-2 in the rhizosphere was significantly lower than that in the CK soil, WPS-2 bacteria was reported to involving in carbon-dioxide fixation and encoding the Calvin cycle [27,28,29,30]. It may explain why the relative abundance of WPS-2 was higher in the CK soil area where there was no green on the soil surface.
For fungal communities, most of the OTUs were classified as Ascomycota, Zygomycota and Basidiomycota at phylum level (Fig. 1B), and was consistent with previous studies [25]. Ascomycota, Basidiomycota and Mortierellomycota were commonly in the rhizosphere of other agricultural crops, such as chives, tea, jujube, etc. [7, 31, 32]. This might be due to the low pH environment and interaction between blueberry plant and rhizosphere.
The Chao Index was used to estimate the number of OTUs. The larger value of index reflects the high degree of community richness. The Shannon Index and the Simpson Index were used to estimate microbial diversity. There were significant differences in Chao 1 indices among the samples from the rhizosphere of three blueberry species and CK soils (Fig. 2A and B). However, the microbial diversities in these soil samples were not different (Appendix Fig. 1A–D). It was suggested that the blueberry plants could change soil microbial abundance, but had no significant effect on the microbial diversity, compared with the CK soil samples [33, 34]. The greater number of microorganisms in blueberry rhizosphere was also important in participating in the interaction between the plants and rhizosphere microbes.
The results of LEfSe analysis indicated the difference in the composition of rhizosphere microorganisms (Fig. 3A, B). It was found that different blueberry species had a similar core rhizobiome with slight difference in microbial diversity. The slight differences might be attributed to the fact that these blueberry species were domesticated from wild blueberry species. For example, highbush blueberries were developed by hybridizing V. corymbosum with several other Vaccinium species [35]. In addition, the domestication may affect on the root-associated microorganisms. Many other factors, including the plant-growing environment, may affect rhizosphere microbial diversity as well. Several studies reported that the plant domestication is associated with compositional changes in the rhizosphere microbiome [36]. A meta-analysis on microbiome in barley, lettuce, common bean, and Arabidopsis revealed that the members of Bacteroidetes were more abundant in the roots of wild plant relatives, while Proteobacteria and Actinobacteria consistently decreased with the rhizosphere of corresponding domesticated accessions [37].
It was found that different blueberry species had a similar core rhizobiome with slight difference in microbial diversity. The slight differences might be attributed to the fact that these blueberry species were domesticated from wild blueberry species. For example, highbush blueberries were developed by hybridizing V. corymbosum with several other Vaccinium species [35]. In addition, the domestication may affect on the root-associated microorganisms. Many other factors, including the plant-growing environment, may affect rhizosphere microbial diversity as well. Several studies reported that the plant domestication is associated with compositional changes in the rhizosphere microbiome [36]. A meta-analysis on microbiome in barley, lettuce, common bean, and Arabidopsis revealed that the members of Bacteroidetes were more abundant in the roots of wild plant relatives, while Proteobacteria and Actinobacteria consistently decreased with the rhizosphere of corresponding domesticated accessions [37].
Promoting Effects of Isolated iPSB Strains on Blueberry Plant Growth and Seed Germination
The stability of rhizosphere microenvironment is important for plant growth [38]. The microbial diversity and microenvironment of rhizosphere could be affected by addition of exogenous microbial strains [5]. In addition, the original rhizosphere microbes which are PGPR (plant growth-promoting rhizobacteria) can be enhanced as microbial fertilizer without affecting the stability of the original plant rhizosphere microorganism system. The PGPR from plant rhizosphere soil could not only support plant growth, but also minimize the impact of exogenous microbial strains on the original microbial system in plant rhizosphere. iPSBs, as an important part of PGPR, play an essential role in releasing and preserving P in rhizosphere systems for plant uptake and growth [39].
A total of 18 iPSB strains were isolated from the blueberry plant rhizosphere samples. They were phyla Proteobacteria and classified into Buttiauxella, Paraburkholderia, and Pseudomonas (Table 2). Paraburkholderia and Pseudomonas were reported as the common genre in iPSB [40, 41]. Genus Buttiauxella, a member of the Enterobacteriaceae family isolated from mollusks (slugs and snails), annelids (earthworms), soil and drinking water, was reported as iPSB in 1996 [42, 43].
Different strains had different inorganic phosphate-solubilizing activity. Our results showed that the higher phosphorus-solubilizing capacity and auxin production in blueberry rhizosphere belonged to genus Buttiauxella sp. with labeled as P6, P7, P37, P40 and P42 strains in this study.
The genus Paraburkholderia, identified as a member of the family Burkholderiaceae [44], has been received increasing attention due to their great plant growth-promoting effects on solubilizing insoluble phosphates and nitrogen-fixation [41]. The strains (P12 and P17) belong to genus Paraburkholderia in this study had the same function of dissolving both phosphorus and producing auxin, as well as silicate and nitrogen fixation.
Genus Pseudomonas is an important member of PGPRs with phosphorus- and silicate-solubilizing capacity, auxin production and nitrogen fixation functions [45,46,47,48]. Among the strains isolated in this study, the strains in this genus were P8 with good phosphorous solubilizing capability, P32 with good auxin production capability, P10, P11, P21 and P26 with silicate solubilizing capability, and P22 with good nitrogen fixation capability.
Effects of different iPSB strains on seed germination were studied to identity the best PGPR strains for blueberry plants. Blueberry seeds are usually difficult to germinate under natural condition due to their dormancy characteristics and small seed size. Also the traditional cultivation method to maintain blueberry plant growth is a labour-intensive and time consuming process [49]. The seed incubated with the isolate strains had better germination, compared with the control treatment (CK). The seed germination with inoculation of the isolated iPSB strains were significantly improved. Therefore, the isolated iPSB strains in this study could be used as microbial fertilizer to increase the seed germination rate of blueberry.
Soil nutrients are significantly different in different types of soils, while different plants have different requirements for soil nutrients [50, 51]. Using the PGPRs with different growth promoting effects to supplement and fortify soil nutrients can not only promote plant growth, but also avoid the negative effects of artificial fertilizer to soil. In general, the isolated microbes in our results could be used as a natural microbial fertilizer instead of traditional chemical fertilizer to promoting blueberry seed germination and growth method. The information of these microbes will also be submitted to culture bank after more plant growth promotion functions are evaluated.
Conclusions
The microbial diversities of the rhizospheres of three different blueberry plant species were analyzed in this study. The genotype-level variations in the distribution of specific microbial taxa were observed. Eighteen iPSB strains were isolated and identified. They could effectively solubilize inorganic phosphorus in the medium and have capability of auxin production. Some of them could also solubilize inorganic silicate and fix nitrogen. According to the results, strain P7, P40, and P42 could be used as the bacterial fertilizer with function of phosphorus solubilization and auxin production. Strain P17 could be added into the bacterial fertilizer for improving the available nitrogen content and soluble silicate in soil. Blueberry seed germinations in the medium inoculated the iPSB strains were significantly enhanced. The results of this study are very helpful for understanding the microbial phyla and genera in the rhizosphere of blueberry plant and developing the PGPR fertilizer for promoting blueberry plant growth.
Data Availability
All data have been submitted in the paper.
References
Massarotto G, Barcellos T, Garcia CS, Brandalize AP, Moura S, Schwambach J, Henriques JAP, Roesch-Ely M (2016) Chemical characterization and cytotoxic activity of blueberry extracts (cv. Misty) cultivated in Brazil. J Food Sci 81:H2076–H2084. https://doi.org/10.1111/1750-3841.13385
Miller K, Feucht W, Schmid M (2019) Bioactive compounds of strawberry and blueberry and their potential health effects based on human intervention studies: a brief overview. Nutrients 11:1510–1520. https://doi.org/10.3390/nu11071510
Yurgel SN, Nearing JT, Douglas GM, Langille MG (2019) Metagenomic functional shifts to plant induced environmental changes. Front Microbiol 10:1682–1691. https://doi.org/10.3389/fmicb.2019.01682
Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Bio Biochem 35:1183–1192. https://doi.org/10.1016/S0038-0717(03)00179-2
Li LJ, Wang ST, Li XZ, Li TZ, He XH, Tan Y (2018) Effects of Pseudomonas chenduensis and biochar on cadmium availability and microbial community in the paddy soil. Sci Total Environ 11(640–641):1034–1043. https://doi.org/10.1016/j.scitotenv.2018.05.287
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327. https://doi.org/10.1111/j.1574-6941.2010.00860.x
Zhang S, Wang Y, Sun L, Qiu C, Ding Y, Gu H, Wang L, Wang Z, Ding Z (2020) Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation. BMC Microbiol 20:103–112. https://doi.org/10.1186/s12866-020-01794-8
Richardson AE, Simpson RJ (2011) Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156:989–996. https://doi.org/10.1104/pp.111.175448
Elser JJ, Bracken ME, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142. https://doi.org/10.1111/j.1461-0248.2007.01113.x
Kochian LV (2012) Plant nutrition: rooting for more phosphorus. Nature 488:466–477. https://doi.org/10.1038/488466a
Bianco C, Defez R (2010) Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl Environ Microbiol 76:4626–4632. https://doi.org/10.1128/AEM.02756-09
Wang XH, Wang CD, Sui JK, Liu ZY, Li Q, Ji C, Song X, Hu YR, Wang CQ, Sa R, Zhang JM, Du JF, Liu XL (2018) Isolation and characterization of phosphofungi, and screening of their plant growth-promoting activities. AMB Express 8:63–71. https://doi.org/10.1186/s13568-018-0593-4
Fujii Y, Akihiro F, Syuntro H (2004) Rhizosphere soil method: a new bioassay to evaluate allelopathy in the field. 3rd Australian New Zealand Soils Conference: Novel Approaches. Australia: Charles Sturt University Press, 490–492.
Wang MJ, Sun HY, Xu L, Xu ZM (2021) Bacterial diversity in tea plant (Camellia sinensis) rhizosphere soil from Qinling Mountains and its relationship with environmental elements. Plant Soil 460:403–415. https://doi.org/10.1007/s11104-020-04822-8
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406e425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Cox JA (1972) Electrochemical method for the determination of phosphate in natural water. Water Research Center Research Report NO. 61. https://hdl.handle.net/2142/90404
da Silva JF, da Silva TR, Escobar IEC, Fraiz ACR, Dos Santos JWM, do Nascimento TR, Dos Santos JMR, Peters SJW, de Melo RF, Signor D (2018) Screening of plant growth promotion ability among bacteria isolated from field-grown sorghum under different managements in Brazilian drylands. World J Microbiol Biotechnol 34:186–193. https://doi.org/10.1007/s11274-018-2568-7
Adrain JM, Westrop SR, Chatterton DEB, Ramskild L (2000) Silurian trilobite alpha diversity and the end-Ordovician mass extinction. Paleobiology 26:625–646. https://www.jstor.org/stable/2666103
Chao A (1984) Non-parametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Lond B Biol Sci 345:101–118. https://doi.org/10.1098/rstb.1994.0091
Camp WH (1945) The North American blueberries with notes on other groups of vacciniaceae. Brittonia 5:203–275. https://doi.org/10.2307/2804880
Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206:1196–1206. https://doi.org/10.3389/fmicb.2014.00491
Yurgel SN, Douglas GM, Comeau AM, Mammoliti M, Dusault A, Percival D, Langille MGI (2017) Variation in bacterial and eukaryotic communities associated with natural and managed wild blueberry habitats. Phytobiomes 1:102–113. https://doi.org/10.1094/PBIOMES-03-17-0012-R
Yurgel SN, Douglas GM, Dusault A, Percival D, Langille MG (2018) Dissecting community structure in wild blueberry root and soil microbiome. Front Microbiol 9:1187–1196. https://doi.org/10.3389/fmicb.2018.01187
Li J, Mavrodi OV, Hou J, Blackmon C, Babiker EM, Mavrodi DV (2020) Comparative Analysis of Rhizosphere Microbiomes of Southern Highbush Blueberry (Vaccinium corymbosum L.), Darrow's Blueberry (V. darrowii Camp), and Rabbiteye Blueberry (V. virgatum Aiton). Front Microbiol 11:370. https://doi.org/10.3389/fmicb.2020.00370
Pester M, Schleper C, Wagne M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. https://doi.org/10.1016/j.mib.2011.04.007
Wang J, Shi X, Zheng C, Suter H, Huang Z (2021) Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci Total Environ 10;755(Pt 1):142449. https://doi.org/10.1016/j.scitotenv.2020.142449
Ji M, van Dorst J, Bissett A, Brown MV, Palmer AS, Snape I (2016) Microbial diversity at Mitchell Peninsula, eastern Antarctica: a potential biodiversity “hotspot.” Polar Biol 39:237–249. https://doi.org/10.1007/s00300-015-1776-y
Holland-Moritz H, Stuart J, Lewis LR, Miller S, Mack MC, McDaniel SF, Fierer N (2018) Novel bacterial lineages associated with boreal moss species. Environ Microbiol 20:2625–2638
Ward LM, Cardona T, Holland-Moritz H (2019) Evolutionary implications of anoxygenic phototrophy in the bacterial phylum candidatus Eremiobacterota (WPS-2). Front Microbiol 10:1658–1668. https://doi.org/10.3389/fmicb.2019.01658
Gu Y, Wang Y, Wang P, Wang C, Ma J, Yang X, Ma D, Li M (2020) Study on the diversity of fungal and bacterial communities in continuous cropping fields of Chinese chives (Allium tuberosum). Biomed Res Int 2020:3589758. https://doi.org/10.1155/2020/3589758
Wang R, Cao B, Sun Q, Song L (2020) Response of grass interplanting on bacterial and fungal communities in a jujube orchard in Ningxia, northwest China. Heliyon 6:e03489. https://doi.org/10.1016/j.heliyon.2020.e03489
Cline LC, ZaK DR (2015) Soil microbial communities are shaped by plant-driven changes in resource availability during secondary succession. Ecology 96:3374–3385. https://doi.org/10.1890/15-0184.1
Ma Z, Zhao W, Zhao C, Wang D, Liu M, Li D, Liu Q (2018) Plants regulate the effects of experimental warming on the soil microbial community in an alpine scrub ecosystem. PLoS ONE 13:e0195079. https://doi.org/10.1371/journal.pone.0195079
Hancock JF (2006) Highbush blueberry breeders. HortScience 41:20–21
Cardinale M, Grube M, Erlacher A, Quehenberger J, Berg G (2015) Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ Microbiol 17:239–252. https://doi.org/10.1111/1462-2920.12686
Perez-Jaramillo JE, Carrion VJ, De Hollander M, Raaijmakers JM (2018) The wild side of plant microbiomes. Microbiome 6:143. https://doi.org/10.1186/s40168-018-0519-z
Harris SL, Pelaez CA, Shank EA (2019) Monitoring bacterial colonization and maintenance on Arabidopsis thaliana roots in a floating hydroponic system. J Vis Exp 5:147–158. https://doi.org/10.3791/59517
Kaur G, Reddy MS (2015) Effects of phosphate-solubilizing bacteria, rock phosphate and chemical fertilizers on maize–wheat cropping cycle and economics. Pedosphere 25:428–437. https://doi.org/10.1016/S1002-0160(15)30010-2
Gao ZH, Ruan SL, Huang YX, Lv YY, Qiu LH (2019) Paraburkholderia phosphatilytica sp. nov., a phosphate-solubilizing bacterium isolated from forest soil. Int J Syst Evol Microbiol 69:196–202. https://doi.org/10.1099/ijsem.0.003129
Tang A, Haruna AO, Majid NMA, Jalloh MB (2020) Effects of selected functional bacteria on maize growth and nutrient use efficiency. Microorganisms 8:854–861. https://doi.org/10.3390/microorganisms8060854
Ferragut C, Izard D, Gavini F, Lefebvre B, Leclerc H (1981) Buttiauxella, a new genus of the family Enterobacteriaceae. Zbl Bakt Mik Hyg 2:33–44. https://doi.org/10.1016/S0721-9571(81)80016-65
Müller HE, Brenner DJ, Fanning GR, Grimont PAD, Kämpfer P (1996) Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int J Syst Evol Microbiol 46:50–63. https://doi.org/10.1099/00207713-46-1-50
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429–436. https://doi.org/10.3389/fgene.2014.00429
Ahmad M, Zahir ZA, Nazli F, Akram F, Arshad M, Khalid M (2014) Effectiveness of halo-tolerant, auxin producing Pseudomonas and Rhizobium strains to improve osmotic stress tolerance in mung bean (Vigna radiata L.) Braz. J Microbiol 44:1341–1348. https://doi.org/10.1590/s1517-83822013000400045
Alaylar B, Güllüce M, Karadayi M, Isaoglu M (2019) Rapid detection of phosphate-solubilizing bacteria from agricultural areas in Erzurum. Curr Microbiol 76:804–809. https://doi.org/10.1007/s00284-019-01688-7
Fox AR, Soto G, Valverde C, Russo D, Lagares JA, Zorreguieta A, Alleva K, Pascuan C, Frare R, Mercado-Blanco J, Dixon R, Ayub ND (2016) Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ Microbiol 18:3522–3534. https://doi.org/10.1111/1462-2920.13376
Dong X, Lv L, Wang WJ, Liu YZ, Yin CH, Xu QQ, Yan H, Fu JX, Liu XL (2019) Differences in distribution of potassium-solubilizing bacteria in forest and plantation soils in Myanmar. J Environ Res Public Health 16:700–710. https://doi.org/10.3390/ijerph16050700
Debnath SC, Goyali CJ (2020) In vitro propagation and variation of antioxidant properties in Micropropagated Vaccinium berry plants-a review. Molecules 25:788–794. https://doi.org/10.1016/S0038-0717(03)00179-2
Abbas F, Hammad HM, Ishaq W, Farooque AA, Bakhat HF, Zia Z, Fahad S, Farhad W, Cerdà A (2020) A review of soil carbon dynamics resulting from agricultural practices. J Environ Manag 268:110319–111329. https://doi.org/10.1016/j.jenvman.2020.110319
Wang HH, Ren TB, Müller K, Van Zwieten L, Wang HL, Feng HL, Xu CS, Yun F, Ji XM, Yin QY, Shi HZ, Liu GS (2021) Soil type regulates carbon and nitrogen stoichiometry and mineralization following biochar or nitrogen addition. Sci Total Environ 753:141645. https://doi.org/10.1016/j.scitotenv.2020.141645
Funding
This study was supported by the Foundation of Young Scientist Empowerment Project of Shaanxi Association of Science and Technology (20200206), Natural Science Basic Research Program of Shaanxi (2021JQ-755), and the Foundation of Shaanxi University of Technology (SLG1809), China.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by MW, HS and ZX. The first draft of the manuscript was written by MW and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.”
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, M., Sun, H. & Xu, Z. Analysis of Blueberry Plant Rhizosphere Bacterial Diversity and Selection of Plant Growth Promoting Rhizobacteria. Curr Microbiol 79, 331 (2022). https://doi.org/10.1007/s00284-022-03031-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03031-z